Abstract
HAMP domains convert an extracellular sensory input into an intracellular signaling response in a wide variety of membrane-embedded bacterial proteins. These domains are almost invariably found adjacent to the inner leaflet of the cell membrane. We therefore examined the interaction of peptides corresponding to either AS1 or AS2 of four different, well-characterized HAMP domains with several membrane model systems. The proteins included an Archaeoglobus fulgidus protein (Af1503), the Escherichia coli osmosensor EnvZEc, the E. coli nitrate/nitrite sensor NarXEc, and the aspartate chemoreceptor of E. coli (TarEc). Far-UV CD and NMR spectroscopy were used to monitor the induction of secondary structure upon association with neutral or acidic large unilamellar vesicles (LUVs) and bicelles. We observed significant increases in α-helicity within AS1 from NarXEc and TarEc but not in AS1 from the other proteins. To characterize these interactions further, we determined the solution structure of AS1 from TarEc associated with acidic bicelles. The bulk of AS1 formed an amphipathic α-helix, whereas the N-terminal control cable, the region between TM2 and AS1, remained unstructured. We observed that the conserved prolyl residue found in AS1 of many membrane-adjacent HAMP domains defined the boundary between the unstructured and helical regions. In addition, two positively charged residues that flank the hydrophobic surface of AS1 are thought to facilitate electrostatic interactions with the membrane. We interpret these results within the context of the helix-interaction model for HAMP signaling and propose roles for AS1-membrane interactions during the membrane assembly and transmembrane communication of HAMP-containing receptors.
Keywords: HAMP domain, signal transduction, transmembrane communication, AS1-membrane interactions, helix interaction model
1. Introduction
HAMP domains (recently reviewed in [1]) are found adjacent to the inner leaflet of the cytoplasmic membrane within many prokaryotic membrane-spanning receptors [2], including well-characterized sensor histidine kinases (SHKs) and methyl-accepting chemotaxis proteins (MCPs) that serve as chemoreceptors. These domains have been identified within more than 5500 different proteins [2]. The first HAMP domain was identified by genetic analysis of the region joining the second transmembrane helix (TM2) of the serine chemoreceptor of Escherichia coli (TsrEc) and the cytoplasmic domain responsible for signal propagation [3]. A subsequent bioinformatic analysis demonstrated that similar linker regions exist within four protein families: histidine kinases, adenlyate cyclases, methyl-accepting chemotaxis proteins and phosphatases [4]. Although HAMP domains are poorly conserved at the sequence level, they remain highly conserved at the level of secondary structure, consisting of two amphipathic sequences (AS1 and AS2) that form α-helices joined by a non-helical connector (CTR) [3, 5, 6].
Two models have been proposed for how HAMP domains receive sensory input from periplasmic or membrane-embedded stimulus-sensing domains. The crankshaft and gearbox model is based on the high-resolution structures of the HAMP domain of the Archaeoglobus fulgidus protein (Af1503) [7, 8], which reveal that AS1 and AS2 of each subunit of a homodimeric protein form a parallel four-helix bundle. This model proposes that the C-terminal transmembrane helix (TM2) undergoes an axial rotation that causes a concerted rotation of AS1. The second model suggests that a small (~1-3 Å) piston-type displacement of TM2 perpendicular to the plane of the membrane occurs upon binding of a ligand to the periplasmic domain of various SHKs and chemoreceptors [9-17].
The second model has led to three different explanations for how these piston-type displacements are coupled to alterations in HAMP domain configuration. The first proposes that a piston-type displacement of a relatively rigid connection between TM2 and AS1 facilitates a scissor or pivoting-type motion within the HAMP domain [18]. The second, supported by extensive mutational evidence within an intact chemoreceptor (TsrEc) [19-21], proposes that displacements of TM2 modulate the overall structural stability of the HAMP domain bundle via changing tension of a sequence connecting TM2 to AS1, referred to as the control cable [1, 19, 22]. The third explanation proposes that two HAMP conformations are in equilibrium: one in which the hydrophobic face of AS1 interacts with the inner leaflet of the membrane, and another in which AS1 interacts with AS2 [6]. However, none of these previous studies directly assessed the influence of the membrane on transmembrane communication by SHKs and chemoreceptors.
The work presented here was undertaken to determine whether AS1 is capable of association with a phospholipid bilayer. We chose four well-characterized HAMP domains and used circular dicroism (CD) and solution NMR to monitor the ability of their AS1 or AS2 peptides to interact with various model membrane systems. The A. fulgidus Af1503 protein was chosen because it was used for the high-resolution structures [7, 8]. The E. coli EnvZ osmosensor (EnvZEc) and the nitrate/nitrite-sensing histidine kinase (NarXEc) were selected because their HAMP domains have been extensively characterized [23-26]. Finally, the E. coli aspartate chemoreceptor (TarEc) was analyzed because it is a well-characterized chemoreceptor [9-13, 18] and extensive genetic studies have been performed with the closely related serine chemoreceptor TsrEc, which serve as the basis for the dynamic bundle model of HAMP signaling [1, 19-21].
2. Materials and Methods
2.1 Materials
Synthetic peptides corresponding to either AS1 or AS2 (denoted AS1p and AS2p, respectively) were analyzed in the presence of different membrane model systems. The initial high-resolution three-dimensional structure of Af1503 HAMP (S278-V328) [7] served as the template for peptide design. Residues that form the flexible connector (G297-A309) were excluded from AS1p-Af1503 and AS2p-Af1503. For EnvZEc, NarXEc and TarEc, similar peptides were constructed based on previously published [7, 23, 27] residue alignments. The resulting eight peptides are shown in Table 1. They were obtained from PolyPeptide Group (Strasbourg, France) and used without further purification.
Table 1.
Properties of the AS1- and AS2-containing peptides used in the study.
| Receptora | GIb | Peptide | Sequence | Net Charge | Hydrophobic moment (μH)c | ΔGwif (kcal/mol)d |
|---|---|---|---|---|---|---|
| Af1503 (278-296) | 1484730 | AS1p-Af1503 | STITRPIIELSNTADKIAE | −1 | 6.28 | −0.71 |
| EnvZEc (180-198) | 947272 | AS1p-EnvZEc | RIQNRPLVDLEHAALQVGK | +1 | 3.84 | −1.51 |
| NarXEc (177-195) | 945788 | AS1p-NarXEc | RLLQPWRQLLAMASAVSHR | +3 | 6.19 | −6.35 |
| TarEc (214-232) | 946399 | AS1p-TarEc | RMLLTPLAKIIAHIREIAG | +2 | 5.79 | −4.32 |
| Af1503 (310-328) | 1484730 | AS2p-Af1503 | DEIGILAKSIERLRRSLKV | +2 | 5.42 | −0.29 |
| EnvZEc (211-229) | 947272 | AS2p-EnvZEc | SEVRSVTRAFNHMAAGVKQ | +2 | 3.46 | −1.51 |
| NarXEc (208-226) | 945788 | AS2p-NarXEc | EMAMLGTALNNMSAELAES | −3 | 5.57 | −2.25 |
| TarEc (246-263) | 946399 | AS2p-TarEc | EMGDLAQSVSHMQRSLTD | −2 | 4.81 | −0.29 |
The receptor that served as the source of AS1- and AS2-containing peptides. The position of the residues within the primary structure is provided in parentheses.
The “GenInfo Identifier” sequence identification number.
The hydrophobic moment (μH) of the peptides according to Eisenberg [58].
The energy of interfacial partitioning (ΔGwif) calculated within MPEx [59]. Significantly different values are indicated in bold font.
Large unilamellar vesicles (LUVs) and phospholipid bicelles were used as membrane model systems. 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (POPG) were used to produce LUVs. 1,2-dihexanoyl-d22-sn-glycero-3-phosphocholine (DHPC), 1,2-dimyristoyl-d54-sn-glycero-3-phosphocholine (DMPC) and 1,2-dimyristoyl-d54-sn-glycero-3-phospho-(1’-rac-glycerol) (DMPG) were used to produce phospholipid bicelles. Phospholipids were obtained from Avanti Polar Lipids (Alabaster, AL, USA).
2.2 Preparation of samples containing LUVs for CD measurements
LUVs with a diameter of 100 nm were used for circular dichroism (CD) measurements because their large size and low curvature was deemed appropriate for studies of peptide-membrane interaction [28]. In order to produce neutral and acidic LUVs, POPC or a 4:1 mixture of POPC/POPG, respectively, were initially dissolved in chloroform to produce 20 mM stock solutions. The solutions were subsequently dried under a flow of nitrogen gas and stored under vacuum overnight to ensure that no solvent remained. These dried lipid films were soaked in 100 mM sodium phosphate buffer (pH 7.2) and vortexed for 10 minutes to obtain a more defined size distribution multilamellar vesicles. These solutions were then subjected to five freeze-thaw cycles to decrease lamellarity. Finally, to obtain uniform LUVs, the samples were extruded approximately 20 times through a polycarbonate microfilter with a pore size of 100 nm.
Peptide samples were prepared by dilution from the stock solutions of 20 mM LUVs, 1 mM AS1-, and 1 mM AS2-containing peptides to a final mixture containing 50 μM peptide, 50 mM sodium phosphate buffer (pH 7.2), and 1 mM neutral or acidic LUVs. This low concentration of LUVs was chosen to diminish disturbances due to light scattering. Samples containing a mixture of 25 μM AS1 and 25 μM AS2 peptides were also prepared for analysis. To study the nature of the interaction between the peptide and the LUVs, 150 mM KF was added to appropriate samples [29]. KF was used because chloride salts are known to interfere with far-UV CD spectra [30].
2.3 Preparation of phospholipid bicelles for CD and NMR measurements
Small phospholipid bicelles with a q-ratio between 0.25-0.5 form isotropic solutions that are especially suitable for high-resolution membrane-interaction studies by NMR [31-35]. The q-ratio is the ratio of long-chained phospholipids [DMPC and DMPG] to short-chained phospholipids [DHPC].
Samples for pulsed-field-gradient NMR (PFG NMR) were prepared in D2O to avoid severe baseline distortions in spectra originating from the water signal. Initially, a solution of 50 mM sodium phosphate (pH 7.2) was dried and subsequently dissolved in D2O. DMPC or a 4:1 mixture DMPC/DMPG was lyophilized in this solution and subsequently mixed with the desired amount of a 1 M stock solution of DHPC in D2O. This mixture was vortexed until a clear solution was obtained. The final concentration of phospholipid was maintained at 150 mM, and the q-ratio was maintained at 0.5. AS1 or AS2 peptides were then added to the phospholipid-bicelle solution to yield a peptide concentration of 500 μM. For measurement of peptide diffusion in the absence of bicelles (Dfree), AS1 or AS2 peptides were also added to 50 mM sodium phosphate (pH 7.2) in D2O to a final concentration of 500 μM.
For far-UV CD measurements, samples were prepared in the same manner except that H2O was used instead of D2O, resulting in solutions containing 500 μM peptide in 50 mM sodium phosphate (pH 7.2) or a mixture of 500 μM peptide and 150 mM neutral (DMPC) or acidic (4:1 mixture DMPC/DMPG) bicelles (q = 0.5) in 50 mM sodium phosphate (pH 7.2).
To prepare acidic phospholipid bicelles for 2D 1H-1H NMR analysis, similar mixtures were made with slightly altered phospholipid ratios to arrive at a final mixture containing 300 mM [DMPC/DMPG 9:1]/DHPC bicelles with a q-ratio of 0.25 in 50 mM sodium phosphate (pH 7.2) in H2O. These changes in q-ratio, total lipid concentration, and charge of the phospholipid bicelle were made to assure collection of usable NMR spectra with acceptable line broadening. 2 mM AS1p-TarEc was dissolved in this bicelle solution. Finally, 10% D2O was added to achieve field/frequency lock stabilization during NMR experiments.
2.4 Far-UV circular dicroism (Far-UV CD) spectroscopy
Far-UV CD spectra were acquired on a Chirascan CD spectrometer. A 1 mm quartz cell was used for the samples with 50 μM peptide concentration, and a 0.1 mm quartz cell was used for the samples with 500 μM peptide concentration. The temperature was maintained at 25 °C with a TC 125 temperature control. Wavelengths ranging from 190 to 260 nm were measured with a 0.5-nm step resolution. Spectra were collected and averaged over ten measurements. Background spectra of the sodium phosphate buffer, LUVs, or bicelles without peptide were subtracted where appropriate. The CD spectra were further analyzed with Dichroweb [36] using the CONTIN method [37] with reference set 7, optimized for the range 190-240 nm.
2.5 NMR spectroscopy
All 1H-NMR experiments were performed on a Bruker Avance spectrometer, equipped with a triple-resonance probe head and operating at a 1H frequency of 600 MHz. For translational diffusion analysis, a standard sample of 0.01% H2O in D2O and 1 mg/ml GdCl3 was used for gradient calibration at 25 °C. Diffusion coefficients were measured using a modified Stejskal-Tanner spin-echo experiment [38-40] with a fixed diffusion time and gradient length with a gradient strength that increased linearly over 32 steps. The attenuation of the signal, as a result of the increasing gradient, is described by the Stejskal-Tanner equation [39]. The intensity of the signal was measured and plotted against the corresponding gradient strength according to this equation, and the diffusion coefficient was deduced from the slope of the resulting curve. The linearity of the gradient was calibrated as described previously [41]. The diffusion coefficient for HDO was measured and divided by the standard diffusion of HDO in D2O (1.9 10-9 m2/s) [42]. This factor was then multiplied by all measured diffusion coefficients, to correct for viscosity differences induced by the sample. To estimate the amount of peptide bound to the bicelle, a two-state model was used. In this model, it is assumed that the peptide is either bound to the bicelle, and thus diffuses with the rate of the bicelle (Dbicelle), or the peptide is free, and thus diffuses with the diffusion rate of the peptide in buffer (Dfree). Dcomplex is the diffusion time of the peptide in the presence of bicelles, and x is the fraction of peptide molecules bound to the phospholipid bicelle.
| Equation 1. |
Two-dimensional TOCSY [43], and NOESY [44] spectra were recorded for AS1p-TarEc in the presence of q=0.25 300 mM acidic phospholipid bicelles. The TOCSY spectra were recorded with mixing times of 30, 60 and 85 ms, and the NOESY spectra were recorded with mixing times of 100, 150, 250 and 300 ms. The 150 ms and the 250 ms NOESY spectra were used for assigning distance constraints. Spectra were typically collected as 2048−4096 × 512 data point matrices using 32−64 scans. Water suppression was achieved using excitation sculpting. The spectra were processed with Topspin version 2.1, and spectral analysis was performed with Sparky 3 [45]. All NMR experiments were performed at 25 °C.
2.6 Structure calculation
The spectra collected from AS1p-TarEc were partly obscured by large peaks from the phospholipid bicelles. Therefore, cross-peaks from both the 150 and 250 ms mixing time NOESY had to be used to derive distance constraints for calculation of a solution structure for AS1p-TarEc. In all, 133 distance constraints were assigned (59 intraresidue, 41 sequential, and 33 medium-range). The cross-peak intensities were initially converted to distances using routines in CYANA 2.0 [46]. These distances were subsequently altered manually in several stages as described previously [47]. Structures were generated using CYANA, applying standard annealing algorithms. Analyses of the structures, including secondary structure and backbone dihedral angles, were performed with PROCHECK NMR [48]. A total of 100 structures were calculated, and a final ensemble of 25 structures was selected, on the basis of the CYANA target function, to represent the final solution structure. The coordinates of the final ensemble of structures and the distance constraints have been deposited in the Protein Data Bank as entry 2L9G. The chemical shift assignments have been deposited within the BMRB as entry 17450.
3. Results
3.1 LUVs induce α-helical content in AS1 peptides from the NarXEc and TarEc HAMP domains
The secondary structure within the various AS1 and AS2 peptides was investigated using far-UV CD spectroscopy. The peptides were analyzed under three different experimental conditions: buffered aqueous solution of sodium phosphate at pH 7.2, the same buffered solution with the addition of 1 mM LUVs composed of POPC, and finally, the same buffered solution with the addition of 1 mM LUVs composed of a 4:1 mixture of POPC and POPG (Figure 1). These conditions were selected to study whether secondary structure was induced by the presence of LUVs and to investigate whether the charge on the LUV surface is important for structural induction. The CD data were then analyzed with the CONTIN method to estimate the amount of helical secondary structure (Figure 2).
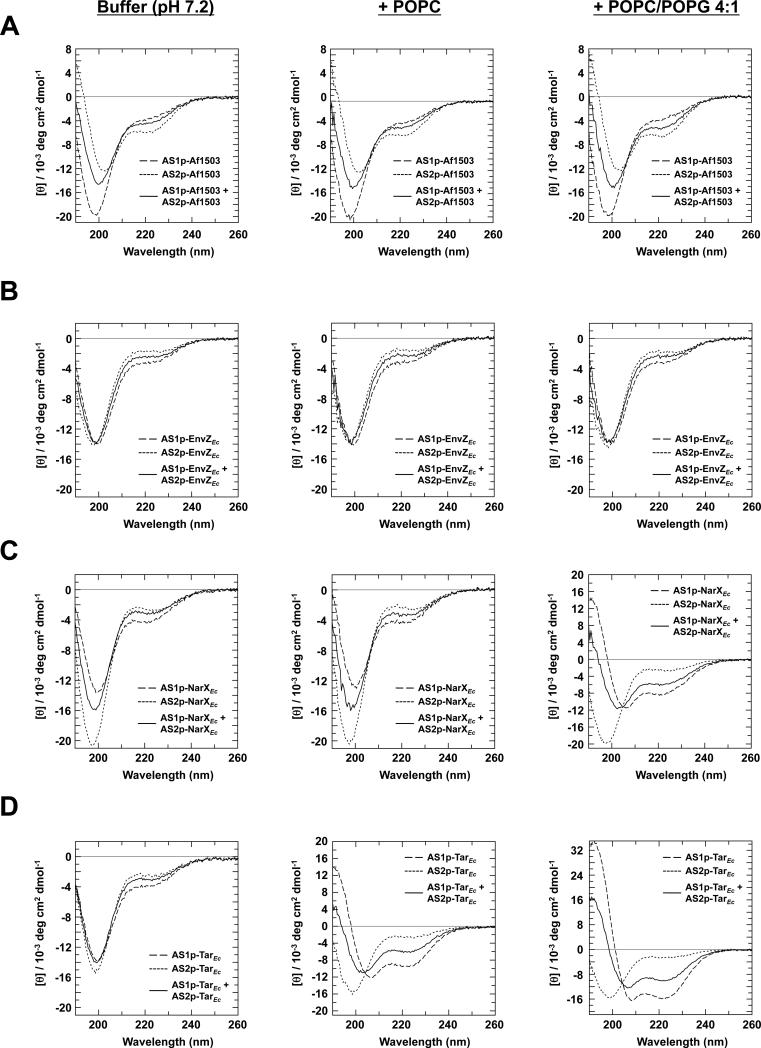
Figure 1.
Far-UV CD spectra of the AS1- and AS2-containing peptides in the presence of LUVs. The peptides analyzed are constituents of the HAMP domains from (A) Af1503, (B) EnvZEc, (C) NarXEc, and (D) TarEc. Samples contained a final concentration of 50 μM peptide (AS1p-, AS2p- or a 1:1 mix) in 50 mM sodium phosphate buffer (pH 7.2) (left panel), buffer with an additional 1 mM POPC (center panel), or buffer with an additional 1 mM POPC/POPG 4:1 (right panel).
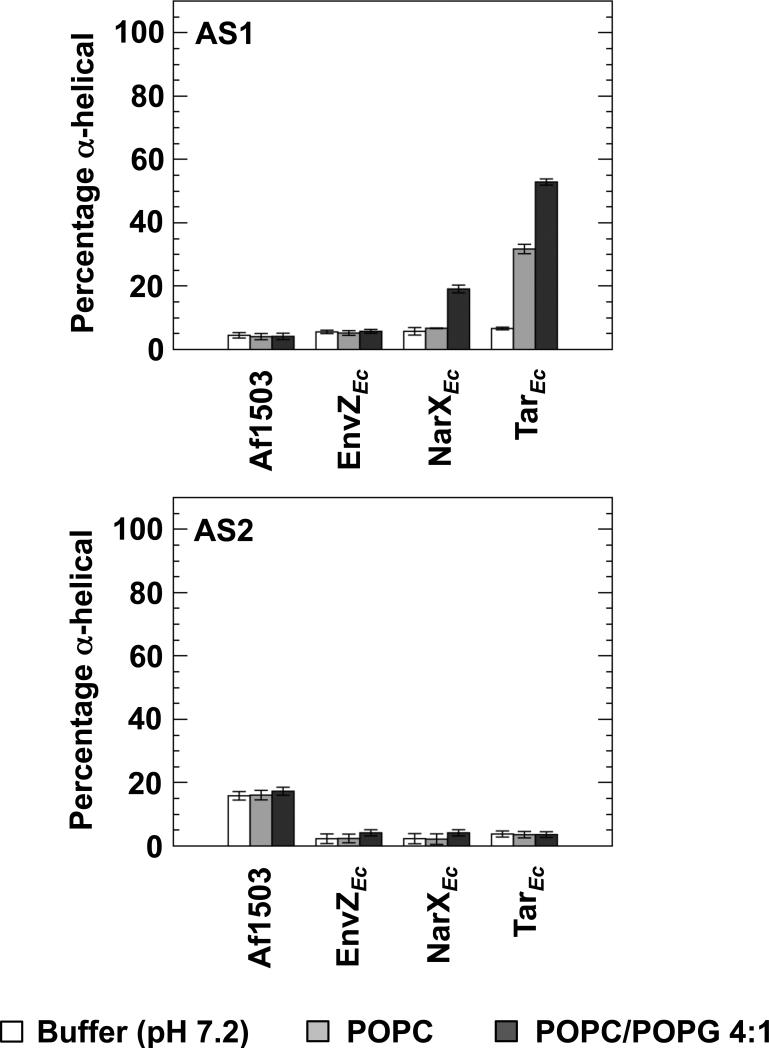
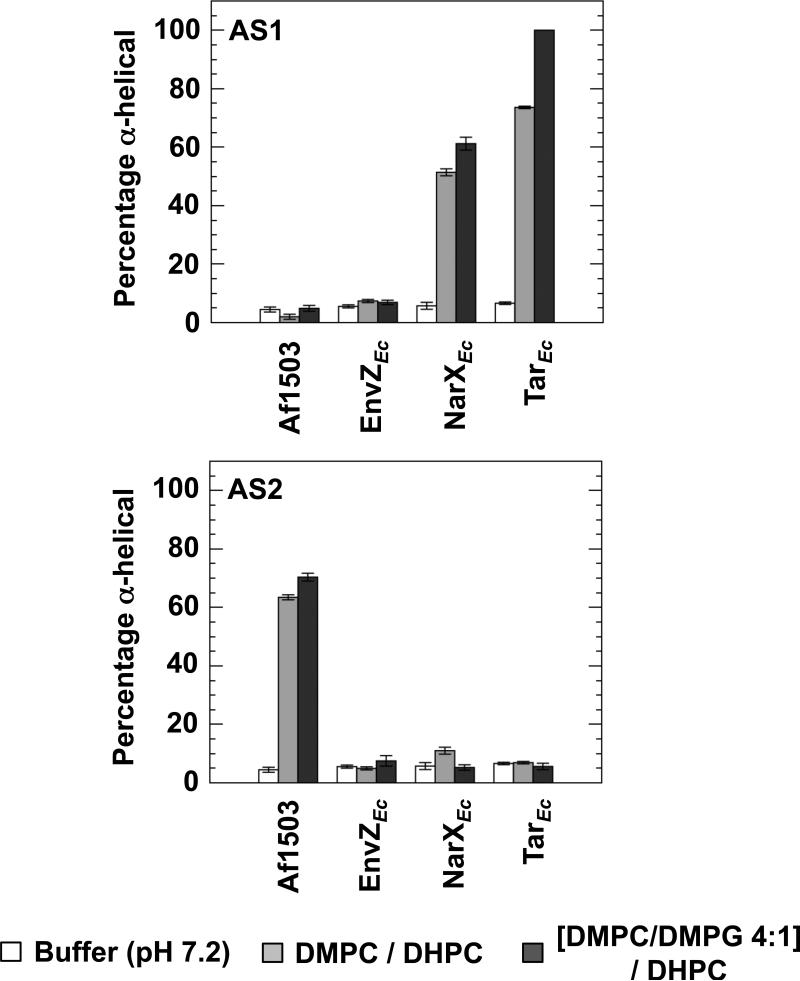
Figure 2.
Estimation of the extent of secondary structure in the presence of LUVs. The far-UV CD spectra were further analyzed with Dicroweb [36], using the CONTIN method [37]. The α-helical content of all 8 peptides in 50 mM sodium phosphate buffer (pH 7.2) (white), buffer with an additional 1 mM POPC (light grey), or buffer with an additional 1 mM POPC/POPG 4:1 (dark grey) are shown.
In phosphate buffer, random-coil features remained dominant for all the AS1 peptides. Upon addition of LUVs consisting of POPC, we observed a 5-fold induction of α-helical content in AS1p-TarEc. Addition of LUVs consisting of a 4:1 mixture of POPC/POPG caused a 3.4-fold induction of α-helical content in AS1p-NarXEc and a futher induction of α-helical content in AS1p-TarEc to 8 times the amount observed in phosphate buffer. We observed no significant induction of α-helical charcater in any of the AS2 peptides. However, AS2p-Af1503 exhibited a consistent intrinsic (~ 16%) α-helical character under all three conditions. The amount of helix, as determined from the analysis of CD data, can be interpreted as the peptide possessing this extent of α-helical structure, but it is not possible to discern whether only part of the peptide is structured or if this number represents an average over time. It is also worth noting that it is much easier to quantify differences in structural content than to determine absolute structural content. Nevertheless, a significant part of this AS2 peptide is on average helical, regardless of solvent.
Even though the Af1503 HAMP domain possesses extensive secondary structure [7, 8], our results indicate that the AS1p-Af1503 peptide is predominately unordered on its own. We therefore asked whether the AS1 and AS2 peptides when present together would take on increased helical structure. However, the far-UV CD spectra from the mixtures of all four cognate pairs of AS1 and AS2 peptides were simply composites of the average spectral properties of the two peptides measured individually (Figure 1). This result is perhaps not surprising, because the intervening CTR has been implicated in stabilization of the HAMP bundle [21]. Notably, an intact EnvZEc HAMP domain containing a single point mutation (R218K) introduced to aid in purification has also been shown to be unstructured [27].
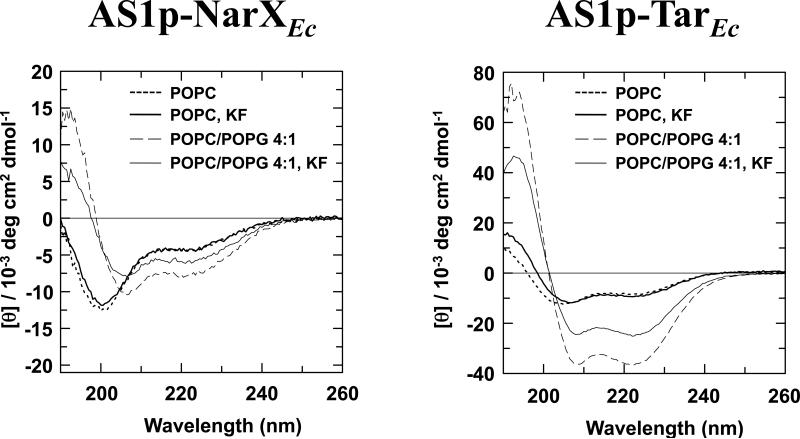
To investigate the nature of the association between the LUVs and AS1p-NarXEc, or LUVs and AS1p-TarEc, we monitored the influence of addition of 150 mM KF to the samples (Figure 3). In the peptide samples with neutral LUVs, no difference was seen in the CD spectra, suggesting that AS1p-TarEc interacts with neutral LUVs predominately via hydrophobic interactions. In contrast, addition of KF to the peptide samples with acidic LUVs resulted in a decrease of the helical content of AS1p-NarXEc and AS1p-TarEc. Raising the ionic strength of the buffer should yield a similar result to lowering the surface charge of the LUV. Thus, the α-helical induction of AS1p-NarXEc and the extra α-helical induction of AS1p-TarEc seen with acidic LUVs are probably caused by electrostatic interactions between the lipids and the peptide.
Figure 3.
Effect of increasing ionic strength on peptide-membrane interactions. 150 mM KF was added to samples containing AS1p-NarXEc (left panel) and AS1p-TarEc (right panel) in the presence of 50 mM potassium phosphate buffer (pH 7.2) and either 1 mM POPC or 1 mM POPC/POPG 4:1.
3.2 AS1p-NarXEc, AS1p-TarEc, and AS2p-Af1503 interact with phospholipid bicelles
To study the interaction of the peptides with membranes in further detail, translational-diffusion measurements were used to quantify the extent to which the peptides interact with membrane. Here, isotropic phospholipid bicelles were used as a membrane mimetic, since they are more suitable for NMR measurements [35]. The peptides were analyzed under three different experimental conditions: buffered aqueous solution of sodium phosphate at pH 7.2, the same buffered solution with the addition of 150 mM DMPC/DHPC bicelles (q=0.5), and finally, the same solution with the addition of 150 mM [DMPC/DMPG 4:1]/DHPC bicelles (q=0.5). All samples were dissolved in D2O. The derived translational-diffusion cofficients were then used to estimate how much peptide binds to the membrane (Table 2).
Table 2.
Diffusion coefficients of the AS1- and AS2-containing peptides employed in this study.
| Diffusion coefficient (10-10 m2/s)a | |||||||
|---|---|---|---|---|---|---|---|
| Buffer |
DMPC/DHPC |
[DMPC/DMPG 4:1]/DHPC |
|||||
| Peptide | Dfree | Dcomplex | Dbicelle | x (%)b | Dcomplex | Dbicelle | x (%)b |
| AS1p-Af1503 | 1.6 ± 0.07 | 1.2 ± 0.07 | 0.38 ± 0.01 | 36 | 0.96 ± 0.1 | 0.36 ± 0.01 | 52 |
| AS2p-Af1503 | 1.6 ± 0.02 | 0.39 ± 0.01 | 0.37 ± 0.01 | 98 | 0.40 ± 0.01 | 0.37 ± 0.01 | 98 |
| AS1p-EnvZEc | 1.6 ± 0.05 | 1.2 ± 0.04 | 0.38± 0.02 | 36 | 1.1 ± 0.04 | 0.39 ± 0.01 | 43 |
| AS2p-EnvZEc | 1.6 ± 0.04 | 1.0 ± 0.05 | 0.35 ± 0.01 | 48 | 1.1 ± 0.03 | 0.37 ± 0.01 | 40 |
| AS1p-NarXEc | 1.5 ± 0.05 | 0.40 ± 0.01 | 0.38 ± 0.01 | 98 | 0.35 ± 0.01 | 0.36 ± 0.01 | 100 |
| AS2p-NarXEc | 1.6 ± 0.04 | 1.0 ± 0.02 | 0.37 ± 0.01 | 49 | 0.92 ± 0.02 | 0.37 ± 0.01 | 58 |
| AS1p-TarEc | 1.4 ± 0.04 | 0.38 ± 0.01 | 0.37 ± 0.01 | 99 | 0.35 ± 0.01 | 0.36 ± 0.01 | 100 |
| AS2p-TarEc | 1.7 ± 0.06 | 1.2 ± 0.04 | 0.38 ± 0.01 | 42 | 1.2 ± 0.05 | 0.37 ± 0.01 | 40 |
Diffusion coefficients in D2O normalized according to the diffusion of HDO in order to account for differences of viscosity.
Estimation of the percentage of peptide bound to the phospholipid bicelle as calculated by equation 1. Significantly different values are indicated in bold font.
The results in Table 2 show that AS1p-NarXEc exhibited a diffusion coefficient of 1.5·10-10 m2/s in phosphate buffer, a value consistent with the translational motion of a peptide of this size [49]. The diffusion coefficient was reduced to 0.40·10-10 m2/s in the presence of neutral bicelles and to 0.36·10-10 m2/s in the presence of acidic bicelles, values comparable to the coefficients for the bicelles alone (0.38·10-10 m2/s and 0.35·10-10 m2/s, respectively). AS1p-TarEc diffused at 1.4·10-10 m2/s in phosphate buffer, and this value fell to 0.38·10-10 m2/s in the presence of neutral bicelles and to 0.35·10-10 m2/s in the presence of acidic bicelles. AS2p-Af1503 was also seen to interact strongly with the bicelles. For the other AS1- and AS2-containing peptides examined, all showed a slight reduction in the diffusion coefficient upon addition of bicelles, which suggests that a smaller proportion of the peptide is interacting with the bicelles, but none were found to associate with the bicelles extensively.
3.3 Phospholipid bicelles induce α-helical content in AS1p-NarXEc, AS1p-TarEc and AS2p-Af1503
AS2p-Af1503 bound completely to the phospholipid bicelles despite showing no additional induction of secondary structure in the presence of LUVs. To examine the efficacy of bicelles in inducing structural changes, far-UV CD measurements were performed in samples of the same composition used in the PFG-NMR experiments, with the only difference being that these samples were dissolved in H2O instead of D2O. From these spectra (Figure 4), the extent of helical secondary structure was estimated (Figure 5).
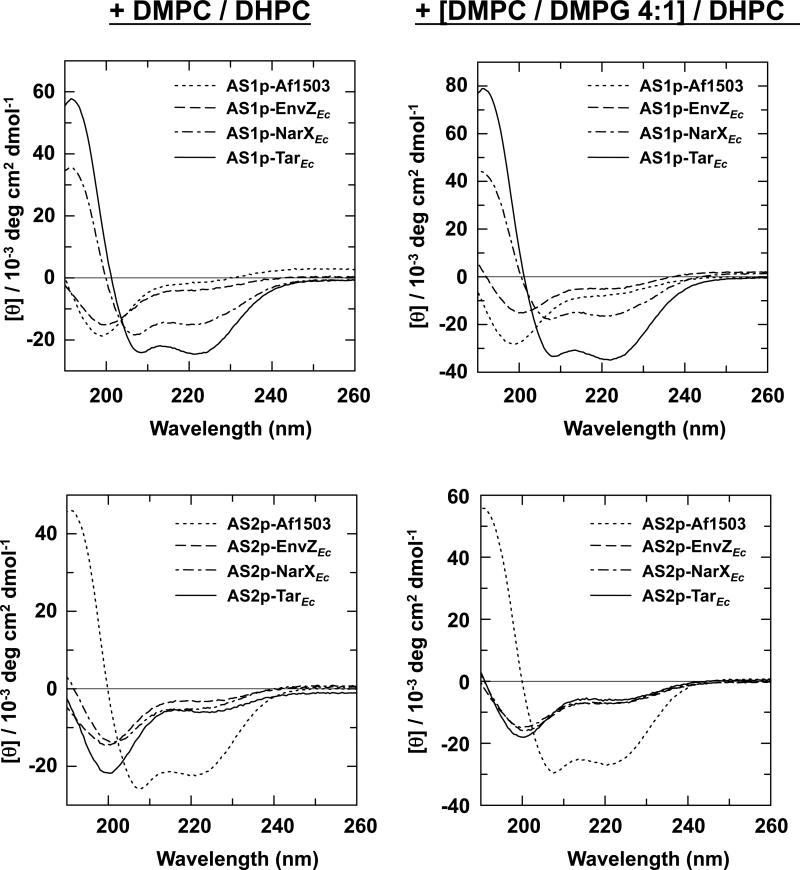
Figure 4.
Far-UV CD spectra of the AS1- and AS2-containing peptides in the presence of neutral and acidic phospholipid bicelles. Samples contained a final concentration of 500 μM peptide, 50 mM potassium phosphate (pH 7.2), and 150 mM DMPC/DHPC (q=0.5) (left panel) or 150 mM [DMPC/DMPG 4:1]/DHPC (q=0.5) (right panel).
Figure 5.
Estimation of the extent of secondary structure from the far-UV CD spectra in the presence of phospholipid bicelles. The α-helical content was determined for all 8 peptides in 50 mM sodium phosphate buffer pH 7.2 (white), buffer with an additional 150 mM DMPC/DHPC (q=0.5) (light grey), or buffer with an additional 150 mM [DMPC/DMPG 4:1]/DHPC (q=0.5) (dark grey).
The results were similar to those obtained when LUVs were employed. Of the eight peptides examined, only AS1p-NarXEc, AS1p-TarEc and AS2p-Af1503 showed significant secondary structure in the presence of bicelles. Addition of neutral bicelles caused a 9-fold induction of helical character in AS1p-NarXEc. Thus, AS1p-NarXEc interacted with neutral bicelles but not with neutral LUVs. Although some differences exist between LUVs and bicelles, it is important to note that the concentration of LUVs was only 1 mM, whereas the concentration of bicelles was 150 mM. For AS1p-TarEc, an 11-fold induction in helical character was observed with neutral bicelles. In the presence of acidic bicelles, a slight additional induction of helicial character, to 11-fold over phosphate buffer, was observed with AS1p-NarXEc, and AS1p-TarEc became essentially completely α-helical. AS2p-Af1503 exhibited a 4-fold induction of helical character in the presence of neutral bicelles and a 5-fold increase of helical character in the presence of acidic bicelles.
Although more helical structure was induced by bicelles than by LUVs, the results with both membrane systems agree qualitatively and clearly demostrate that only two of the AS1 peptides, AS1p-NarXEc and AS1p-TarEc, adopted helical structure in the presence of a bilayer. The only AS2-containing peptide seen to contain any significant amount of helical structure was AS2p-Af1503.
3.4 Solution structure of AS1p-TarEc in the presence of acidic bicelles
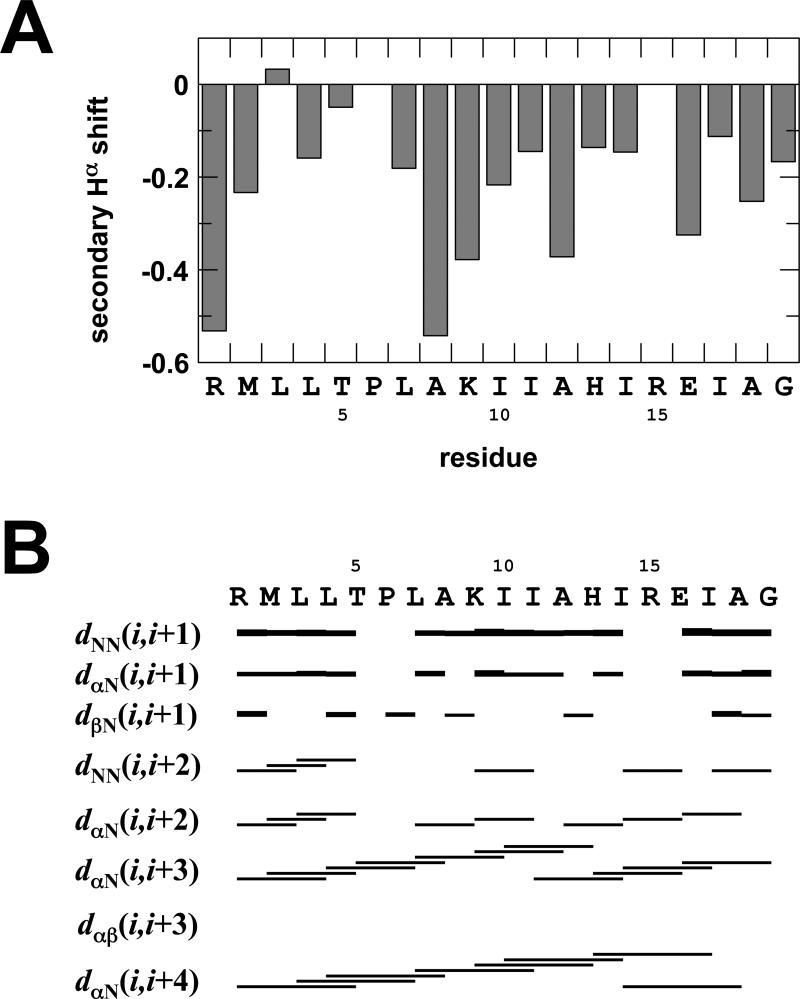
To assess the nature of the structural change seen with AS1p-TarEc in the presence of membranes, an NMR structure of the membrane-associated peptide was determined. Two-dimensional TOCSY and NOESY spectra were used to obtain resonance assignments in the presence of 300 mM 10% negatively charged phospholipid bicelles with a q-ratio of 0.25. These bicelles are small, soluble lipid aggregates that have been used extensively for solution studies of membrane-associated peptides [50-53]. A combination of the sequential-assignment strategy [54] and the main chain-directed approach (MCD) [55, 56] was utilized to identify some initial spin systems in the TOCSY spectrum and to identify local NOE connectivity patterns. The assignment proceeded by alternating between the two assignment methods. Assignments for all non-proline HN and Hα-protons were found except for Arg15, whose protons probably underwent rapid exchange with water. Based on this assignment, secondary chemical shifts [57] for the Hα-protons were calculated (Figure 6A). In the representation in Figure 6, an α-helix is defined as at least three sequential amino acids possessing a secondary Hα shift of less than -0.1 [57]. Based on this criterion, the residues comprising the control cable, (i.e. Arg1 - Pro6) possess no secondary structure, whereas chemical shifts representative of α-helical structure were observed for the residues between Leu7 and Gly19, which is the part of the primary structure that corresponds to AS1 proper [20].
Figure 6.
Secondary structure of AS1p-TarEc as determined by NMR. (a) Secondary Hα chemical shifts of AS1p-TarEc in the presence of acidic bicelles. (b) A summary of the sequential and medium-range NOE-derived distances from the NOESY spectra.
In total, 133 NOEs were assigned in the two-dimensional NOESY spectra, with mixing times of 150 ms and 250 ms. Intraresidue-, sequential-, and medium-range constraints were found for all residues except Pro6 and Arg15. Many NOEs typical for α-helical structure (i.e. Hα-HN(i, i+3), Hα-HN(i, i+4), Hα-HN(i, i+2) and HN-HN(i, i+1)) were identified. The typical pattern of these NOE connectivities (Figure 6B) and the secondary Hα shifts shown in Figure 6A provide strong evidence for the presence of α-helical character.
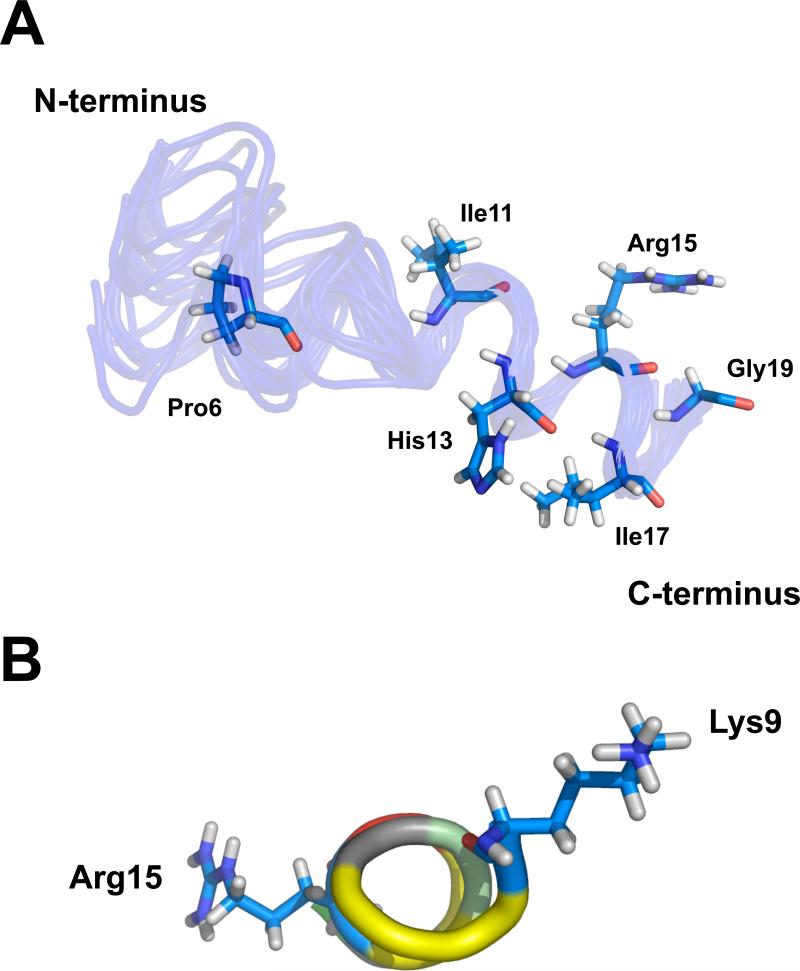
A solution structure for the peptide was calculated on the basis of these distance constraints. From the final round of calculations, an ensemble of 25 structures was selected to represent the solution structure (Figure 7A). These structures give a low target function and modest distance violations in CYANA (Table S1). In Figure 7A, the location of the side-chains of Pro6, Ile11, His13, Arg15, Ile17 and Gly19 from the average structure are shown within the ensemble. The helical portion of the structure, starting with Lys9, is shown in Figure 7B.
Figure 7.
(a) Ensemble of the 25 structures of AS1p-TarEc in 10% negatively charged phospholipid bicelles with the lowest CYANA target function. The side-chains of Pro6, Ile11, His13, Arg15, Ile17 and Gly19 from the average structure in the ensemble are shown. (b) AS1 of TarEc adopts an amphipathic α-helical structure in the presence of acidic bicelles. The position of the hydrophobic (yellow), postively charged (blue), negatively charged (red), and partially positively charged residues (grayish green) are indicated. The side chains of Lys9 and Arg15 that flank this region are indicated as well.
4. Discussion
To assess whether AS1 segments from four different HAMP domains are capable of association with a phospholipid bilayer, we monitored their interaction with LUVs and bicelles. We observed that only AS1 from NarXEc and TarEc became structured in the presence of either membrane mimetic. However, differences were seen when the spectra with LUVs were compared to those with bicelles. AS1p-NarXEc was found to associate only with acidic LUVs but with both neutral and acidic bicelles, whereas AS1p-TarEc became structured in the presence of all membrane models examined. This altered behavior is likely due to the different concentrations of lipids used and demonstrates why it is important to use complementary membrane models when analyzing lipid-peptide interactions. Nevertheless, these results show that out of the four HAMP domains investigated, only two possess AS1 segments that extensively associate with membranes.
To better understand the selectivity of AS1-membrane association, we compared various physiochemical properties of the peptides. We began by calculating the net charge of each peptide at pH 7.2 (Table 1) because electrostatic potential was important for association of the AS1 segments with acidic LUVs. AS1 of EnvZEc, NarXEc, and TarEc are all positively charged at pH 7.2. However, AS1 of EnvZEc does not associate with the acidic LUVs to a significant extent, suggesting that the helix-inducing association is more complex than a simple electrostatic interaction.
We also compared the hydrophobic moment (μH) according to Eisenberg [58] and did not observe a significant difference between the various AS1-containing peptides (Table 1). Finally, the free energies of partitioning from unfolded peptide in solution to an α-helical peptide associated with a bilayer interface (ΔGwif) were calculated using Membrane Protein Explorer (MPEx) [59]. AS1p-NarXEc and AS1p-TarEc were found to possess the most favorable ΔGwif values, suggesting that a more complex mechanism facilitates helix-inducing associations with phospholipid membranes.
Based on these observations, we considered that the positioning of individual residues might be critical for association. We constructed helical wheel projections, which revealed that the amphipathic nature of AS1p-NarXEc and AS1p-TarEc is more pronounced than those of the other peptides (Figure S1). Two positively charged residues are projected to flank the hydrophobic surface within these AS1 segments but not within those that fail to become structured in the presence of membranes.
To visualize the nature of these interactions in detail, we calculated an ensemble of 25 structures of AS1p-TarEc associated with acidic bicelles (Figure 7A). Based on α-proton chemical shifts (Figure 6A) and the pattern of NOE connectivites (Figure 6B), the residues N-terminal to Pro6 are unstructured whereas those C-terminal to Pro6 form an extended α-helix. This unstructured region, which correlates with the region recently termed the control cable within full-length receptors, is located between the aliphatic core of TM2 and the first critical packing residues within AS1 [1, 19, 22]. The change in helical register between TM2 and AS1 [5, 18] is supported by sulfhydryl-reactivity experiments with TarSt. This region also tolerates a variety of mutations that do not disrupt TarEc function [22]. When taken together, these results strongly suggest that the region between TM2 and AS1 is not a contiguous helix within these well-characterized chemoreceptors.
Two physiological roles for AS1-membrane interactions have been previously suggested. First, AS1-membrane interactions have been predicted to participate in transmembrane communication [6]. Therefore, the similar behavior of AS1 peptides from NarXEc and TarEc may not be surprising because of the evidence that they share a common mechanism of transmembrane communication. Within TarEc and other enteric chemoreceptors, a small downward displacement TM2 relative to the plane of the membrane is induced by binding of an attractant ligand [9-13, 19]. In the case of NarXEc, recent high-resolution structures of a soluble form of the periplasmic domain in the presence and in the absence of nitrate have shown a slight (~ 1Å) displacement downward of the N-terminal helices (PD1 and PD1') within the periplasmic four-helix bundle relative to the C-terminal helices (PD4 and PD4') [14]. When the intact receptor is embedded within the cytoplasmic membrane, PD4-TM2 and PD4'-TM2' may slide upward relative to the plane of the membrane. The two other HAMP domains are from receptors, Af1503 and EnvZEc, whose mechanism of stimulus detection is not well understood. In addition, structural predictions of these receptors [60, 61] do not possess the periplasmic four-helix bundle shared by NarXEc and TarEc.
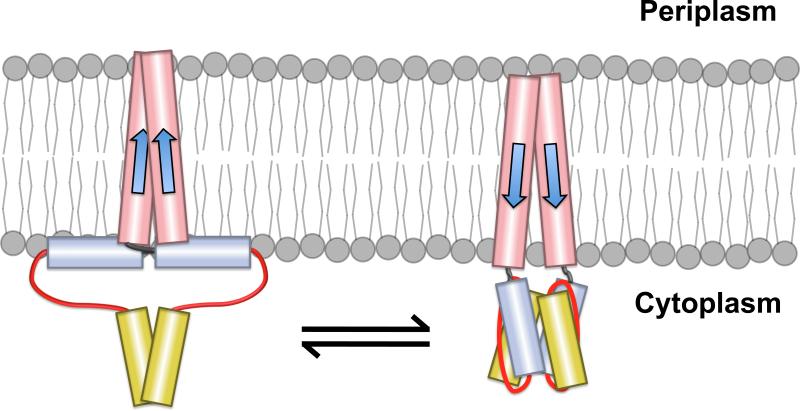
The helix-interaction model [6] proposes that the amphipathic nature of AS1 should allow it to align parallel to the membrane, with its hydrophobic face in contact with the hydrophobic interior of the phospholipid bilayer and its positively charged residues in association with the polar headgroups (Figures 7B and S1). Displacements of TM2 into the cytoplasm would destabilize AS1-membrane interactions, which, in turn, would facilitate interaction between AS1 and its cognate helical partner (Figure 8). However, it is important to note that no direct evidence for the helix interaction model has been observed within full-length receptors.
Figure 8.
Modulation of AS1-membrane interactions by displacements of TM2. The helix interaction model proposes that AS1 (blue) is oriented nearly parallel to the inner leaflet of the cell membrane in one conformation (left panel) [6]. This would allow the hydrophobic surface of AS1 to interact with the hydrophobic core of the membrane and the flanking positively charged residues to interact with the negatively charged membrane surface as depicted in Figure 7B. In the alternative conformation, AS1 would not interact with the membrane, but rather with its cognate helical partner, AS2 (yellow) (right panel). Displacements of TM2 are predicted to bias this equilibrium by altering the position of AS1 relative to the inner leaflet of the membrane (right panel). The intrinsic properties, such as length and flexibility, of the control cable (black) connecting TM2 (red) and AS1 are also expected to contribute to the baseline equilibrium between these conformations.
A second possibility is that AS1-membrane association could also be required for proper membrane assembly or subcellular localization of SHKs or chemoreceptors. AS1-membrane associations have been shown to serve as critical determinants of membrane topology [62-64]. In addition, chemoreceptors, including TarEc, exist predominantly in membrane-associated clusters of signaling complexes at the cell poles (recently reviewed in [65]). A recent study has shown that prolyl substitutions within the control cable or AS1, resulting in the presence of two prolyl residues within this region, caused TsrEc receptors to be defective in polar cluster formation [20]. In the structure presented here, Pro6 serves as a critical residue for separating the structured and unstructured regions of AS1p-TarEc. It should also be noted that NarXEc-TarEc chimeras that possess the NarXEc HAMP domain, including AS1, facilitate chemotactic responses to the NarX-specific ligands, nitrate and nitrite. This suggests that AS1 of NarXEc can properly localize these chimeras to the cell poles [66, 67]. To date, no evidence exists for polar localization of Af1503 or EnvZEc. Therefore, in the case of NarXEc and TarEc, AS1-membrane interactions could be critical for proper membrane integration of monomeric receptors, dimerization of these monomers, or formation of higher-order complexes. Further experimentation must be performed within full-length receptors to differentiate these possibilities.
In summary, the results of this study underscore the point that not all HAMP domains are created equal. HAMP domains serve as a universal coupler between various classes of domains and, thus, different subclasses of HAMP domains would be expected to exist [2]. These differences may explain some of the discrepancies observed between the bodies of evidence that exist for HAMP signaling within various different membrane-spanning receptors. Although we examined only four HAMP domains, it appears that correlations are beginning to emerge between the propensity for AS1-membrane interactions and the mechanisms of transmembrane signaling and subcellular localization of HAMP-containing receptors.
Supplementary Material
Highlights.
AS1-containing peptides from NarXEc and TarEc become structured in the presence of membrane mimetics.
AS1 from TarEc forms an amphipathic α-helix upon association with acidic bicelles.
Residues comprising the control cable of TarEc remain unstructured.
A conserved prolyl residue separates the unstructured and helical regions.
The propensity for AS1-membrane interactions correlates with signaling mechanisms and subcellular localization.
Acknowledgements
Members of the von Heijne group (Stockholm University) provided valuable support and discussion during the early stages of this manuscript. We thank an anonymous reviewer for critical and insightful comments that helped improve the final output. L.M. was supported by the Swedish Research Council and the Center for Biomembrane Research (CBR). R. R. D. was supported by a Kirschstein National Research Service Award from the National Institutes of Health (AI075773) and the German Research Foundation (SFB807 – Transport and Communication across Biological Membranes).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: HAMP domain (domain present in histidine kinases, adenylyl cyclases, methyl-accepting proteins and phosphatases); SHK, sensor histidine kinase; MCP, methyl-accepting chemotaxis protein; TM1, first transmembrane domain; TM2, second transmembrane domain; AS1, first amphipathic sequence; AS2, second amphipathic sequence; AS1p, AS1 containing peptide; AS2p, AS2 containing peptide; CTR, connector domain; POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; POPG, 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1'-rac-glycerol); LUV, large unilamellar vesicle; DHPC, 1,2-dihexanoyl-d22-sn-glycero-3-phosphocholine; DMPC, 1,2-dimyristoyl-d54-sn-glycero-3-phosphocholine; DMPG, 1,2-dimyristoyl-d54-sn-glycero-3-phospho-(1’-rac-glycerol); Far-UV CD, Far-ultraviolet circular dicroism; PFG-NMR, Pulsed field gradient-nuclear magnetic resonance; TOCSY, Total correlation spectroscopy; NOESY, Nuclear Overhauser effect spectroscopy; PD1, periplasmic domain 1; PD4, periplasmic domain 4.
References
- 1.Parkinson JS. Signaling mechanisms of HAMP domains in chemoreceptors and sensor kinases. Annu Rev Microbiol. 2010;64:101–122. doi: 10.1146/annurev.micro.112408.134215. [DOI] [PubMed] [Google Scholar]
- 2.Dunin-Horkawicz S, Lupas AN. Comprehensive analysis of HAMP domains: implications for transmembrane signal transduction. J Mol Biol. 2010;397:1156–1174. doi: 10.1016/j.jmb.2010.02.031. [DOI] [PubMed] [Google Scholar]
- 3.Ames P, Parkinson JS. Transmembrane signaling by bacterial chemoreceptors: E. coli transducers with locked signal output. Cell. 1988;55:817–826. doi: 10.1016/0092-8674(88)90137-7. [DOI] [PubMed] [Google Scholar]
- 4.Aravind L, Ponting CP. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol Lett. 1999;176:111–116. doi: 10.1111/j.1574-6968.1999.tb13650.x. [DOI] [PubMed] [Google Scholar]
- 5.Butler SL, Falke JJ. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry. 1998;37:10746–10756. doi: 10.1021/bi980607g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams SB, Stewart V. Functional similarities among two-component sensors and methyl-accepting chemotaxis proteins suggest a role for linker region amphipathic helices in transmembrane signal transduction. Mol Microbiol. 1999;33:1093–1102. doi: 10.1046/j.1365-2958.1999.01562.x. [DOI] [PubMed] [Google Scholar]
- 7.Hulko M, Berndt F, Gruber M, Linder JU, Truffault V, Schultz A, Martin J, Schultz JE, Lupas AN, Coles M. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126:929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 8.Ferris HU, Dunin-Horkawicz S, Mondéjar LGM, Hulko K, Hantke J, Martin JE, Schultz K, Zeth AN, Lupas M. Coles, The mechanisms of HAMP-mediated signaling in transmembrane receptors. Structure. 2011;19:378–385. doi: 10.1016/j.str.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isaac B, Gallagher GJ, Balazs YS, Thompson LK. Site-directed rotational resonance solid-state NMR distance measurements probe structure and mechanism in the transmembrane domain of the serine bacterial chemoreceptor. Biochemistry. 2002;41:3025–3036. doi: 10.1021/bi015759h. [DOI] [PubMed] [Google Scholar]
- 11.Miller AS, Falke JJ. Side chains at the membrane-water interface modulate the signaling state of a transmembrane receptor. Biochemistry. 2004;43:1763–1770. doi: 10.1021/bi0360206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Draheim RR, Bormans AF, Lai RZ, Manson MD. Tryptophan residues flanking the second transmembrane helix (TM2) set the signaling state of the Tar chemoreceptor. Biochemistry. 2005;44:1268–1277. doi: 10.1021/bi048969d. [DOI] [PubMed] [Google Scholar]
- 13.Draheim RR, Bormans AF, Lai RZ, Manson MD. Tuning a bacterial chemoreceptor with protein-membrane interactions. Biochemistry. 2006;45:14655–14664. doi: 10.1021/bi061259i. [DOI] [PubMed] [Google Scholar]
- 14.Cheung J, Hendrickson WA. Structural analysis of ligand stimulation of the histidine kinase NarX. Structure. 2009;17:190–201. doi: 10.1016/j.str.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falke JJ, Erbse AH. The piston rises again. Structure. 2009;17:1149–1151. doi: 10.1016/j.str.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore JO, Hendrickson WA. Structural analysis of sensor domains from the TMAO-responsive histidine kinase receptor TorS. Structure. 2009;17:1195–1204. doi: 10.1016/j.str.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, Hendrickson WA. Structural characterization of the predominant family of histidine kinase sensor domains. J Mol Biol. 2010;400:335–353. doi: 10.1016/j.jmb.2010.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swain KE, Falke JJ. Structure of the conserved HAMP domain in an intact, membrane-bound chemoreceptor: a disulfide mapping study. Biochemistry. 2007;46:13684–13695. doi: 10.1021/bi701832b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Q, Ames P, Parkinson JS. Mutational analyses of HAMP helices suggest a dynamic bundle model of input-output signalling in chemoreceptors. Mol Microbiol. 2009;73:801–814. doi: 10.1111/j.1365-2958.2009.06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Q, Ames P, Parkinson JS. Biphasic Control Logic of HAMP Domain Signaling in the Escherichia coli Serine Chemoreceptor. Mol Microbiol. 2011 doi: 10.1111/j.1365-2958.2011.07577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ames P, Zhou Q, Parkinson JS. Mutational analysis of the connector segment in the HAMP domain of Tsr, the Escherichia coli serine chemoreceptor. J Bacteriol. 2008;190:6676–6685. doi: 10.1128/JB.00750-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright GA, Crowder RL, Draheim RR, Manson MD. Mutational analysis of the transmembrane helix 2-HAMP domain connection in the Escherichia coli aspartate chemoreceptor tar. J Bacteriol. 2011;193:82–90. doi: 10.1128/JB.00953-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Appleman JA, Stewart V. Mutational analysis of a conserved signal-transducing element: the HAMP linker of the Escherichia coli nitrate sensor NarX. J Bacteriol. 2003;185:89–97. doi: 10.1128/JB.185.1.89-97.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins LA, Egan SM, Stewart V. Mutational analysis reveals functional similarity between NARX, a nitrate sensor in Escherichia coli K-12, and the methyl-accepting chemotaxis proteins. J Bacteriol. 1992;174:3667–3675. doi: 10.1128/jb.174.11.3667-3675.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalman LV, Gunsalus RP. Nitrate- and molybdenum-independent signal transduction mutations in narX that alter regulation of anaerobic respiratory genes in Escherichia coli. J Bacteriol. 1990;172:7049–7056. doi: 10.1128/jb.172.12.7049-7056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida T, Phadtare S, Inouye M. The design and development of Tar-EnvZ chimeric receptors. Methods Enzymol. 2007;423:166–183. doi: 10.1016/S0076-6879(07)23007-1. [DOI] [PubMed] [Google Scholar]
- 27.Kishii R, Falzon L, Yoshida T, Kobayashi H, Inouye M. Structural and functional studies of the HAMP domain of EnvZ, an osmosensing transmembrane histidine kinase in Escherichia coli. J Biol Chem. 2007;282:26401–26408. doi: 10.1074/jbc.M701342200. [DOI] [PubMed] [Google Scholar]
- 28.Maler L, Graslund A. Artificial membrane models for the study of macromolecular delivery. Methods Mol Biol. 2009;480:129–139. doi: 10.1007/978-1-59745-429-2_9. [DOI] [PubMed] [Google Scholar]
- 29.Magzoub M, Eriksson LE, Graslund A. Conformational states of the cell-penetrating peptide penetratin when interacting with phospholipid vesicles: effects of surface charge and peptide concentration. Biochim Biophys Acta. 2002;1563:53–63. doi: 10.1016/s0005-2736(02)00373-5. [DOI] [PubMed] [Google Scholar]
- 30.Kelly SM, Jess TJ, Price NC. How to study proteins by circular dichroism. Biochim Biophys Acta. 2005;1751:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Ram P, Prestegard JH. Magnetic field induced ordering of bile salt/phospholipid micelles: new media for NMR structural investigations. Biochim Biophys Acta. 1988;940:289–294. doi: 10.1016/0005-2736(88)90203-9. [DOI] [PubMed] [Google Scholar]
- 32.Sanders CR, 2nd, Schwonek JP. Characterization of magnetically orientable bilayers in mixtures of dihexanoylphosphatidylcholine and dimyristoylphosphatidylcholine by solid-state NMR. Biochemistry. 1992;31:8898–8905. doi: 10.1021/bi00152a029. [DOI] [PubMed] [Google Scholar]
- 33.Struppe J, Whiles JA, Vold RR. Acidic phospholipid bicelles: a versatile model membrane system. Biophys J. 2000;78:281–289. doi: 10.1016/S0006-3495(00)76591-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vold RR, Prosser RS. Magnetically Oriented Phospholipid Bilayerd Micelles for Structural Studies of Polypeptides. does the Ideal Bicelle Exist? J. Magn. Reson. 1996;113:267–271. [Google Scholar]
- 35.Vold RR, Prosser RS, Deese AJ. Isotropic solutions of phospholipid bicelles: a new membrane mimetic for high-resolution NMR studies of polypeptides. J Biomol NMR. 1997;9:329–335. doi: 10.1023/a:1018643312309. [DOI] [PubMed] [Google Scholar]
- 36.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Provencher SW, Glockner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 38.Callaghan PT, Komlosh ME, Nyden M. High magnetic field gradient PGSE NMR in the presence of a large polarizing field. J Magn Reson. 1998;133:177–182. doi: 10.1006/jmre.1998.1424. [DOI] [PubMed] [Google Scholar]
- 39.Stejskal EO, Tanner JE. Spin Diffusion Measurements: Spin Echoes in the Presence of a Time-Dependent Field Gradient. J. Chem. Phys. 1965;42:288–292. [Google Scholar]
- 40.Von Meerwall EK, M. Effect of Residual Field Gradients on Pulsed Gradient NMR Diffusion Measurments. J. Magn. Reson. 1989;83:309–323. [Google Scholar]
- 41.Damberg P, Jarvet J, Graslund A. Accurate measurement of translational diffusion coefficients: a practical method to account for nonlinear gradients. J Magn Reson. 2001;148:343–348. doi: 10.1006/jmre.2000.2260. [DOI] [PubMed] [Google Scholar]
- 42.Longsworth LG. The Mutual Diffusion of Light and Heavy Water. J. Phys. Chem. 1960;64:1914–1917. [Google Scholar]
- 43.Braunschweiler L, Ernst RR. Coherence Transfer by Isotropic Mixing: Application to Proton Correlation Spectroscopy. J. Magn. Reson. 1983;53:521–528. [Google Scholar]
- 44.Jeener J, Meier BH, Bachmann P, Ernst RR. Investigation of Exchange Processes by two-dimensional NMR Spectroscopy. J. Chem. Phys. 1979;71:4546. [Google Scholar]
- 45.Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: 2004. [Google Scholar]
- 46.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 47.Andersson A, Maler L. NMR solution structure and dynamics of motilin in isotropic phospholipid bicellar solution. J Biomol NMR. 2002;24:103–112. doi: 10.1023/a:1020902915969. [DOI] [PubMed] [Google Scholar]
- 48.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 49.Danielsson J, Jarvet J, Damberg P, Gräslund A. Translational diffusion measured by PFG-NMR on full length and fragments of the Alzheimer Aβ(1-40) peptide. Determination of hydrodynamic radii of random coil peptides of varying length. Magn. Reson. Chem. 2002;40:S89–S97. [Google Scholar]
- 50.Andersson A, Almqvist J, Hagn F, Maler L. Diffusion and dynamics of penetratin in different membrane mimicking media. Biochim Biophys Acta. 2004;1661:18–25. doi: 10.1016/j.bbamem.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 51.Lind J, Ramo T, Klement ML, Barany-Wallje E, Epand RM, Epand RF, Maler L, Wieslander A. High cationic charge and bilayer interface-binding helices in a regulatory lipid glycosyltransferase. Biochemistry. 2007;46:5664–5677. doi: 10.1021/bi700042x. [DOI] [PubMed] [Google Scholar]
- 52.Lindberg M, Biverstahl H, Graslund A, Maler L. Structure and positioning comparison of two variants of penetratin in two different membrane mimicking systems by NMR. Eur J Biochem. 2003;270:3055–3063. doi: 10.1046/j.1432-1033.2003.03685.x. [DOI] [PubMed] [Google Scholar]
- 53.Marcotte I, Auger M. Bicelles as model membranes for solid- and solution-state NMR studies of membrane peptides and proteins. Concpets Magn. Reson. 2005;24A:17–37. [Google Scholar]
- 54.Wuthrich K. NMR of Proteins and Nucleic Acids. John Wiley & Sons, Inc.; 1986. [Google Scholar]
- 55.Di Stefano DL, Wand AJ. Two-dimensional 1H NMR study of human ubiquitin: a main chain directed assignment and structure analysis. Biochemistry. 1987;26:7272–7281. doi: 10.1021/bi00397a012. [DOI] [PubMed] [Google Scholar]
- 56.Englander SW, Wand AJ. Main-chain-directed strategy for the assignment of 1H NMR spectra of proteins. Biochemistry. 1987;26:5953–5958. doi: 10.1021/bi00393a001. [DOI] [PubMed] [Google Scholar]
- 57.Wishart DS, Sykes BD. Chemical shifts as a tool for structure determination. Methods Enzymol. 1994;239:363–392. doi: 10.1016/s0076-6879(94)39014-2. [DOI] [PubMed] [Google Scholar]
- 58.Eisenberg D, Weiss RM, Terwilliger TC. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature. 1982;299:371–374. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- 59.Snider C, Jayasinghe S, Hristova K, White SH. MPEx: a tool for exploring membrane proteins. Protein Sci. 2009;18:2624–2628. doi: 10.1002/pro.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Egger LA, Inouye M. Purification and characterization of the periplasmic domain of EnvZ osmosensor in Escherichia coli. Biochem Biophys Res Commun. 1997;231:68–72. doi: 10.1006/bbrc.1996.6007. [DOI] [PubMed] [Google Scholar]
- 62.Kimbrough TG, Manoil C. Role of a small cytoplasmic domain in the establishment of serine chemoreceptor membrane topology. J Bacteriol. 1994;176:7118–7120. doi: 10.1128/jb.176.22.7118-7120.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seligman L, Manoil C. An amphipathic sequence determinant of membrane protein topology. J Biol Chem. 1994;269:19888–19896. [PubMed] [Google Scholar]
- 64.Seligman L, Bailey J, Manoil C. Sequences determining the cytoplasmic localization of a chemoreceptor domain. J Bacteriol. 1995;177:2315–2320. doi: 10.1128/jb.177.9.2315-2320.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ward SM, Bormans AF, Manson MD. Mutationally altered signal output in the Nart (NarX-Tar) hybrid chemoreceptor. J Bacteriol. 2006;188:3944–3951. doi: 10.1128/JB.00117-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ward SM, Delgado A, Gunsalus RP, Manson MD. A NarX-Tar chimera mediates repellent chemotaxis to nitrate and nitrite. Mol Microbiol. 2002;44:709–719. doi: 10.1046/j.1365-2958.2002.02902.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.