Abstract
We utilized a series of pyrimidine analogues modified at O2, N-3, and N4/O4 to determine if two B family DNA polymerases, human DNA polymerase α and herpes simplex virus I DNA polymerase, choose whether or not to polymerize pyrimidine dNTPs using the same mechanisms they use for purine dNTPs. Removing O2 of a pyrimidine dNTP vastly decreased incorporation by these enzymes and also compromised fidelity in the case of C analogues, while removing O2 from the templating base had more modest effects. Removing the Watson-Crick hydrogen bonding groups of N-3 and N4/O4 greatly impaired polymerization, both of the resulting dNTP analogues as well as polymerization of natural dNTPs opposite these pyrimidine analogues when present in the template strand. Thus, the Watson-Crick hydrogen bonding groups of a pyrimidine clearly play an important role in enhancing correct dNTP polymerization, but are not essential for preventing misincorporation. These studies also indicate that DNA polymerases recognize bases extremely asymmetrically, both in terms of whether they are a purine or pyrimidine and whether they are in the template or are the incoming dNTP. The mechanistic implications of these results regarding how polymerases discriminate between right and wrong dNTPs are discussed.
Keywords: Fidelity, misincorporation, kinetics, base-pair, nucleotide
Accurate replication of genomic DNA is critical for cellular survival and proliferation. Despite the complexity of the cellular DNA replication process, the overall error rates are remarkably low. Three discrete processes contribute to the accuracy of replication. First, replicative DNA polymerases typically exhibit relatively low error frequencies of 10−3 to 10−6 errors per dNTP inserted (1). Second, misincorporation events decrease the rate of elongation significantly, thereby allowing 3′-5′ exonucleolytic proofreading to occur and decreasing the error rate by 100-fold or so. Finally, postreplicative repair enzymes reduce the overall error frequency to roughly 10−9 errors per nucleotide inserted (2).
The mechanisms by which various polymerases efficiently distinguish between correct and incorrect dNTP substrates during polymerization are not yet fully understood. Furthermore, different polymerases appear to utilize significantly different mechanisms (3-4). Some low-fidelity enzymes, such as human primase, herpes primase, and Y-family DNA polymerases, appear to largely utilize the Watson-Crick hydrogen bonding groups on the incoming (d)NTPs and the templating base to help identify correct and incorrect incorporation events (5-10). On the other hand, various studies have shown that A and B family DNA polymerases do not require Watson-Crick hydrogen bond formation for dNTP incorporation (11-12). B family polymerases use specific functional groups on the base of an incoming purine dNTP to prevent misincorporation and to enhance correct incorporation (4, 13-16). The methods used by A family polymerases are less well understood; while some studies indicate that shape selectivity may be critical for correct dNTP incorporation, others imply that shape is not important (16-26). For example, Kool and coworkers showed that some A family polymerases efficiently incorporate 2,4-dihalotoluene dNTPs in a manner consistent with the enzyme using shape as a primary determinant (27). However, these enzymes also readily incorporate purine dNTP opposite a templating T and dITP opposite a templating C (19, 28), even though the shapes of purine and hypoxanthine significantly vary from the shapes of adenine and guanine, respectively.
DNA polymerase α (pol α1) is a key polymerase (along with primase, pol δ, and pol ε) required during nuclear DNA replication (29). Pol α is a B-family polymerase that typically exhibits low processivity, polymerizing ~12 nucleotides before dissociating, and moderate fidelity, making 10−3-10−5 errors per dNTP polymerized (30-31). Biologically, pol α is responsible for the initial extension of primase-synthesized RNA primers in all new DNA strands. Pol α lacks 3′-5′ exonuclease activity; therefore, the incorporation of an incorrect dNTP results in the dissociation of pol α and subsequent association of a processive replicative polymerase with exonuclease activity (pol δ or pol ε) (29, 32).
Pol α requires neither the formation of Watson-Crick hydrogen bonds nor a correctly shaped base pair for the rapid incorporation of a dNTP (16, 33-34). Instead, recent work has shown that during incorporation of dATP and related purine analogues, pol α employs a combination of positive and negative selectivity to ensure accuracy of replication. N-1 and N-3 serve as negative selectors and help prevent misincorporation, while N-1 and N6 act as positive selectors and enhance correct incorporation (4). Thus, a combination of positive and negative selectivity provides accuracy for pol α during dNTP incorporation, rather than shape or hydrogen bonding patterns. The HSV-1 polymerase, another B-family DNA polymerase, uses the same general mechanisms as pol α, although the precise roles of N-1 and N6 vary between the two enzymes (14).
While the chemical features of purine bases have been examined with respect to their roles in correct incorporation and preventing misincorporation by pol α and herpes DNA polymerase, the contributions of the different functional groups on pyrimidines have not been examined. Accordingly, we examined the role of O2, N-3, and N4/O4 of pyrimidine dNTPs and template bases for incorporation and fidelity with these two enzymes.
EXPERIMENTAL PROCEDURES
Materials
All reagents were of the highest quality commercially available. Unlabeled natural dNTPs were from Sigma, and radiolabeled dNTPs were from Perkin-Elmer. 2-pyrimidinone dNTP (dZeb) was from Trilink. Protected phosphoramidites were from Glen Research. Synthetic DNA oligonucleotides were purchased from IDT or Biosearch. The two subunit p180-p70 polymerase complex was expressed in baculovirus-infected SF9 cells at the Tissue Culture Core Facility of the University of Colorado Health Sciences Center and purified as previously described (35), with the exception that the enzyme was stored in 50% glycerol, 1 mM ethylenediaminetetraacetic acid, 50 mM Tris-HCl, pH 8.8, and 1 mM dithiothreitol. HSV-1 DNA polymerase (UL30/UL42 complex) was expressed and purified as previously described (7).
5′-End Labeling of Primers and Annealing of Primer-Templates
DNA primers were 5′-32P-labeled using polynucleotide kinase (New England Biolabs) and [γ-32P]ATP. The labeled primer was gel purified and annealed to the template as previously described (36-37).
Polymerization Assays
Assays were performed under steady-state conditions essentially as previously described (4, 13, 14) using either herpes simplex I DNA polymerase or pol α. Briefly, assays typically contained 1 μM 5′-[32P]-primer/template, 50 mM Tris-HCl (pH 8.0), 50 mM MgCl2, 1 mM dithiothreitol, 0.1 mg/mL bovine serum albumin, 5% glycerol, and varying concentrations of natural or analogue dNTPs, in a total volume of 5-10 μL. Reactions were initiated by adding enzyme, incubated at 37 °C for 5-30 min, and quenched with an equal volume of gel loading buffer (formamide/0.05% xylene cyanol and bromophenol blue). Products were separated using denaturing gel electrophoresis (20% polyacrylamide, 8 M urea) and imaged using a Typhoon Phosphorimager (Molecular Dynamics). Kinetic parameters were determined by fitting data to the Michaelis-Menten equation using KaleidaGraph 4.0. All rates were normalized to the same final enzyme concentration (1 nM herpes pol, 5 nM pol α). The reported discrimination values were determined by comparing the efficiency of incorporation for the analogue (Vmax/KM) to the efficiency of incorporation for the corresponding natural nucleotide (Vmax/KM normalized to 1).
Synthesis of Nucleotide Analogues
C-2′-deoxyribonucleosides (6AmPy, 6ClPy, 6MePy, and 6OPy1) were synthesized as previously described (38). 2OPy1 nucleoside and 5′-dimethoxytrityl-3′-phosphoramidites and oligonucleotides were synthesized as described using established procedures (39) on an Applied Biosystems 394 automatic DNA synthesizer (40, 41).
Synthesis of Nucleoside 5′-Triphosphates
Nucleosides were converted to nucleoside 5′-triphosphates as previously described (39). Crude nucleoside triphosphates were purified by loading them onto an anion-exchange column (Sephadex-DEAE A-25, Aldrich, pre-equilibrated in TEAB and then washed with H2O) followed by elution with a 0-1 M TEAB gradient. Fractions were identified by MALDI mass spectrometry (negative M-1 ion) with a THAP matrix. Collected fractions were evaporated and purified by reverse-phase (C18) HPLC using a gradient of 0-50% MeCN in 20 mM triethylammonium acetate.
The purity was determined by an appearance of a single peak in HPLC (gradient from 0% to 60 % MeCN in 20 mM TEAB in water) and the triphosphates identified by MALDI MS and 31P NMR.
6AmPy dNTP: MS (MALDI, negative ion) 449 (M-1) calcd 449; 31P NMR (400MHz,D2O) δ-9.8 (bs, 2P, α-P, γ-P), −22.3 (m, 1P, β-P). 6ClPy dNTP: MS (MALDI, negative ion) 468 (M-1) calcd 468; 31P NMR (400MHz,D2O) δ-9.5 (bs, 2P, α-P, γ-P), –23.0 (m, 1P, β-P). 6MePy dNTP: MS (MALDI, negative ion) 448 (M-1) calcd 448; 31P NMR (400MHz,D2O) δ-10.4 (bs, 2P, α-P, γ-P), –22.9 (m, 1P, β-P). 6OPy dNTP: MS (MALDI, negative ion) 450 (M-1) calcd 450; 31P NMR(400MHz,D2O) δ- δ-10.0 (bs, 2P, α-P, γ-P), –21.1 (m, 1P, β-P).
RESULTS
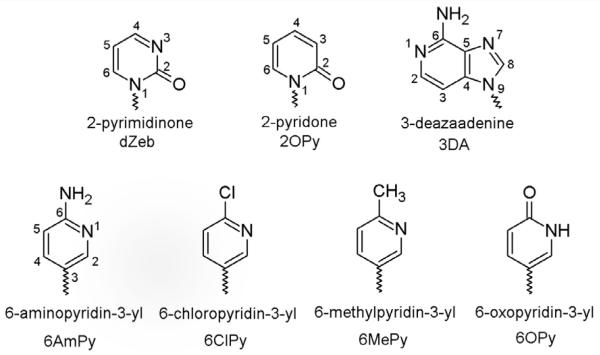
Previous studies have shown that B family DNA polymerases readily polymerize purine dNTP analogues whose base size, shape, and Watson-Crick hydrogen-bonding pattern do not closely resemble those of canonical bases, while the canonical bases are only very rarely misincorporated (4, 16, 33). The incorporation rate of purine dNTPs opposite correct and incorrect template bases depends upon the effects of N-1, N-3, and N6 via a combination of positive and negative selectivity. Herein, we probed the roles of O2, N-3, and N4/O4 of pyrimidines to identify the similarities and differences between the interaction of B family polymerases with purines and pyrimidines (Figure 1).
Figure 1.
Structures, names, and abbreviations of base analogue used.
The steady-state kinetic parameters for incorporation of both natural and analogue dNTPs were measured on synthetic primer-templates of defined sequence across from both natural and analogue templating bases. Discrimination values reflect the efficiency of correct dNTP incorporation compared to the incorporation efficiency of the tested dNTP (Vmax/Km). Table 1 shows the data for natural nucleotide incorporation by pol α and herpes pol. Both enzymes discriminated against incorrect natural dNTPs by 500 to >10,000-fold (14, 16).
Table 1.
Natural Base Incorporations
| dNTP | DNA-N | Vmax | Km | Vmax/Km | Discriminationa |
|---|---|---|---|---|---|
| (primer elongation min−1) (μM) (elongation μM−1 min−1) | |||||
| a)Pol α | |||||
| dA | DNA-T | 3.4 (0.1) | 0.31 (0.05) | 11 | 1 |
| dT | DNA-A | 3.2 (0.3) | 1.7 (0.4) | 1.8 | 1 |
| dU | DNA-A | 4.8 (0.4) | 2.2 (0.4) | 2.1 | 0.86 |
| dC | DNA-G | 9.6 (0.2) | 1.2 (0.1) | 8.4 | 1 |
| dG | DNA-C | 1.15 (0.09) | 0.11 (0.3) | 10 | 1 |
| b)Herpes pol | |||||
| dA | DNA-T | 1.6 (0.05) | 0.41 (0.06) | 3.9 | 1 |
| dT | DNA-A | 1.7 (0.06) | 1.3 (0.2) | 1.3 | 1 |
| dC | DNA-G | 1.2 (0.02) | 1.1 (0.1) | 1.1 | 1 |
| dG | DNA-C | 6.5 (1.4) | 2.6 (0.8) | 2.5 | 1 |
Discrimination values reflect the efficiency of dNTP incorporation compared to the incorporation efficiency of the corresponding natural base pair. Values in parentheses are the standard deviations.
Role of Watson-Crick hydrogen bonding groups
We initially examined the effect of removing N4 from cytosine in the incoming dNTP using the base analogue dZebTP. Table 2a shows that pol α polymerized dZebTP 16-fold less efficiently opposite a templating G than it polymerized dCTP. Removing N4 also had effects on fidelity since pol α discriminated against polymerizing dZebTP opposite a templating A by only 140-fold, somewhat less than the 3700-fold for misincorporation of dCTP. To examine the effects of removing the Watson-Crick hydrogen bonding groups at both N-3 and N4/O4, we utilized the base analogue 2-pyridone. Pol α strongly discriminated against polymerizing both 2-pyridone dNTP opposite the natural bases, as well as the natural dNTPs opposite 2-pyridone. Thus, the Watson-Crick hydrogen bonding groups of a pyrimidine clearly play an important role in enhancing correct dNTP polymerization by pol α, but are not essential for preventing misincorporation.
Table 2.
Effects of Watson-Crick Hydrogen Bonding Groups
| dNTP | DNA-N | Vmax | Km | Vmax/Km | Discriminationa |
|---|---|---|---|---|---|
| (primer elongation min−1) (μM) (elongation μM−1 min−1) | |||||
| a)Pol α | |||||
| dZeb | DNA-A | 2.1 (0.3) | 162 (61) | 0.013 | 140 |
| dZeb | DNA-G | 4.6 (0.2) | 8.5 (1.5) | 0.54 | 16 |
| dC | DNA-A | 0.22 (0.01) | 432 (65) | 0.0005 | 3700 |
| d2OPy | DNA-A | 4.5 (0.5) | 684 (242) | 0.0041 | 450 |
| d2OPy | DNA-T | 2.6 (1.4) | 335 (173) | 0.0042 | 2600 |
| d2OPy | DNA-C | nd | nd | nd | >10000 |
| d2OPy | DNA-G | 0.50 (0.3) | 161 (17) | 0.0015 | 5600 |
| dA | DNA-2OPy | nd | nd | nd | >10000 |
| dT | DNA-2OPy | nd | nd | nd | >10000 |
| dC | DNA-2OPy | nd | nd | nd | >10000 |
| dG | DNA-2OPy | nd | nd | nd | >10000 |
| b)Herpes pol | |||||
| d2OPy | DNA-A | 0.48 (0.07) | 430 (180) | 0.0011 | 1200 |
| d2OPy | DNA-T | 0.29 (0.01) | 120 (30) | 0.0025 | 1900 |
| d2OPy | DNA-C | nd | nd | nd | >20000 |
| d2OPy | DNA-G | 0.21 (0.02) | 350 (80) | 0.0006 | 1850 |
Discrimination values reflect the efficiency of dNTP incorporation compared to the incorporation efficiency of the corresponding natural base pair. nd: not detectable. Values in parentheses are the standard deviations.
To determine if other B family DNA polymerases utilize the Watson-Crick hydrogen bonding groups of a pyrimidine similarly to pol α, we examined HSV-1 DNA polymerase. Table 2b shows that HSV-1 pol also strongly discriminated against polymerizing 2-pyridone dNTP across from a natural template base. Since the amino acids that comprise the active sites of B-family DNA polymerases are remarkably well conserved, these data suggest that this family of enzymes will generally discriminate against pyrimidine dNTP analogues lacking both Watson-Crick hydrogen bonding groups.
Role of the Minor Groove Hydrogen Bond Acceptor on a Pyrimidine dNTP
The importance of O2, a hydrogen bond acceptor that lies in the minor groove (Figure 2), for generation of base pairs involving pyrimidines was examined using a series of analogues that lacked O2. So that N-3 and N4/O4 (if present) maintained the hybridization states appropriate for either C or T and to avoid adding a positive charge to the ring, N-1 was also converted to carbon such that the nucleosides were C-glycosides. Three of the bases, 6AmPy, 6MePy, and 6ClPy, are C analogues that can form two, one, and one Watson-Crick hydrogen bonds with G, respectively, while 6OPy is a T analogue that can form two Watson-Crick hydrogen bonds with A (Figure 1). As a T analogue, 6OPy also lacks the methyl group found at C-5 of T. To first test the possibility that the absence of this methyl group was significant, we compared polymerization of dUTP and dTTP (Table 1). The similar kinetic parameters for dUTP and dTTP polymerization indicate that the absence of the methyl has little if any effect on polymerization.
Figure 2.
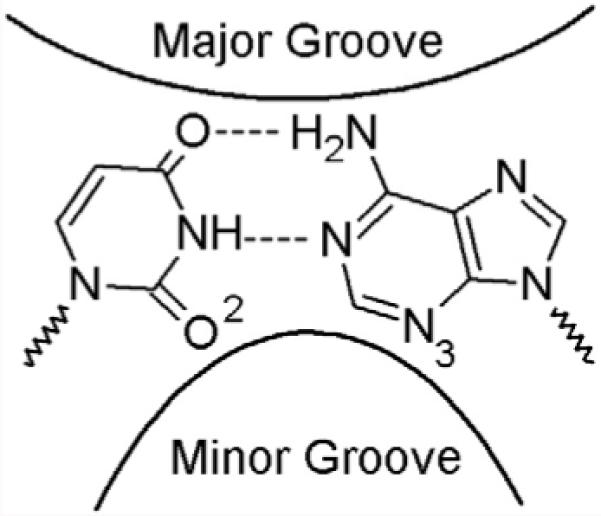
Major and minor grooves in DNA.
Pol α strongly discriminated against polymerizing 6OPy dNTP opposite a templating A (120-fold discrimination, Table 3a) even though this base can form two Watson-Crick hydrogen bonds. On the other hand, the loss of O2 did not result in major changes in the rate of misincorporation opposite a template T, C, or G compared to the natural dTTP, indicating that O2 is not a major determinant in preventing misincorporation of dTTP. HSV-1 DNA polymerase likewise discriminated against polymerization of 6OPy dNTP, with the only difference between HSV-1 DNA polymerase and pol α being the larger effect of losing O2 with the HSV-1 enzyme (610-fold opposite a template A, Table 3b).
Table 3.
Effects of O2 in dTTP
| dNTP | DNA-N | Vmax | Km | Vmax/Km | Discriminationa |
|---|---|---|---|---|---|
| (primer elongation min−1) (μM) (elongation μM−1 min−1) | |||||
| a)Pol α | |||||
| d6OPy | DNA-A | 2.27 (0.07) | 146 (13) | 0.016 | 120 |
| d6OPy | DNA-T | nd | nd | nd | >10000 |
| d6OPy | DNA-C | L | L | 0.018 | 600 |
| d6OPy | DNA-G | nd | nd | nd | >10000 |
| b)Herpes pol | |||||
| d6OPy | DNA-A | 0.42 (0.12) | 200 (110) | 0.0022 | 610 |
| d6OPy | DNA-T | nd | nd | nd | >20000 |
| d6OPy | DNA-C | nd | nd | nd | >20000 |
| d6OPy | DNA-G | nd | nd | nd | >20000 |
Discrimination values reflect the efficiency of dNTP incorporation compared to the incorporation efficiency of the corresponding natural base pair. nd: not detectable. L: the rate increased linearly with increasing dNTP concentration, thus making it impossible to calculate Km or Vmax. Vmax/Km was calculated from the slope of the line. Values in parentheses are the standard deviations.
Removing O2 from dCTP had larger and more diverse effects than the corresponding modification of dUTP. We synthesized and tested three analogues, 6ClPy dNTP, 6MePy dNTP and 6AmPy dNTP, capable of forming one, one and two Watson-Crick hydrogen bonds with G, respectively (Table 4). Pol α did not detectably polymerize 6AmPy dNTP opposite G, even though two Watson-Crick hydrogen bonds could be formed. Likewise, it did not detectably polymerize 6ClPy dNTP or 6MePy dNTP opposite a templating G. Second, pol α misincorporated d6APy dNTP at higher rates than it did dCTP, suggesting that O2 of dCTP analogues plays a greater role in fidelity than does O2 of dTTP. HSV-1 DNA polymerase strongly discriminated against 6AmPy, 6ClPy, and 6MePy dNTPs opposite any template base, more so than did pol α and analogous to this enzymes stronger discrimination against 6OPy dNTP polymerization.
Table 4.
Effects of O2 in dCTP
| dNTP | DNA-N | Vmax | Km | Vmax/Km | Discriminationa |
|---|---|---|---|---|---|
| (primer elongation min−1) (μM) (elongation μM−1 min−1) | |||||
| a)Pol α | |||||
| d6AmPy | DNA-A | 0.83 (0.03) | 99 (9) | 0.0084 | 220 |
| d6AmPy | DNA-T | L | L | 0.037 | 270 |
| d6AmPy | DNA-C | L | L | 0.0096 | 1100 |
| d6AmPy | DNA-G | nd | nd | nd | >10000 |
| d6MPy | DNA-A | 0.31 (0.05) | 675 (210) | 0.0005 | 4000 |
| d6MPy | DNA-T | nd | nd | nd | >10000 |
| d6MPy | DNA-C | nd | nd | nd | >10000 |
| d6MPy | DNA-G | nd | nd | nd | >10000 |
| d6ClPy | DNA-A | 1.03 (0.03) | 43 (3) | 0.024 | 76 |
| d6ClPy | DNA-T | nd | nd | nd | >10000 |
| d6ClPy | DNA-C | nd | nd | nd | >10000 |
| d6ClPy | DNA-G | nd | nd | nd | >10000 |
| dC | DNA-A | 0.22 (0.01) | 432 (65) | 0.0005 | 3700 |
| dC | DNA-T | 0.18 (0.01) | 67 (8) | 0.0027 | 4100 |
| dC | DNA-C | nd | nd | nd | >10000 |
| b)Herpes pol | |||||
| 6AmPy | DNA-A | 0.075 (0.007) | 120 (40) | 0.00063 | 2100 |
| 6AmPy | DNA-T | nd | nd | nd | >20000 |
| 6AmPy | DNA-C | nd | nd | nd | >20000 |
| 6AmPy | DNA-G | 0.043 (0.04) | 110 (40) | 0.0004 | 2800 |
| 6ClPy | DNA-A | 0.71 (0.04) | 1900 (200) | 0.00037 | 3700 |
| 6ClPy | DNA-T | nd | nd | nd | >20000 |
| 6ClPy | DNA-C | nd | nd | nd | >20000 |
| 6ClPy | DNA-G | nd | nd | nd | >20000 |
| 6MePy | DNA-A | L | L | 0.00011 | 12000 |
| 6MePy | DNA-T | nd | nd | nd | >20000 |
| 6MePy | DNA-C | nd | nd | nd | >20000 |
| 6MePy | DNA-G | nd | nd | nd | >20000 |
Discrimination values reflect the efficiency of dNTP incorporation compared to the incorporation efficiency of the corresponding natural base pair. nd: not detectable. L: the rate increased linearly with increasing dNTP concentration, thus making it impossible to calculate Km or Vmax. Vmax/Km was calculated from the slope of the line. Values in parentheses are the standard deviations.
Role of the Minor Groove Hydrogen Bond Acceptor on the Templating Base
We extended these studies to determine how removing O2 from a templating pyrimidine would affect polymerization of a natural dNTP. The loss of O2 from a templating T significantly reduced polymerization of dATP even though two Watson-Crick hydrogen bonds can still form (Table 5). Polymerization of non-cognate dNTPs was not affected, indicating that O2 of the template T does not play an important role in fidelity. These data also indicate that removing O2 from T in either the incoming dNTP or the templating base has very similar effects.
Table 5.
Effects of O2 in the template
| dNTP | DNA-N | Vmax | Km | Vmax/Km | Discriminationa |
|---|---|---|---|---|---|
| (primer elongation min−1) (μM) (elongation μM−1 min−1) | |||||
| Pol α | |||||
| dA | DNA-6OPy | 0.38 (0.03) | 4 (1) | 0.093 | 120 |
| dT | DNA-6OPy | 0.11 (0.01) | 66 (16) | 0.0016 | 6900 |
| dG | DNA-6OPy | 0.10 (0.01) | 71 (16) | 0.0014 | 7700 |
| dC | DNA-6OPy | nd | nd | nd | >10000 |
| dA | DNA-6AmPy | 0.8 (0.1) | 642 (184) | 0.0012 | 8100 |
| dT | DNA-6AmPy | 0.12 (0.01) | 73 (20) | 0.0016 | 6400 |
| dC | DNA-6AmPy | nd | nd | nd | >10000 |
| dG | DNA-6AmPy | 0.12 (0.01) | 0.16 (0.33) | 0.74 | 14 |
Discrimination values reflect the efficiency of dNTP incorporation compared to the incorporation efficiency of the corresponding natural base pair. nd: not detectable. Values in parentheses are the standard deviations.
Removing O2 from a templating C had relatively small effects on the efficiency of dGTP polymerization (14-fold, Table 5), in contrast to the huge effect observed upon removing O2 from the incoming dCTP (undetectable polymerization opposite G, Table 4a). Misincorporation opposite a templating 6AmPy (Table 5) was not significantly different than misincorporation opposite a templating C, a result that stands in contrast to the decreased fidelity of 6AmPy dNTP polymerization. Thus, the effects of removing O2 from C are highly asymmetric depending upon whether one looks at the incoming dCTP or templating C, and quite different than the effects of removing O2 from T.
The role of the minor groove hydrogen bond acceptor in a purine, N-3, was likewise examined by measuring dNTP incorporation opposite 3-deazaadenine (3DA). Table 6 shows that this loss of N-3 impaired incorporation of the cognate dTTP, but did not greatly impact incorporation of noncognate dNTPs. Thus, purine N-3 plays decidedly different roles depending upon whether the purine nucleotide is the dNTP or the templating nucleotide. In the dNTP, N-3 has minimal impact on correct dNTP incorporation, but plays a major role in preventing misincorporation (4), while in the template, N-3 is important for correct incorporation, but has minimal effect in preventing misincorporation. (Table 6).
Table 6.
Effects of removing N-3 from a templating dA.
| dNTP | DNA-N | Vmax | Km | Vmax/Km | Discriminationa |
|---|---|---|---|---|---|
| (primer elongation min−1) (μM) (elongation μM−1 min−1) | |||||
| Pol α | |||||
| dA | DNA-3DA | 0.13 (0.02) | 134 (66) | 0.0010 | 1900 |
| dT | DNA-3DA | 33 (2) | 178 (30) | 0.037 | 50 |
| dC | DNA-3DA | nd | nd | nd | >10000 |
| dG | DNA-3DA | nd | nd | nd | >10000 |
Discrimination values reflect the efficiency of dNTP incorporation compared to the incorporation efficiency of the corresponding natural base pair. nd: not detectable. Values in parentheses are the standard deviations.
DISCUSSION
The effects of removing heteroatoms from pyrimidine bases in both the template and incoming dNTP on two B family DNA polymerases were examined. Unlike the very small effects of removing N-3 from a purine dNTP (4), removing the functionally equivalent O2 of a pyrimidine dNTP greatly decreased incorporation by B family DNA polymerases and also compromised fidelity in the case of C analogues. Removing O2 from the templating base had more modest effects. Finally, removing the Watson-Crick hydrogen bonding groups from pyrimidines greatly impaired polymerization of the resulting dNTPs, as well as polymerization of natural dNTPs opposite these bases.
Removing O2 from pyrimidine dNTPs had surprisingly large effects on incorporation by both HSV-1 polymerase and pol α (Tables 2, 3). Even with analogues that could still form two Watson-Crick hydrogen bonds, correct incorporation was significantly impaired. The effects on fidelity varied from no effect to moderate increases in misincorporation (as defined by the polymerization of the analogue dNTP opposite a templating base with which it cannot form any normal Watson-Crick hydrogen bonds).
These very large effects of removing O2 from an incoming pyrimidine dNTP contrast with the effects of removing N-3 from a purine dNTP (4). Both groups reside within the minor groove in similar locations and are hydrogen bond acceptors (Figure 2). Yet, whereas converting N3 of dATP or dGTP to a carbon results in only small effects on correct polymerization and either significantly increases or has no effect on polymerization of an incorrect purine dNTP, removing O2 of a pyrimidine dNTP cripples polymerization generally.
While not as dramatic as the differences observed for polymerization of purine dNTPs versus pyrimidine dNTPs, comparing the effects of removing O2 from dUTP and dCTP showed significantly different effects. Removing O2 from dCTP more severely impacted correct incorporation and in the case of pol α, had a greater effect on fidelity. These differences provide further support to the idea that the mechanism(s) by which DNA polymerases recognize each of the canonical base-pairs are non-identical. Previous studies showed that the identical modification to different bases can have vastly different effects (28, 42). Additional support from this idea comes from biophysical studies by Waksman and coworkers, who found that ternary E-DNA-dNTP structures of KlenTaq varied accordingly to the identity of the dNTP-templating base examined (43). Furthermore, the enzyme showed different dynamics during replication of each canonical base-pair (44). Thus, it is probably inappropriate to consider a DNA polymerase as a “simple” machine that simply pairs an incoming dNTP with the templating base. Rather, the polymerase may actively read the templating base and then alter the chemistry of base-pair recognition depending upon the templating base.
The effects of removing O2 from a templating U were very similar to the effects of removing O2 from the incoming dUTP during pol α-catalyzed DNA synthesis. In contrast, whereas removing O2 from an incoming dCTP severely impacted both correct incorporation and fidelity, eliminating O2 from a templating C had but modest effects on polymerization of a correct dGTP and no significant effects on misincorporation of noncognate dNTPs. In the case of adenine, removing N-3 from a templating A produced a greater impact on polymerization efficiency (50-fold decrease, Table 6) than did removing N-3 from the incoming dATP (5-fold decrease, (4)) during generation of an A-T base-pair. These differences provide further evidence that DNA polymerases can recognize nucleotides very asymmetrically depending upon whether they are in the template or are the incoming dNTP, and that minor groove chemistry plays a major role in this asymmetry.
Potentially, three alternative mechanisms could explain these very different results on the effects of removing purine N-3 and pyrimidine O2. The different steric effects of altering N-3 and O2 could explain the differences. Whereas replacing N-3 of a purine with C has only a small steric effect, removing O2 of a pyrimidine has a large effect. Alternatively, the polymerases may differentially interact with the minor groove side of incoming purine and pyrimidine dNTPs, thereby causing the vastly different effects. The active sites of the B family polymerases are remarkably well conserved, especially the group of amino acids that surround the incipient base-pair between the incoming dNTP and the templating base. The B family polymerase from RB69 has been extensively studied structurally and provides potential clues for these differences (45-47). While there are no obvious hydrogen bonding groups in RB69 polymerase that could interact with O2 or N-3, the electron deficient edge of a tyrosine sits squarely in the minor groove of the incipient base-pair (Y567 in RB69 polymerase, Y957 in pol α and Y818 in HSV-1 DNA polymerase (4, 45, 47)). In the ternary RB69 polymerase-DNA-dTTP complex, this tyrosine interacts with O2 of the incoming dTTP (45). It is unknown if this tyrosine adopts a similar conformation during polymerization of other dNTPs, or if it adopts different conformations at different points during the catalytic cycle. Importantly, differential interaction of this tyrosine with a purine N-3 versus a pyrimidine O2 would explain the differences between removing the purine N-3 and pyrimidine O2. Likewise, the different effects of removing O2 from an incoming dCTP versus a templating C could be mediated by this tyrosine; for example, it could adopt different positions depending upon the identity of the templating base. This would significantly alter its interactions with the electron rich N-3 of a purine or O2 of a pyrimidine and, depending upon the functional role of these interactions, alter correct and/or incorrect dNTP polymerization. Finally, these differential results could reflect the electronic changes in the aromatic rings caused by removal of purine N-3 and pyrimidine O2. For example, removing N-3 will decrease the π electron density of the ring. The altered electron density could impact both stacking onto the neighboring bases as well as interactions with two conserved amino acids that interact with the π faces of the templating and dNTP bases (asparagines and serine), thereby affecting polymerization. Importantly, the remarkably well-conserved chemistry of this region of the active site among different B-family polymerases (48) suggests that the results observed for HSV-1 polymerase and pol α will be similar in other members of this family.
The Watson-Crick hydrogen bonding groups of a pyrimidine clearly play an important role in enhancing correct dNTP polymerization, but are not essential for preventing misincorporation. In the absence of any Watson-Crick hydrogen bonding groups (i.e., 2-pyridone), neither pol α nor HSV-1 polymerase efficiently polymerized a canonical dNTP. Adding back N-3 to the pyridine significantly increased polymerization of the resulting dZebTP opposite a templating G, demonstrating the key role of this group in promoting efficient polymerization. As noted earlier, however, the ability to form two Watson-Crick hydrogen bonds did not by itself ensure efficient polymerization. Additionally, the efficient polymerization of dZebTP provides further evidence that a correctly shaped base-pair is not needed for efficient dNTP incorporation by pol α.
These results have significant implications for the design of novel base pairs that are incorporated efficiently and accurately by polymerases, particularly if one would like to design bases that are “purine-like” and “pyrimidine-like”. Efficient and accurate replication of any novel base-pair requires that the chemical features of the two bases satisfy the requirements of the polymerase. However, the data presented herein show that polymerases can exhibit distinctly different requirements for purine and pyrimidine dNTPs (e.g., the minor groove H-bond acceptors N-3 and O2 on purines and pyrimidines). Furthermore, the chemical requirements can vary depending upon whether the base is on the incoming dNTP or on the template strand. For example, while accurate and efficient incorporation of dCTP absolutely requires O2 of dCTP, this is not true for a templating C. These large differences in chemical requirements for even the different natural bases imply that different unnatural bases will also have diverse and potentially unpredictable requirements, thereby greatly complicating efforts to rationally design novel base-pairs.
Acknowledgments
This work was supported by grants from the NIH (GM54194 and AI59764) to R.D.K. and by grants from the Academy of Sciences of the Czech Republic (Z4 055 905), the Ministry of Education (LC 512) and the Grant Agency of the ASCR (IAA400550902) to M.H.
Footnotes
Abbreviations used: 6AmPy, 6-aminopyridin-3-yl; 6ClPy, 6-chloropyridin-3-yl; 3DA, 3-deazaadenine; MALDI, matrix-assisted laser desorption ionization; 6MePy, 6-methylpyridin-3-yl; 2OPy, 2-pyridone; 6OPy, 6-oxopyridin-3-yl; pol α, DNA polymerase α; pol δ, DNA polymerase δ; pol ε, DNA polymerase ε; SD, standard deviation; TEAB, triethylammonium bicarbonate; THAP, 2′,4′,6′-trihydroxyacetophenone; Tris-HCl, tris(hydroxymethyl)-aminomethane-hydrochloric acid; dZeb, 2-pyrimidinone (zebularine)
REFERENCES
- 1.Kunkel TA, Bebenek K. Recent Studies on the Fidelity of DNA Synthesis. Biochim. Biophys. Acta. 1988;951:1–15. doi: 10.1016/0167-4781(88)90020-6. [DOI] [PubMed] [Google Scholar]
- 2.Roberts JD, Kunkel TA. In: DNA Replication in Eukaryotic Cells: Concepts, Enzymes and Systems. Depamphilis M, editor. Cold Spring Harbor Laboratory Press; Plainview, NY: 1996. pp. 217–247. [Google Scholar]
- 3.Washington M, Helquist S, Kool ET, Prakash L, Prakash S. Requirement of Watson-Crick Hydrogen Bonding for DNA Synthesis by Yeast DNA Polymerase eta. Mol. Cell. Biol. 2003;23:5107–5112. doi: 10.1128/MCB.23.14.5107-5112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beckman J, Kincaid K, Hocek M, Spratt T, Engels J, Cosstick R, Kuchta RD. Human DNA Polymerase α Uses a Combination of Positive and negative Selectivity to Polymerize Purine dNTPs with High Fidelity. Biochemistry. 2007;46:448–460. doi: 10.1021/bi061243s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramirez-Aguilar KA, Kuchta RD. Herpes Simplex Virus-1 PrimasesA Polymerase with Extraordinarily Low Fidelity. Biochemistry. 2004;43:9084–9091. doi: 10.1021/bi049335+. [DOI] [PubMed] [Google Scholar]
- 6.Moore CL, Chiaramonte M, Higgins T, Kuchta RD. Synthesis of Nucleotide Analogs That Potently and Selectively Inhibit Human DNA Primase. Biochemistry. 2002;41:14066–14075. doi: 10.1021/bi026468r. [DOI] [PubMed] [Google Scholar]
- 7.Moore CL, Zivkovic A, Engels JW, Kuchta RD. Human DNA primase uses Watson-Crick hydrogen bonds to distinguish between correct and incorrect nucleoside triphosphates. Biochemistry. 2004;43:12367–12374. doi: 10.1021/bi0490791. [DOI] [PubMed] [Google Scholar]
- 8.Arezi B, Kuchta RD. Eukaryotic DNA primase. Trends Biochem. Sci. 2000;25:572–576. doi: 10.1016/s0968-0004(00)01680-7. [DOI] [PubMed] [Google Scholar]
- 9.Choi JY, Lim S, Eoff RL, Guengerich FP. Kinetic Analysis of Base-Pairing Preference for Nucleotide Incorporation Opposite Template Pyrimidines by Human DNA Polymerase iota. J. Mol. Biol. 2009;389:264–274. doi: 10.1016/j.jmb.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolfle WT, Washington MT, Kool ET, Spratt TE, Helquist SA, Prakash L, Prakash S. Evidence for a Watson-Crick hydrogen bonding requirement in DNA synthesis by human DNA polymerase kappa. Mol. Cell Biol. 2005;25:7137–7143. doi: 10.1128/MCB.25.16.7137-7143.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuchta RD, Beckman J, Kincaid K, Moore C, Loi M, Timblin G. Mechanisms by which DNA and RNA polymerases discriminate between right and wrong (d)NTPs. FASEB J. 2006;20:A512–A513. [Google Scholar]
- 13.Patro JN, Urban M, Kuchta RD. Role of the 2-Amino Group of Purines during dNTP Polymerization by Human DNA Polymerase alpha. Biochemistry. 2009;48:180–189. doi: 10.1021/bi801823z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavanaugh NA, Urban M, Beckman J, Spratt TE, Kuchta RD. Identifying the Features of Purine dNTPs that Allow Accurate and Efficient DNA Replication by Herpes Simplex Virus I DNA Polymerase. Biochemistry. 2009;48:3554–3564. doi: 10.1021/bi8022202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kincaid K, Beckman J, Zivkovic A, Halcomb RL, Engels JW, Kuchta RD. Exploration of factors driving incorporation of unnatural dNTPS into DNA by Klenow fragment (DNA polymerase I) and DNA polymerase alpha. Nucleic Acids Res. 2005;33:2620–2628. doi: 10.1093/nar/gki563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiaramonte M, Moore CL, Kincaid K, Kuchta RD. Facile polymerization of dNTPs bearing unnatural base analogues by DNA polymerase alpha and Klenow fragment (DNA polymerase I) Biochemistry. 2003;42:10472–10481. doi: 10.1021/bi034763l. [DOI] [PubMed] [Google Scholar]
- 17.Henry AA, Olsen AG, Matsuda S, Yu CZ, Geierstanger BH, Romesberg FE. Efforts to expand the genetic alphabet: Identification of a replicable unnatural DNA self-pair. J. Am. Chem. Soc. 2004;126:6923–6931. doi: 10.1021/ja049961u. [DOI] [PubMed] [Google Scholar]
- 18.Leconte AM, Hwang GT, Matsuda S, Capek P, Hari Y, Romesberg FE. Discovery, characterization, and optimization of an unnatural base pair for expansion of the genetic alphabet. J. Am. Chem. Soc. 2008;130:2336–2343. doi: 10.1021/ja078223d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trostler M, Delier A, Beckman J, Urban M, Patro JN, Spratt TE, Beese LS, Kuchta RD. Discrimination between Right and Wrong Purine dNTPs by DNA Polymerase I from Bacillus stearothermophilus. Biochemistry. 2009;48:4633–4641. doi: 10.1021/bi900104n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee HR, Helquist SA, Kool ET, Johnson KA. Importance of hydrogen bonding for efficiency and specificity of the human mitochondrial DNA polymerase. J. Biol. Chem. 2008;283:14402–14410. doi: 10.1074/jbc.M705007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim TW, Delaney JC, Essigmann JM, Kool ET. Probing the active site tightness of DNA polymerase in subangstrom increments. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15803–15808. doi: 10.1073/pnas.0505113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunkel TA, Bebenek K. DNA replication fidelity. Ann. Rev. Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 23.Kool ET. Active site tightness and substrate fit in DNA replication. Ann. Rev. Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 24.Guckian KM, Kool ET. Highly precise shape mimicry by a difluorotoluene deoxynucleoside, a replication-competent substitute for thymidine. Angew. Chem. Int. Edit. 1997;36:2825–2828. doi: 10.1002/anie.199728251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moran S, Ren RXF, Kool ET. A thymidine triphosphate shape analog lacking Watson-Crick pairing ability is replicated with high sequence selectivity. Proc. Natl. Acad. Sci. U.S.A. 1997;94:10506–10511. doi: 10.1073/pnas.94.20.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moran S, Ren RXF, Rumney S, Kool ET. Difluorotoluene, a nonpolar isostere for thymine, codes specifically and efficiently for adenine in DNA replication. J. Am. Chem. Soc. 1997;119:2056–2057. doi: 10.1021/ja963718g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim TW, Delaney JC, Essigman JM, Kool ET. Probing the Active Site Tightness of DNA Polymerase in Subangstrom Increments. Proc. Natl. Acad. Sci. USA. 1998;102:15803–15808. doi: 10.1073/pnas.0505113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patro JN, Urban M, Kuchta RD. Interaction of Human DNA Polymerase α and DNA Polymerase I from Bacillus stearothermophilus with Hypoxanthine and 8-Oxoguanine Nucleotides. Biochemistry. 2009;48:8271–8278. doi: 10.1021/bi900777s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kornberg A, Baker T. In: DNA Replication. 2nd ed. Freeman WH, editor. San Francisco: 1992. [Google Scholar]
- 30.Thompson HC, Sheaff RJ, Kuchta RD. Recognition of RNA and DNA Primers by DNA Polymerase α. Nucleic Acids Res. 1995;23:4109–4115. doi: 10.1093/nar/23.20.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong Q, Copeland WC, Wang TSF. Mutational Studies of Human DNA Polymerase α. J. Biol. Chem. 1993;268:24175–24182. [PubMed] [Google Scholar]
- 32.Pavlov YI, Frahm C, Nick McElhinny SA, Niimi A, Suzuki M, Kunkel TA. Evidence that Errors Made by DNA Polymerase α are Corrected by DNA Polymerase δ. Curr. Biol. 2006;16:202–207. doi: 10.1016/j.cub.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Ogawa AK, Wu Y, McMinn DL, Liu J, Schultz PG, Romesberg F. Efforts toward the Expansion of the Genetic Alphabet: Information Storage and Replication with Unnatural Hydrophobic Base Pairs. J. Am. Chem. Soc. 2000;122:3274–3287. [Google Scholar]
- 34.Wu Y, Ogawa AK, Berger M, McMinn DL, Schultz PG, Romesberg F. Efforts toward Expansion of the Genetic Alphabet: Optimization of Interbase Hydrophobic Interactions. J. Am. Chem. Soc. 2000;122:7621–7632. [Google Scholar]
- 35.Cavanaugh NA, Kuchta RD. Initiation of new DNA Strands by the Herpes Primase-Helicase Complex and either Herpes DNA Polymerase or Human DNA Polymerase α. J. Biol. Chem. 2009;284:1523–32. doi: 10.1074/jbc.M805476200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuchta RD, Mizrahi V, Benkovic PA, Johnson KA, Benkovic SJ. Kinetic Mechanism of DNA Polymerase I (Klenow) Biochemistry. 1989;26:8410–8417. doi: 10.1021/bi00399a057. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratories; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 38.Joubert N, Pohl R, Klepetarova B, Hocek M. Modular and practical synthesis of 6-substituted pyridin-3-yl C-nucleosides. J. Org. Chem. 2007;72:6797–6805. doi: 10.1021/jo0709504. [DOI] [PubMed] [Google Scholar]
- 39.Urban M, Joubert N, Hocek M, Alexander ER, Kuchta RD. Herpes Simplex Virus-1 DNA Primase: A Remarkably Inaccurate yet Selective Polymerase. Biochemistry. 2009;48:10866–10881. doi: 10.1021/bi901476k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beaucage SL, Caruthers MH. Deoxunucleoside Phosphoramidites: A New Class of Key Intermediates for Deoxypolynucleotide Synthesis. Tetrahedron Lett. 1981;22:1859–1862. [Google Scholar]
- 41.McBride LJ, Caruthers MH. An Investigation of Several Deoxynucleoside Phosphoramidites Useful for Synthesizing Deoxynucleotides. Tetrahedron Lett. 1983;24:245–248. [Google Scholar]
- 42.Henry AA, Yu C, Romesberg FE. Determinants of Unnatural Nucleobase Stability and Polymerase Recognition. J. Am. Chem. Soc. 2003;125:9638–9646. doi: 10.1021/ja035398o. [DOI] [PubMed] [Google Scholar]
- 43.Li Y, Kong Y, Korolev S, Waksman G. Crystal structures of the Klenow fragment of Thermus aquaticus DNA polymerase I complexed with deoxyribonucleoside triphosphates. Protein Sci. 1998;7:1116–1123. doi: 10.1002/pro.5560070505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rothwell PJ, Waksman G. A pre-equilibrium before nucleotide binding limits fingers subdomain closure by Klentaq1. J Biol Chem. 2007;282:28884–28892. doi: 10.1074/jbc.M704824200. [DOI] [PubMed] [Google Scholar]
- 45.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol α family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 46.Shamoo Y, Steitz TA. Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell. 1999;99:155–166. doi: 10.1016/s0092-8674(00)81647-5. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Sattar AK, Wang CC, Karam JD, Konigsberg WH, Steitz TA. Crystal structure of a pol α family replication DNA polymerase from bacteriophage RB69. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 48.Wang TS, Wong SW, Korn D. Human DNA polymerase α: Predicted functional domains and relationships with viral DNA polymerases. FASEB J. 1989;3:14–21. doi: 10.1096/fasebj.3.1.2642867. [DOI] [PubMed] [Google Scholar]