Abstract
Progenitor cells are thought to be an important cell of origin of human malignancies. However, there has not been any single gene that can define mammary bipotential progenitor cells, and as such it has not been possible to use genetic methods to introduce oncogenic alterations into these cells in vivo to study tumorigenesis from them. Keratin 6a is expressed in a subset of mammary luminal epithelial cells and body cells of terminal end buds. By generating transgenic mice using the Keratin 6a (K6a) gene promoter to express tva, which encodes the receptor for avian leukosis virus subgroup A (ALV/A), we provide direct evidence that K6a+ cells are bipotential progenitor cells. These K6a+ cells were readily induced to form mammary tumors by intraductal injection of RCAS (an ALV/A-derived vector) carrying the gene encoding the polyoma middle T antigen. Tumors in this K6a-tva line were unique in that they resemble the normal breast-like subtype of human breast cancer, providing a model for preclinical testing of targeted therapy for patients with normal-like breast cancers. These observations also provide direct in vivo evidence for the hypothesis that the cell of origin affects mammary tumor phenotypes.
Keywords: Keratin 6, TVA, RCAS, PyMT, mammary gland, normal-like breast cancer
Introduction
Human breast cancers are heterogeneous in several aspects including histopathological features and gene expression profiles. These tumors arise from the breast epithelium as a result of genetic mutations and epigenetic changes. However, the breast epithelium is comprised of cells at different stages of differentiation – a small number of mammary stem cells and bipotential progenitor cells give rise to larger populations of more differentiated ductal (or luminal) epithelial cells and myoepithelial (basal) cells (Bai and Rohrschneider 2010, Shackleton et al 2006, Stingl et al 2006). These differentiated progeny constitute the bulk of the mammary epithelium. Stem and progenitor cells have been suggested to be the cells of origin for leukemia and solid cancers including those arising in the colon, prostate, and brain (Visvader 2011). However, it has been controversial whether breast cancers arise from stem cells, progenitor cells, or their differentiated progeny, or whether they may arise from different subsets of cells, in which case differences in the cell of origin could be at least partly responsible for the phenotypic heterogeneity of breast cancer (Li and Rosen 2005, Lim et al 2009, Molyneux et al 2010).
Mouse models can be very powerful in addressing these issues. Many mouse models have been made to target oncogenic lesions primarily to the more differentiated luminal mammary cells through the use of the promoter of the gene encoding whey acidic protein (WAP) or the MMTV LTR, both of which are known to be more highly expressed in the bulk, more differentiated mammary epithelial cells (Vargo-Gogola and Rosen 2007). These studies have discovered that differentiated luminal cells may be the origin of ErbB2+ breast cancer (Andrechek et al 2004, Henry et al 2004, Jeselsohn et al 2010, Li et al 2003). However, it has not been possible to introduce genetic alterations selectively into the precursor populations of mammary cells in vivo to investigate their tumor susceptibility or their influence on the phenotypes of the resulting tumors. This was largely a technical issue: To manipulate cancer genes for in vivo tumor studies, a single gene promoter typically has to be used to direct the transgenic expression of an oncogene (or Cre for deletion of a tumor suppressor gene). s-SHIP, axin2, and LRP5 have recently been reported to enrich for mammary stem cells (Badders et al 2009, Bai and Rohrschneider 2010, Zeng and Nusse 2010), while CD24 and Sca1 could separate mammary cells into different differentiation subsets based on levels of their expression (Sleeman et al 2006, Welm et al 2002); but none could define mammary progenitor cells.
The keratin 6 (K6 or Krt6) gene family is comprised of three members, K6a, K6b, and K6hf (or Krt75) (Wojcik et al 2001). Only K6a is expressed in the mammary gland, and only in a very small fraction of mammary luminal epithelial cells based on co-staining with keratin 8 and estrogen receptor (Grimm et al 2006, Li et al 2003, Smith et al 1990). K6a is found in more mammary cells when the ductal tree expands, such as at the onset of puberty or early pregnancy (Grimm et al 2006, Smith et al 1990). This distribution pattern is consistent with the distribution of mammary progenitors. Furthermore, K6a has been found by qPCR to be more highly expressed in the population of mammary cells that are enriched for cells with colony-forming potential in collagen gel (Stingl et al 2006), but it has not been shown what percentage of the cells in this population express K6a, and whether the K6a+ cells in this population actually can form a colony or have other progenitor properties. Here, we report that K6a+ cells in the mammary gland are indeed bipotential progenitor cells and that they can be induced to form mammary tumors that resemble the normal-like breast cancer subtype in patients.
Results
Generation of K6a-tva BAC transgenic mice
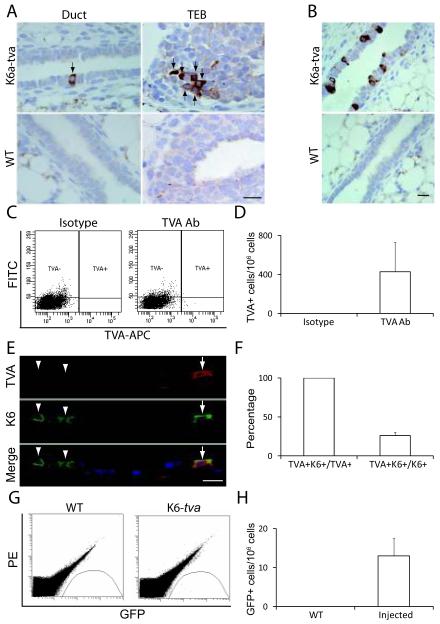
To conclusively determine whether K6a+ cells are progenitor cells, it was necessary to isolate live K6a+ cells away from the bulk of the mammary epithelium and test them in commonly used progenitor cell assays such as colony-forming and transplantation assays. K6a is a cytoskeleton protein rather than a surface protein, so it was not amenable to antibody-mediated fluorescence-activated cell sorting (FACS) of live cells. To isolate live K6a+ cells, we placed tva – a gene encoding the cell surface receptor for an avian leukosis virus, subgroup A (ALV/A) (Bates et al 1993) – behind the K6a promoter in a BAC transgenic construct (Supplemental Figure 1). Pronuclear injection of this transgenic construct into fertilized eggs from FVB/N mice resulted in one founder line. In this line, TVA was detected in a small fraction of cells among the luminal layer of ducts and the body cells of terminal end buds (TEBs) at five weeks of age: Among 29 mammary sections made from 29 transgenic mice, seven sections were found to have a few positive cells in the luminal layer and among body cells (Figure 1A). By flow cytometry analysis, the TVA+ cells were estimated to be present at a frequency of approximately 400 per million mammary cells (Figure 1C & D). These TVA+ mammary cells, like K6a+ cells, could be increased significantly by daily subcutaneous administration of estrogen and progesterone for 5 days, which stimulated ductal expansion; TVA+ cells could now be detected in every section (Figure 1B), and at 6~7-fold greater frequency based on flow cytometry (Supplemental Figure 2). To confirm that TVA+ cells also express K6a, mammary sections from three of the estrogen/progesterone-treated 7-week-old K6a-tva mice were co-stained for TVA and pan-K6. All TVA+ cells were also stained for K6, confirming that tva expression is restricted to K6+ cells, though only a subset (26 ± 4%) of K6+ cells were also positive for TVA (Figure 1E & F). To verify further that the tva expression is restricted to the K6+ cell population, we also stained sections of hyperplastic mammary glands from three 12-week-old K6a-tva/MMTV-Wnt1 bi-transgenic mice as well as sections of mammary tumors arising in this line of mice. Again, all TVA+ cells were also positive for K6, while 65 ± 9% and 22 ± 5% of K6+ cells in these hyperplastic and cancerous mammary tissues, respectively, also stained for TVA (Supplemental Figure 3A and B). In addition, we detected TVA in the K6+ populations of cells in a number of non-mammary tissues including the skin (in the outer root sheath of the hair follicle), esophagus, trachea, papilla of the dorsal tongue, and sole of the foot (Supplemental Figure 3C), but not aberrantly in K6-negative tissues including connective, muscle, and nervous tissues (data not shown). As expected of K6, after topical treatment with phorbol ester 12-O-tetradecanoylphorbol-13 acetate (TPA), TVA was also induced in the epidermis in the skin, ear, and foot, and it continued to be restricted to the K6+ cell population (Supplemental Figure 3C and data not shown). Collectively, these data demonstrate that tva expression is restricted to the K6+ subset of cells in this K6a-tva transgenic line. The lack of detectable TVA in some of the K6+ cells in these tissues may be due to lower TVA detection sensitivity relative to that for K6, shorter TVA half-life, or the loss of transcriptional regulatory elements in this BAC construct.
Figure 1.
Generation of the K6a-tva BAC transgenic mouse line. (A) Immunohistochemical staining for TVA in mammary glands from 5-week-old K6a-tva or wild-type (WT) mice. Scale bar = 20μm. (B) Immunohistochemical staining for TVA in mammary glands from 7-week-old K6a-tva or WT mice treated with estrogen and progesterone. Scale bar = 20μm. (C & D) Flow cytometry analysis of K6a-tva mammary cells following anti-TVA staining. APC-labeled secondary antibody was used to detect rabbit IgG against TVA. Normal rabbit IgG was used as isotype control. The FITC channel was used for autofluorescence measurement. Quantitation is shown by a bar graph. (E & F) Co-immunofluorescent staining for TVA and K6 in mammary glands from 7-week-old K6a-tva mice (n = 3). Mice were treated with daily injection of estradiol (1 μg) and progesterone (1 mg) for 5 days. All TVA+ cells (arrow) were also positive for K6, but only some of K6+ cells (arrowhead) were stained for TVA. The bar graph shows the percentage of K6+ cells among TVA+ cells and the percentage of TVA+ cells among K6+ cells. Scale bar = 20μm. (G & H) Flow cytometry detection of GFP+ cells in RCAS-GFP-infected K6a-tva mammary glands. Single cell suspensions were made from WT mammary glands or from K6a-tva mammary glands infected with RCAS-GFP (107 IUs per gland). The PE channel was used as autofluorescence control. Quantitation of RCAS-GFP-infected cells is shown by a bar graph.
The expression of tva was also expected to cause these K6a+ cells to be susceptible to infection by ALV/A (Du et al 2006), a useful feature for virus-mediated lineage-tracing and introduction of oncogenes for tumor induction. To confirm that the TVA produced in K6a-tva mice was capable of mediating infection by RCAS (a modified version of ALV/A), we injected RCAS-GFP intraductally into mammary glands in nine K6a-tva mice at five weeks of age (107 IU per gland). Two and a half days after injection, we collected mammary glands from these infected mice, prepared single cell suspensions, and quantified the number of GFP+ cells by flow cytometry (Figure 1G). 13 ± 4 GFP+ cells were detected per million mammary cells (Figure 1H). This infected cell number is much lower than the 0.3% achieved in MMTV-tva mice (Du et al 2006). This difference was expected, since the pool of RCAS-susceptible cells in K6a-tva mice is much smaller than that in MMTV-tva mice. As anticipated, no positive signal was detected in non-transgenic mice (Figure 1G & H).
Mammary K6a+ cells are bipotential progenitor cells, but not stem cells
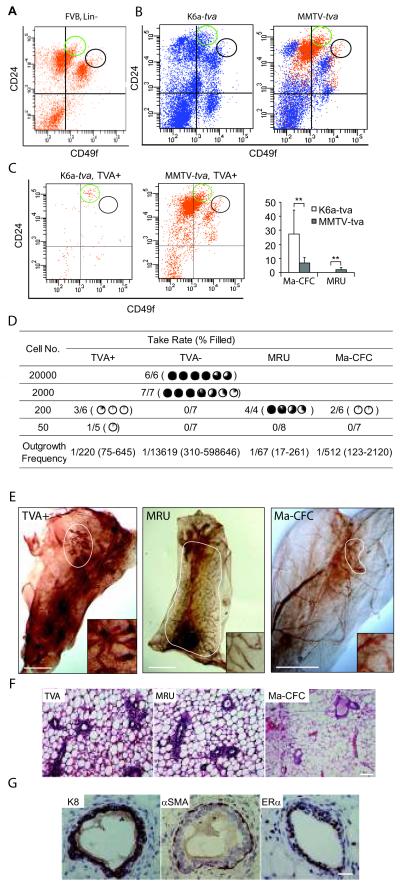
Mouse mammary stem cells are enriched in the CD24medium/CD49fhigh population (MRU), whereas progenitor cells are enriched in the CD24high/CD49flow (Ma-CFC) population (Stingl et al 2006). To establish the relationship between K6a-expressing cells and these subsets of mammary cells, we first prepared single cell suspensions from mammary glands from 5-week-old FVB mice, stained them with Lin (CD45, Ter119, CD31, and CD140), CD24, and CD49f (Figure 2A), and established the FACS gating and positioning of MRU and Ma-CFC, as previously reported (Stingl et al 2006). Next, we stained single cell suspensions from 5-week-old K6a-tva and MMTV-tva mice for TVA, CD24, and CD49f; the Lin staining was omitted so as to accommodate the FACS channel for TVA detection (Figure 2B). As shown in Figure 2C, the TVA+ cells from K6a-tva mice were enriched in the Ma-CFC group (27.5 ± 16.8%), but were rarely found in the MRU population. This result suggests that K6a-expressing cells are enriched for progenitor cells but not stem cells, consistent with a previous report of qPCR analyses of different subsets of mammary cells for K6a mRNA (Stingl et al 2006). In accord, 50% of these TVA+ cells were also positive for CD61 (Supplemental Figure 4), a marker that has been reported to be enriched on mammary progenitor cells (Asselin-Labat et al 2007, Vaillant et al 2008). Among TVA+ from MMTV-tva mice, 6.9 ± 3.8% and 2.0 ± 1.5% were in Ma-CFC and MRU sub-populations, respectively (Figure 2C), and 32 ± 18% were positive for CD61 (Supplemental Figure 4), confirming that the MMTV LTR may also be active in some progenitor and stem cells.
Figure 2.
TVA+ cells in K6a-tva transgenic mammary glands are enriched for progenitor cells, but not stem cells. (A) A representative flow cytometry profile showing the location of Ma-CFC (CD24highCD49flow) and MRU (CD24mediumCD49fhigh) populations. Single cell suspensions were prepared from 9-week-old FVB mouse mammary glands and stained for Lin (CD45, Ter119, CD31, and CD140), CD24, and CD49f. The Lin+ cells were gated out, and the Ma-CFC (green circle) and MRU (black circle) populations were then identified as reported (Stingl et al 2006). This profile was used to draw the location of Ma-CFC and MRU in the rest of the study presented in this figure. (B&C) Single cell suspensions from 5-week-old K6a-tva (n = 6) and MMTV-tva mice (n=7) were stained for CD24, CD49f, and TVA. Lin staining was omitted in order to accommodate the additional channel needed for detection of TVA. The CD24/CD49f staining profiles for total live cells (B) and TVA+ live cells (C) are shown. The bar graph shows the distribution of TVA+ cells in the Ma-CFC and MRU populations. **: P < 0.01. (D) Limiting dilution transplantation of TVA+, TVA−, MRU, and Ma-CFC cells. Single cell suspensions from 5-week-old K6a-tva mice were stained for TVA, CD24, and CD49f. Part of these stained cells were sorted for TVA+ and TVA− cells, and the rest were sorted for MRU and Ma-CFC. (In the absence of Lin staining, the Ma-CFC and MRU cells populations in this sort contained approximately 9% and 29% contamination by Lin+ cells, respectively.) These four groups of sorted cells were transplanted at the indicated dilutions into #4 cleared fat pads of 3-week-old MMTV-tva mice. Outgrowths were recorded 8 weeks after transplantation. The degree of ductal outgrowth relative to the fat pad is shown using the percentage pie. Outgrowth frequencies and the 95% CI, shown in parentheses, were estimated using the method described in Supplemental Materials and Methods. (E) Neutral red whole mount staining of outgrowths from 200 transplanted cells from the TVA+, MRU, and Ma-CFC populations. The outgrowths are highlighted by a circle. The insets show a 5 × enlarged view of the outgrowths. Scale bar = 2 mm. (F) H&E staining of the outgrowths from (E). Scale bar = 50 μm. (G) Immunohistochemical staining of the TVA+ outgrowths from (E) for the markers indicated. Scale bar = 20 μm.
The mammosphere-forming assay is a surrogate assay for breast stem cells (Dontu et al 2003). In this assay, if undifferentiated mammary cells express tva and are susceptible to infection by RCAS-GFP, they would be expected to develop into green mammospheres. We first confirmed that RCAS-GFP infection of mammary cells prepared from 5-week-old MMTV-tva mice resulted in green mammospheres frequently (61 ± 7%) (mean ± SD) (Supplemental Figure 5A), suggesting that the majority of the mammosphere-initiating cells in MMTV-tva mice were TVA+ and susceptible to infection by RCAS. In a parallel experiment, we found that cells from 5-week-old K6-tva mice failed to form green mammospheres (0/1,152 mammospheres in 18 culturing wells), but generated 3 ± 3 much smaller spheres of green cells per culture well (Supplemental Figure 5A). These observations suggest that the TVA+ cells from K6a-tva mice are susceptible to ex vivo infection in the mammosphere assay and have limited progenitor potential, but lack the self-renewal and robust proliferation properties defined in this assay.
To further evaluate the progenitor property of the TVA+ cells in K6a-tva mice, mammary cells from 5-week-old K6a-tva and MMTV-tva mice as well as wild-type mice were seeded in a colony-forming assay in the presence of RCAS-GFP for the first 16 hours to infect the original TVA+ cells. 48% of the resulting colones in the MMTV-tva group were green (Supplemental Figure 5B). Although the TVA+ cells in the K6-tva group were 500-fold less than that in the MMTV-tva group, the frequency of green colonies in the K6-tva group reached 4%. Therefore, the relative colony-forming efficiency of TVA+ cells in the K6a-tva group appears to be much higher than that in MMTV-tva mice (although we could not be certain that the TVA+ cells in these two groups had the same plating efficiency). Furthermore, we found that these green colonies from K6-tva mice contained keratin 8 (K8)+ cells as well as cells positive for α-smooth muscle actin (αSMA) (Supplemental Figure 5C & D), suggesting that the infected K6+ cells can give rise to both epithelial and myoepithelial progeny. The green colonies from MMTV-tva mice contained K8+ cells, but fewer αSMA+ cells (Supplemental Figure 5C & D).
Next, we used cleared fat pad transplantation as an in vivo assay to examine the differentiation status of the TVA+ cells from K6a-tva mice. TVA+ and TVA− as well as Ma-CFC and MRU mammary cells were isolated by FACS from 5-week-old K6a-tva mice, and were transplanted into cleared fat pads in 3-week-old MMTV-tva mice. While none of the fat pads transplanted with 200 TVA− cells (n=7) formed any outgrowth, among the six fat pads transplanted with 200 TVA+ cells from K6a-tva mice, two displayed a 5% outgrowth and another one a 15% outgrowth (P<0.05, Pearson’s χ2 test). Among the five fat pads transplanted with 50 TVA+ cells from K6a-tva mice, one had a 5% outgrowth, but no outgrowth from the transplantations of 50 TVA− cells (n=7) (P<0.05) (Figure 2D and E). These outgrowths were radial in pattern as expected. (Outgrowths from residual ducts of incompletely cleared hosts would have been unidirectional.) We further confirmed that the outgrowths were from transplanted TVA+ cells by staining them for TVA – very few TVA+ cells were detected in these growths and they were usually in the terminal end buds, similar to what we find in the mammary glands of K6a-tva mice (Supplemental Figure 6). Approximately half of the ductal cells would have been positive for TVA if the outgrowths came from residual host ducts, based on our previous characterization of this MMTV-tva line (Du et al 2006). Our observation of very limited outgrowths from transplanted K6a-tva-expressing cells were in sharp contrast to much more extensive outgrowths (≥40%) observed in the fat pads transplanted with the stem cell-enriched MRU cells, but similar to the outgrowths (3% and 6%) from two of the six transplantations using 200 Ma-CFC cells (Figure 2D and E). Nevertheless, these partial outgrowths were normal in histology (Figure 2F), and contained both K8+ epithelial cells and αSMA+ myoepithelial cells, as well as both ERα+ and ERα− cells (Figure 2G). Therefore, this small subset of K6a+ mammary cells exhibit restricted – but frequently detectable – ductal outgrowth potential, suggesting that they are mammary bipotential progenitor cells, but not stem cells. Combined analysis of the data from our small transplantation experiments indicates violation of the single hit Poisson model. For purposes of estimating the outgrowth-generating cell frequency, we incorporated all of the data into a generalized linear model and predicted the frequency at a dose of 200 cells (Figure 2D). The frequency of outgrowth-generating cells was estimated to be 1 per 220 FACS-isolated K6a-tva-expressing cells (approximate 95% CI: 1/75 to 1/645), 1 per 1/512 Ma-CFC cells (approximate 95% CI: 1/123 to 1/2,120), and 1 per 67 MRU cells (approximate 95% CI 1/17 to 1/261) – the estimated stem cell frequency in this MRU control is comparable to what has been reported (Stingl et al 2006). The estimated frequency of outgrowth-generating cells among TVA− cells was 1/13,619 (approximate 95% CI: 1/310 to 1/598,646). Of note, transplantation assays using FACS-isolated cells probably results in a severe underestimation of the absolute outgrowth-generating potential among K6a-tva-expressing cells as well as other subsets, since FACS is known to damage cells and decrease their viability (Mollet et al 2008).
Induction of mammary tumors in K6a-tva transgenic mice
We have previously reported that RCAS can be used to introduce oncogenes into TVA+ cells for tumor induction (Du et al 2006). To test whether tva-expressing K6a+ cells can be induced to form unique tumors, we first injected RCAS virus expressing the gene encoding PyMT (RCAS-PyMT) into five 5-week-old K6a-tva mice (107 IU per gland, 3 glands per mouse). We chose this viral protein for its potent role in activating multiple oncogenic signaling pathways including those mediated by Src, the Shc adapter protein, and phosphatidylinositol 3-kinase (PI3K) (Dilworth 2002), increasing the likelihood that tumor evolution will occur rapidly even with infection of small numbers of cells. One week after infection by RCAS-PyMT, a few foci of atypical ductal hyperplasia or ductal carcinoma in situ (DCIS)-like lesions could be detected on hematoxylin & eosin (H&E)-stained sections of infected glands from each of the infected mice (Supplemental Figure 7A). These lesions contained heterogeneous cell types, including K8+, αSMA+, and K6+ cells (Supplemental Figure 7B). Of note, the αSMA+ cells were not simply uninfected myoepithelial cells recruited into these lesions, since they were also positive for the HA tag in PyMT (Supplemental Figure 7C). Such cellular heterogeneity in these early lesions is consistent with their origin in bipotential progenitors, though not every lesion may be clonally derived. As expected, early lesions could not be detected in K6a-tva transgenic mice that were not injected with RCAS-PyMT, in K6a-tva mice injected with RCAS-GFP (Supplemental Figure 7A), or in non-transgenic FVB mice injected with RCAS-PyMT.
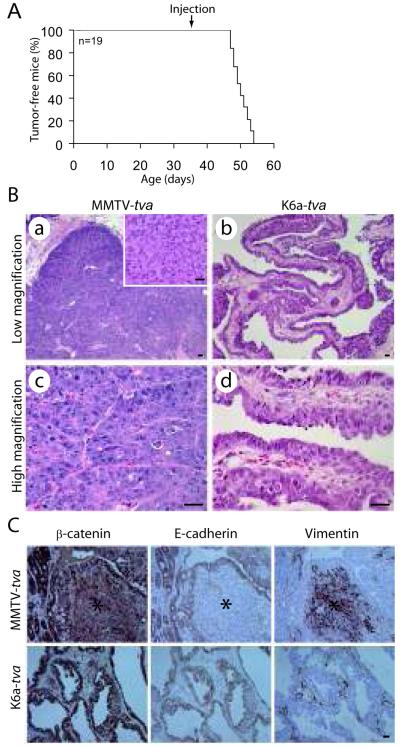
The rapid formation of these premalignant lesions suggests that palpable tumors would also form quickly in this line of mice after infection with RCAS-PyMT. Indeed, all of additional 19 K6a-tva mice infected at 5 weeks of age (107 IUs per gland, three glands per mouse) developed palpable tumors by 8 weeks of age, with a median latency of only 14 days (Figure 3A). This observation of rapid tumorigenesis suggests that the infected K6+ cells did not require additional genetic/epigenetic events to become cancerous. In accord, multiple proviruses were detected in all four tumors analyzed by Southern blotting (Supplemental Figure 8), suggesting an oligoclonal origin. (Of note, no more than 13 cells were infected per gland, so smear on a Southern blot was not anticipated.) However, we cannot exclude the possibility that some or all of these four tumors were monoclonal but they arose from single cells that were infected by multiple viruses.
Figure 3.
RCAS-PyMT induces mammary tumors in K6a-tva mice. (A) Kaplan-Meier tumor-free plot of 19 K6a-tva transgenic mice infected by RCAS-PyMT (107 IU per gland) at 5 weeks of age. Three glands (#2-4) were injected per mouse. (B) Histopathological comparison between mammary tumors arising in RCAS-PyMT-infected K6a-tva vs. MMTV-tva mice. The inset shows an EMT region in a tumor from MMTV-tva mice. Scale bar = 20 μm. (C) The EMT regions in RCAS-PyMT-induced tumors from MMTV-tva mice express vimentin, but not β-catenin or E-cadherin. Immunohistochemistry staining for indicated markers was done on RCAS-PyMT-induced tumors from both MMTV-tva and K6a-tva mice. The regions labeled with a star are the EMT regions. Scale bar = 20 μm.
Pulmonary metastases were found in only one of the 14 tumor-bearing mice that were examined at necropsy. A small percentage of metastases was also noted in tumor-bearing MMTV-tva mice infected by RCAS-PyMT (104 IU per gland; this lower viral dosage was used to achieve a similar rate of infection as in K6a-tva mice). Since the numbers of metastases were small in both groups, a statistical difference could not be established.
RCAS-PyMT-induced tumors are different in K6a-tva mice vs. MMTV-tva mice in histopathology and EMT
RCAS-PyMT-induced tumors (n=10) in MMTV-tva mice were adenocarcinomas, usually with compact acini (Figure 3B-a & c) (Du et al 2006), but they also harbored focal areas of spindle-like cells without glandular structure (inset in Figure 3B-a). These spindle-like areas lacked the epithelial/myoepithelial markers β-catenin, E-cadherin, K8, and K5, but produced the mesenchymal marker vimentin (Figure 3C and Supplemental Figure 9). In contrast, RCAS-PyMT-induced mammary tumors in K6a-tva mice (n=10) were typically papillary and cystic with ample spaces between the papillary fingers (Figure 3B-b & d), though they also harbored focal areas of more compact acini. Spindle-like cell lesions were absent in these tumors, and large areas of vimentin-positive cells were accordingly not detected (Figure 3C). Therefore, we conclude that tumor histopathology and epithelial-to-mesenchymal transition (EMT) are modestly different between RCAS-PyMT-induced tumors in K6a-tva mice vs. MMTV-tva mice.
Since the MMTV LTR appears to be active in a wide range of mammary cells (Figure 2), the cell of origin of tumors arising in MMTV-tva mice is uncertain. Nevertheless, RCAS-PyMT-induced tumors in MMTV-tva mice harbored both epithelial and myoepithelial tumor cells (Du et al 2006), suggesting that they may have an origin in the immature subset of mammary cells. Using immunostaining for K8, K19, K5, K14, and αSMA, we found that both epithelial and myoepithelial cell types were also present in RCAS-PyMT-induced tumors in K6a-tva mice (Supplemental Figure 10), though the myoepithelial content appeared to be lower than the epithelial component. These myoepithelial tumor cells were indeed progeny of infected K6a+ cells, since many (but not all) of the αSMA+ cells were also positive for the HA tag in PyMT (Supplemental Figure 10B). As expected, patchy staining for K6 or, to a lesser degree, TVA was also detected in these tumors (Supplemental Figure 10C and D). The presence of heterogeneous tumor cells is consistent with an origin in bipotential progenitor cells.
RCAS-PyMT-induced tumors in K6a-tva mice uniquely resemble the normal-like breast cancer subset in women
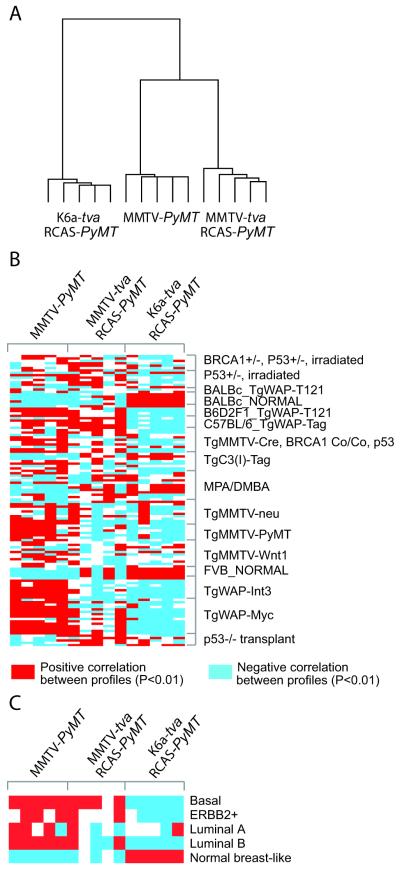
Expression arrays are frequently used to define and compare subtypes of breast cancers. Therefore, we performed Affymetrix array analysis of five RCAS-PyMT-induced tumors each from K6a-tva mice and MMTV-tva mice, as well as five MMTV-PyMT transgene-induced mammary tumors, which are comprised of luminal epithelial tumor cells (Li et al 2003, Rosner et al 2002). Based on unsupervised hierarchical clustering of 1,136 probe sets identified as different by ANOVA (P<0.001), these three sets of tumors were segregated into two distinct clusters – one was comprised of tumors induced by RCAS-PyMT in K6a-tva transgenic mice, while the other was comprised of two sub-clusters, one for RCAS-PyMT-induced tumors in MMTV-tva transgenic mice and the other for tumors arising in MMTV-PyMT transgenic mice (Figure 4A). These results indicate that mammary tumors arising in RCAS-PyMT-infected K6a-tva mice are distinct from those arising in RCAS-PyMT-infected MMTV-tva mice or in MMTV-PyMT mice. Of particular interest, while basal cells were found in RCAS-PyMT-induced tumors in both tva lines but not in the MMTV-PyMT transgene-induced tumors, RCAS-PyMT-induced tumors in MMTV-tva transgenic mice were, nevertheless, more similar to the MMTV-PyMT transgene-induced tumors, possibly reflecting their closer similarity in the cell of origin – both cohorts of tumors must have arisen from cells in which the MMTV LTR is active.
Figure 4.
Expression array analyses of RCAS-PyMT-induced mammary tumors in K6a-tva transgenic mice. (A) Average linkage hierarchical clustering of tumors arising in K6a-tva and MMTV-tva transgenic mice infected by RCAS-PyMT at puberty, and in MMTV-PyMT mice (using genes varying significantly by ANOVA P<0.001). (B) Heat map of the correlations between each mouse tumor profile from part A (columns) and each mouse tumor profile from Herschowitz et al. (2007) (rows) (genes in each dataset being first centered on the mean centroid of profile groups). (C) Heat map of the correlations between each mouse tumor profile from part A and the average expression for each of the five major human tumor molecular profile subtypes (basal, ErbB2+, luminal A, luminal B, and normal-like; averages centered on the mean centroid of the groups). For parts B and C, red indicates significant correlation (P<0.01, Pearson’s) and blue indicates anti-correlation.
Herschkowitz et al. (2007) have reported expression profiles of 13 different mouse models of breast cancer as well as normal mammary glands from FVB and BALB/c mice. Therefore, we compared our expression data with the raw data from that report, by computing the correlation between each of our tumor profiles and each of the Herschkowitz mouse tumor profiles. As expected, the expression profile of tumors from the MMTV-PyMT transgenic mice in our animal facility correlated with that of the tumors from the MMTV-PyMT transgenic mice in the Herschkowitz vivarium. Our RCAS-PyMT-induced tumors in MMTV-tva mice did not resemble any of the tissues used by Herschowitz et al., and our RCAS-PyMT-induced tumors in K6a-tva mice did not resemble the transgenic PyMT-induced tumors or other tumors used by Herschowitz et al. Unexpectedly, the K6a+ cell-derived tumors clustered with normal mammary glands from both FVB and BALB/c mice (Figure 4B), further supporting the observation of their high glandular differentiation.
None of the 13 previously characterized mouse models of human breast cancer resembles the normal-like breast cancer subtype seen in patients (Herschkowitz et al 2007). The close similarity of the expression profiles of our K6a+ cell-derived tumors with that of normal mammary glands suggests that these K6a+ cell-derived tumors may also resemble normal-like breast cancer in women. To test this possibility, we compared our expression arrays to the published database of expression arrays of human breast cancer samples from Hoadley et al. (2007), using only orthologous gene probes that were found in both array platforms. As anticipated, RCAS-PyMT-induced tumors in K6a-tva mice were tightly correlated with this subset of human breast cancers (Figure 4C) (Pearson’s P<1E-20, each K6a-tva tumor). In contrast, neither MMTV-PyMT transgene-induced mammary tumors nor RCAS-PyMT-induced tumors in MMTV-tva mice associated with the normal-like breast cancers (Figure 4C). Collectively, these findings demonstrate that the tumors arising from K6a+ bipotential progenitor cells are unique in that they recapitulate the normal-like breast cancer subtype in patients.
Discussion
Mammary stem and progenitor cells are ill-defined due to the lack of specific markers. Our results from mammosphere, colony-forming, and transplantation assays demonstrate that K6a expression marks mammary bipotential progenitor cells but not stem cells. Previously, cells in the basal compartment have been found to have the potential to generate both basal and luminal lineages (Bai and Rohrschneider 2010, Shackleton et al 2006, Stingl et al 2006, Zeng and Nusse 2010). To our knowledge, this is the first observation of a subset of luminal epithelial cells and body cells that can give rise to both epithelial and basal cell lineages, and our work establishes K6 as the first single marker that can identify mammary bipotential progenitor cells. In addition, the use of Lin− selection is not necessary for isolating K6a+ cells, since K6a is not produced in stromal cells. This single marker appears better for purifying bipotential progenitor cells than combinations of markers that can enrich for progenitor cells, such as Lin−/CD24high/CD49low or Lin−/CD29low/CD24+/CD61+ (Asselin-Labat et al 2007, Stingl et al 2006, Vaillant et al 2008). Of note, we do not yet know whether K6a+ cells constitute only part of the multipotent progenitor cell population in the mammary epithelium. In addition, because a specific antibody for human K6a is not currently available, the significance of K6a as a marker for human breast progenitor cells remains to be determined.
By expressing tva from the K6a promoter, we generated a mouse line in which tumorigenesis from these bipotential progenitor cells can be conveniently studied. We found that these cells are highly susceptible to transformation by PyMT and evolve into tumors that are distinct in histopathology, EMT, and expression profiles from those arising in RCAS-PyMT-infected MMTV-tva mice. More importantly, unlike all 13 other mouse models of breast cancer whose expression profiles have been compared with human breast cancers (Herschkowitz et al 2007), K6a+ cell-derived tumors resemble the normal-like breast cancer subtype in patients. This observation suggests that human normal-like breast cancers may also have an origin in bipotential progenitor cells. This first model of this subtype of human breast cancer provides a valuable preclinical tool for in vivo testing of therapeutic agents that may be especially effective in treating this subset of tumors. Although PyMT is a viral oncogene that is not found in human breast cancers, PyMT transforms breast cells through deregulating pathways (such as PI3K-AKT signaling) that are commonly altered in human breast cancers. However, we do not yet know why these K6a+ cells uniquely give rise to this subtype of breast cancer, or whether introduction of a different oncogenic lesion into this subset of mammary cells would also lead to normal-like breast cancer.
The most likely explanation for the phenotypic differences between RCAS-PyMT-induced tumors in K6a-tva mice vs. MMTV-tva mice is that in MMTV-tva mice, non-K6a+ cells were infected by RCAS-PyMT and evolved into tumors, and that the cell of origin had an impact on these cancer phenotypes. The cell of origin has been previously suggested to affect breast cancer phenotypes: For example, WAP expression is more restricted to the more differentiated subset of luminal cells than the MMTV LTR (Vargo-Gogola and Rosen 2007), and WAP-TAG (SV40 T antigen) transgene-induced mammary tumors differ from MMTV-TAG transgene-induced mammary tumors in gene expression profiles (Desai et al 2002). In addition, by using two different culture media to grow human breast cells from reduction mammoplasties, and then transforming them with a combination of oncogenes followed by transplantation into mice, Ince et al. (Ince et al 2007) found that the resulting tumors had different tumor phenotypes. However, the in vitro selection step used in that study adds uncertainty about the physiological and genetic state of the cells prior to transformation. The observed phenotypic differences in these transgenic mouse model comparisons may also be caused by differences in the level of oncogene expression or the developmental stage when the transgene is expressed (the MMTV LTR becomes active at a much earlier developmental time than the WAP promoter).
These concerns probably do not contribute to the phenotypic differences observed between tumors arising in RCAS-PyMT-infected K6a-tva mice vs. MMTV-tva mice – the PyMT is expected to be expressed at similar levels in infected cells in these two lines of mice, because the RCAS LTR has been reported to be constitutively active (Fisher et al 1999, Hughes 2004) and unaffected by estrogen signaling (Toneff et al 2010), and we have confirmed a similar activity in infected cells in K6a-tva mice vs. in MMTV-tva mice based on quantifying GFP intensity in RCAS-GFP-infected cells in these two lines of mice (734±61 vs. 645±39). In addition, multifocal tumors developed with a short median latency of 14 days in both tva lines following RCAS-PyMT infection at 5 weeks of age, suggesting that no undefined secondary genetic alterations were involved in specifying the tumor phenotypes. Moreover, 1,000-fold less viral particles were used to infect MMTV-tva mice than K6a-tva mice in order to achieve a similar low infection rate (no more than 10 infected cells per gland), minimizing a potential difference in tumor multiplicity. However, there might be a tumor modifier in the BAC transgene or in the genomic site where either transgene was inserted. Such a modifier might affect the potency of PyMT or the cellular response to PyMT – this is a caveat we cannot exclude when a single K6-tva founder transgenic line was used to compare with the single MMTV-tva line.
The identity of the cells that evolved into tumors in RCAS-PyMT-infected MMTV-tva mice is yet to be defined, but the cells of origin may be cells that have not yet committed to the luminal lineage, since basal cells were also found in RCAS-PyMT-induced tumors in MMTV-tva mice. They might be stem cells or a different subset of bipotential progenitor cells that do not express K6a. Of note, basal cells are usually infrequent in tumors arising in conventional MMTV transgenic models (Lindvall et al 2007). While luminal committed progenitor cells or differentiated luminal cells may be the cells of origin in these conventional models, it is also possible that the cells of origin are stem or bipotential progenitor cells – any basal progeny would stop expressing the initiating transgenic oncogene since MMTV LTR is known to be inactive in these cells (Du et al 2006); thus, they would fail to expand.
In summary, we have demonstrated that K6a expression marks mammary bipotential progenitor cells, and that these K6a+ cells can be induced to form distinct normal-like tumors. We further conclude that the cell of origin affects breast cancer phenotypes. This new model offers a tool to study the evolution of normal-like breast cancer and to preclinically test potential therapeutic agents against this subtype, and provides an opportunity to test, in mammary bipotential progenitor cells specifically, oncogenic lesions that are directly implicated in human breast cancer.
Materials and Methods
The MMTV-tva mouse line has been previously reported (Du et al 2006). MMTV-Wnt1 and MMTV-PyMT mice were purchased from the Jackson Laboratory (Bar Harbor, ME). The generation of the K6a-tva mouse line is described in Supplemental Information (SI). All other materials and methods are detailed in SI Text.
Supplementary Material
Acknowledgements
We thank Tammy Tong, Amanda McGrath, Ekaterina Bogoslovskaia, Andrei Lazarev, Meghali Goswami, and Yiqun Zhang for technical assistance, Drs. Jeffrey Rosen and Michael Lewis for stimulating discussions and/or critical review of this manuscript, and Drs. Robert D. Cardiff and Barry Gusterson for advice on pathological comparisons. This work was supported in part by funds from USAMRMC BC030755 and BC073703 (to YL), National Institutes of Health CA113869 and CA124820 (to YL), Project 5 of MMHCC U01 CA084243-07 (to YL; U01 PI: Dr. Jeffrey Rosen), and a developmental project of NCI P50 CA058183 (to Y.L.; SPORE PI: C. Kent Osborne).
Abbreviations
- K6
keratin 6
- tva
tumor virus A
- MMTV
mouse mammary tumor virus
- PyMT
polyoma middle T antigen
- αSMA
α smooth muscle actin
- RCAS
replication-competent, ALV-LTR, splice acceptor, Bryan-RSV pol, subgroup A virus
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
Conflict of interest
The authors declare no conflict of interest.
References
- Andrechek ER, Hardy WR, Laing MA, Muller WJ. Germ-line expression of an oncogenic erbB2 allele confers resistance to erbB2-induced mammary tumorigenesis. Proc Natl Acad Sci U S A. 2004;101:4984–4989. doi: 10.1073/pnas.0306802101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- Badders NM, Goel S, Clark RJ, Klos KS, Kim S, Bafico A, et al. The Wnt receptor, Lrp5, is expressed by mouse mammary stem cells and is required to maintain the basal lineage. PLoS ONE. 2009;4:e6594. doi: 10.1371/journal.pone.0006594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Rohrschneider LR. s-SHIP promoter expression marks activated stem cells in developing mouse mammary tissue. Genes Dev. 2010;24:1882–1892. doi: 10.1101/gad.1932810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates P, Young JA, Varmus HE. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell. 1993;74:1043–1051. doi: 10.1016/0092-8674(93)90726-7. [DOI] [PubMed] [Google Scholar]
- Desai KV, Xiao N, Wang W, Gangi L, Greene J, Powell JI, et al. Initiating oncogenic event determines gene-expression patterns of human breast cancer models. Proc Natl Acad Sci U S A. 2002;99:6967–6972. doi: 10.1073/pnas.102172399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth SM. Polyoma virus middle T antigen and its role in identifying cancer-related molecules. Nat Rev Cancer. 2002;2:951–956. doi: 10.1038/nrc946. [DOI] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Podsypanina K, Huang H, McGrath A, Toneff MJ, Bogoslovskaia E, et al. Introduction of oncogenes into mammary glands in vivo with an avian retroviral vector initiates and promotes carcinogenesis in mouse models. Proc Natl Acad Sci U S A. 2006;103:17396–17401. doi: 10.1073/pnas.0608607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher GH, Orsulic S, Holland E, Hively WP, Li Y, Lewis BC, et al. Development of a flexible and specific gene delivery system for production of murine tumor models. Oncogene. 1999;18:5253–5260. doi: 10.1038/sj.onc.1203087. [DOI] [PubMed] [Google Scholar]
- Grimm SL, Bu W, Longley MA, Roop DR, Li Y, Rosen JM. Keratin 6 is not essential for mammary gland development. Breast Cancer Res. 2006;8:R29. doi: 10.1186/bcr1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry MD, Triplett AA, Oh KB, Smith GH, Wagner KU. Parity-induced mammary epithelial cells facilitate tumorigenesis in MMTV-neu transgenic mice. Oncogene. 2004;23:6980–6985. doi: 10.1038/sj.onc.1207827. [DOI] [PubMed] [Google Scholar]
- Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoadley KA, Weigman VJ, Fan C, Sawyer LR, He X, Troester MA, et al. EGFR associated expression profiles vary with breast tumor subtype. BMC Genomics. 2007;8:258. doi: 10.1186/1471-2164-8-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SH. The RCAS vector system. Folia Biol (Praha) 2004;50:107–119. [PubMed] [Google Scholar]
- Ince TA, Richardson AL, Bell GW, Saitoh M, Godar S, Karnoub AE, et al. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell. 2007;12:160–170. doi: 10.1016/j.ccr.2007.06.013. [DOI] [PubMed] [Google Scholar]
- Jeselsohn R, Brown NE, Arendt L, Klebba I, Hu MG, Kuperwasser C, et al. Cyclin D1 kinase activity is required for the self-renewal of mammary stem and progenitor cells that are targets of MMTV-ErbB2 tumorigenesis. Cancer Cell. 2010;17:65–76. doi: 10.1016/j.ccr.2009.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Welm B, Podsypanina K, Huang S, Chamorro M, Zhang X, et al. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc Natl Acad Sci U S A. 2003;100:15853–15858. doi: 10.1073/pnas.2136825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Rosen JM. Stem/progenitor cells in mouse mammary gland development and breast cancer. J Mammary Gland Biol Neoplasia. 2005;10:17–24. doi: 10.1007/s10911-005-2537-2. [DOI] [PubMed] [Google Scholar]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- Lindvall C, Bu W, Williams BO, Li Y. Wnt signaling, stem cells, and the cellular origin of breast cancer. Stem Cell Rev. 2007;3:157–168. doi: 10.1007/s12015-007-0025-3. [DOI] [PubMed] [Google Scholar]
- Mollet M, Godoy-Silva R, Berdugo C, Chalmers JJ. Computer simulations of the energy dissipation rate in a fluorescence-activated cell sorter: Implications to cells. Biotechnol Bioeng. 2008;100:260–272. doi: 10.1002/bit.21762. [DOI] [PubMed] [Google Scholar]
- Molyneux G, Geyer FC, Magnay FA, McCarthy A, Kendrick H, Natrajan R, et al. BRCA1 Basal-like Breast Cancers Originate from Luminal Epithelial Progenitors and Not from Basal Stem Cells. Cell Stem Cell. 2010;7:403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Rosner A, Miyoshi K, Landesman-Bollag E, Xu X, Seldin DC, Moser AR, et al. Pathway pathology: histological differences between ErbB/Ras and Wnt pathway transgenic mammary tumors. Am J Pathol. 2002;161:1087–1097. doi: 10.1016/S0002-9440(10)64269-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Sleeman KE, Kendrick H, Ashworth A, Isacke CM, Smalley MJ. CD24 staining of mouse mammary gland cells defines luminal epithelial, myoepithelial/basal and non-epithelial cells. Breast Cancer Res. 2006;8:R7. doi: 10.1186/bcr1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GH, Mehrel T, Roop DR. Differential keratin gene expression in developing, differentiating, preneoplastic, and neoplastic mouse mammary epithelium. Cell Growth Differ. 1990;1:161–170. [PubMed] [Google Scholar]
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- Toneff M, Du Z, Dong J, Huang J, Sinai P, Forman J, et al. Somatic expression of PyMT or activated ErbB2 induces estrogen-independent mammary tumorigenesis. Neoplasia. 2010;12:718–726. doi: 10.1593/neo.10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant F, Asselin-Labat ML, Shackleton M, Forrest NC, Lindeman GJ, Visvader JE. The mammary progenitor marker CD61/beta3 integrin identifies cancer stem cells in mouse models of mammary tumorigenesis. Cancer Res. 2008;68:7711–7717. doi: 10.1158/0008-5472.CAN-08-1949. [DOI] [PubMed] [Google Scholar]
- Vargo-Gogola T, Rosen JM. Modelling breast cancer: one size does not fit all. Nat Rev Cancer. 2007;7:659–672. doi: 10.1038/nrc2193. [DOI] [PubMed] [Google Scholar]
- Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- Welm BE, Tepera SB, Venezia T, Graubert TA, Rosen JM, Goodell MA. Sca-1(pos) cells in the mouse mammary gland represent an enriched progenitor cell population. Dev Biol. 2002;245:42–56. doi: 10.1006/dbio.2002.0625. [DOI] [PubMed] [Google Scholar]
- Wojcik SM, Longley MA, Roop DR. Discovery of a novel murine keratin 6 (K6) isoform explains the absence of hair and nail defects in mice deficient for K6a and K6b. J Cell Biol. 2001;154:619–630. doi: 10.1083/jcb.200102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng YA, Nusse R. Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell. 2010;6:568–577. doi: 10.1016/j.stem.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.