Abstract
Introduction
For over 50 years, drugs targeting the folate pathway have significantly impacted disease treatment as anticancer, antimicrobial and immunomodulatory agents. The discovery of novel antifolate agents with improved properties and superior activities remains an attractive strategy, both in academia and the pharmaceutical industry.
Areas covered
This review surveys the patent literature from 2006–2010 for small molecule inhibitors of enzymatic targets in the folate biosynthetic pathway.
Expert opinion
The pursuit of antifolates as anticancer and antimicrobial agents continues to be an active area of research. New patent disclosures reveal novel antifolate scaffolds, antifolates with improved drug-like properties and new strategies to effectively target cancer cells. The continued use of high resolution structural information has guided the discovery of several compounds. Owing to the need for high levels of potency and selectivity, especially in targeting pathogenic species, the use of high resolution crystal structures remains an important tool to guide the design of novel antifolates. Interestingly, the patents disclosing novel compounds were ones where X-ray crystallography was an integral component of the design process. Finally, a variety of new structures have been reported that may play an important role in the future development of therapeutic antifolates.
1. Introduction
The dependence of rapidly dividing cells on a supply of nucleotide precursors offers an attractive series of therapeutic targets for the development of new antiproliferative agents. Perhaps the most widely exploited of these is the folate biosynthetic pathway that is integral to the generation of thymidine. This review will focus on the discovery and development of inhibitors of targets in the folate biosynthetic pathway. In addition, since high resolution structures of these protein targets often contribute to the design of new inhibitors, a review of the relevant structural biology literature is also included. It is clear from the work disclosed over the previous five year period that the development of potent and effective antifolates remains an active area of investigation.
1.1 Drug targets in the folate biosynthetic pathway
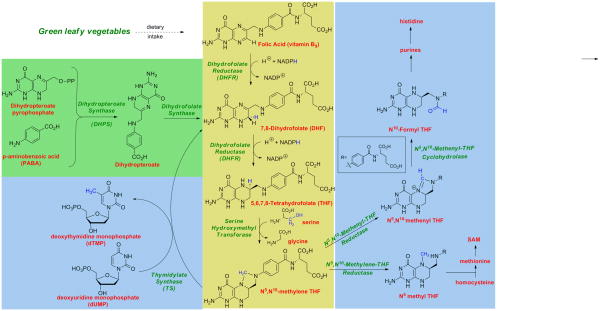
The folate biosynthetic pathway provides tetrahydrofolate cofactors that are a source of C1 units for the synthesis of deoxythymidine monophosphate (dTMP), the biosynthesis of purine nucleotides and the amino acids histidine and methionine (Figure 1). Inhibiting tetrahydrofolate metabolism depletes the cell of dTMP and halts DNA replication. As such, several inhibitors of this pathway, called antifolates, have become successful drugs that inhibit the growth of proliferating malignant mammalian cells or proliferating bacterial and protozoal pathogens.
Figure 1.
The Folate Biosynthetic Pathway. The primary folate biosynthetic pathway is shown in yellow, reflecting the conversion of folic acid to the key 1-carbon donor, 5,10-methylene tetrahydrofolate. De novo synthesis of dihydrofolate in bacteria is shown in green; critical reactions using the folate cofactors are shown in blue.
In humans, ingested folic acid is actively transported into cells. Folic acid is then reduced by the enzyme, dihydrofolate reductase (DHFR), first to dihydrofolate and subsequently to tetrahydrofolate, with an accompanying oxidation of the cofactor, NADPH to NADP+. Serine hydroxymethyltransferase (SHMT) then transfers the side chain of serine to tetrahydrofolate to produce 5,10-methylene tetrahydrofolate and glycine. Finally, thymidylate synthase (TS) uses 5,10-methylene tetrahydrofolate as a cofactor to transfer a methyl group to deoxyuridine monophosphate (dUMP) to form dTMP and dihydrofolate that re-enters the cycle. Additional C1 units are formed by the related metabolites 5-methyltetrahydrofolate and 10-formyltetrahydrofolate that are integral to the synthesis of methionine and purines respectively.
In contrast to the pathway in humans, bacteria, fungi and protozoa possess an endogenous folate biosynthetic pathway and do not depend on the active transport of exogenous folates. In bacteria, fungi and protozoa, an upstream enzyme, dihydropteroate synthase, catalyzes the condensation of p-aminobenzoic acid and 6-hydroxymethyl-7,8-dihydropterin pyrophosphate to form 7,8-dihydropteroate (Figure 1). Dihydrofolate synthase then adds a glutamate moiety to 7,8-dihydropteroate to create 7,8-dihydrofolate, which then acts as a substrate for dihydrofolate reductase. From the point of the creation of dihydrofolate, the tetrahydrofolate cycle containing DHFR, SHMT and TS is the same in bacteria, fungi, protozoa and humans.
1.2 Clinically used antifolates
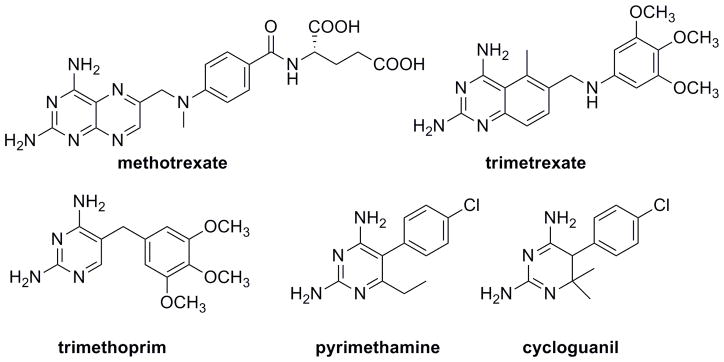
Owing to its essential role in cellular metabolism, DHFR has served as an anticancer, antibacterial and antiprotozoal drug target for several decades. DHFR inhibitors fall into two classes. Classical inhibitors that possess a glutamatetailare zwitterionic and must be actively transported into the cell; once inside, they often become polyglutamylated. Non-classical lipophilic inhibitors cross the cell membrane by passive diffusion. In the 1950s, methotrexate, the only clinically used classical DHFR inhibitor, was developed as a substrate mimic1 (Figure 2). Methotrexate is still used successfully as part of combination therapy, specifically in the treatment of acute lymphoblastic leukemia (ALL) and rheumatoid arthritis. Non-classical inhibitors include trimetrexate that was used in the past to treat pneumocystis pneumonia and trimethoprim that is still used in combination with sulfamethoxazole, an inhibitor of DHPS, to treat methicillin-resistant Staphylococcus aureus (MRSA) and urinary tract infections caused by Escherichia coli. Pyrimethamine and cycloguanil are two DHFR inhibitors used to treat malarial infections, in combination with sulfadoxine and atovaquone, respectively.
Figure 2.
Clinically used DHFR inhibitors include the classical analog methotrexate and the non-classical analogs trimetrexate, trimethoprim, pyrimethamine and cycloguanil.
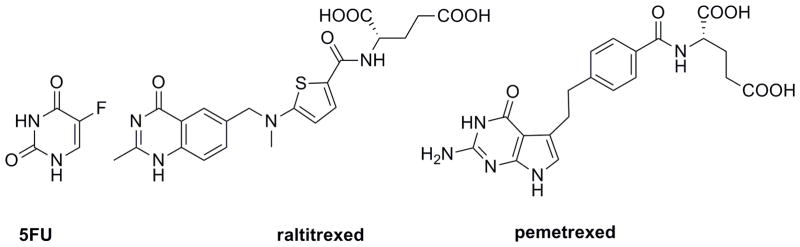
Thymidylate synthase inhibitors are also used as anticancer agents. Substrate mimics include the prodrug, 5-fluorouracil (5FU) (Figure 3). 5FU is converted to 5-fluorodeoxyuridine in the cell, which irreversibly binds a cysteine residue in the TS active site. Raltitrexed and pemetrexed (Figure 3) are mimics of 5,10-methylene tetrahydrofolate and are used in the treatment of colorectal cancer or mesothelioma and non-small cell lung cancer, respectively.
Figure 3.
Clinically used TS inhibitors include 5FU, raltitrexed and pemetrexed.
2. Crystal structures of DHFR and TS published between 2006–2010
High resolution crystal structures of a target enzyme are often critical to the drug discovery process. During 2006–2010, several structures of the targets DHFR and TS from various species were published, speeding the discovery of new antifolates targeting human and pathogenic species. The survey of structures below includes those from human and pathogenic species with the goal of drug discovery.
2.1 Structures of DHFR
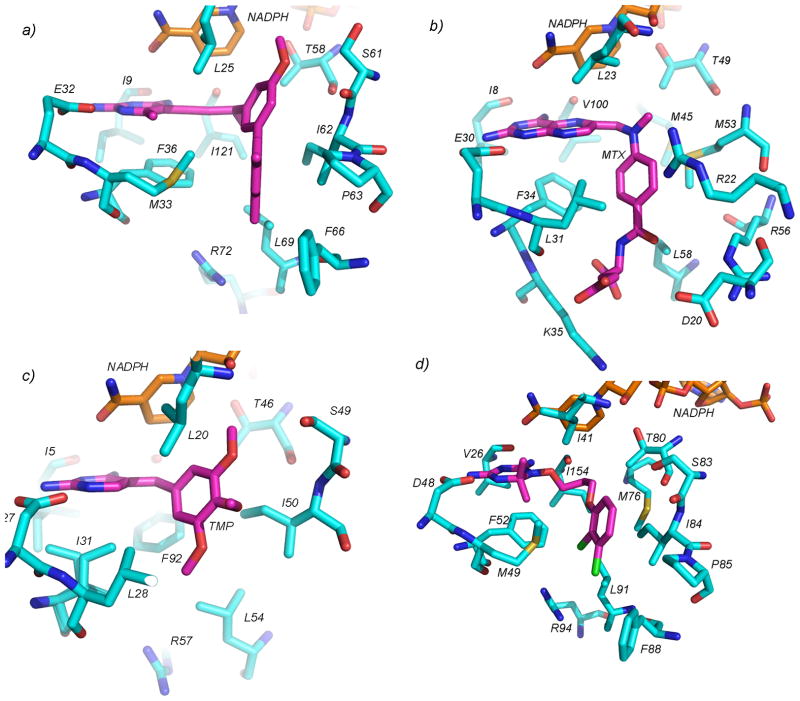
Several new structures of two fungal species of DHFR were published, aiding the design of antifungal antifolates. The structure of DHFR from Candida glabrata, an emerging pathogen resistant to many common azole antifungal agents, was first determined in 2008 bound to NADPH and five propargyl-linked antifolates (Figure 4a; PDB ID: 3CSE, 3EEJ, 3EEK, 3EEL, 3EEM)2, 3. The propargyl-linked antifolates show very high levels of potency (IC50 values of 0.55 nM) for the fungal enzyme and selectivity (2300-fold) over the human enzyme. The propargyl linker forms van der Waals interactions with Leu 25 and Ile 121 and the biphenyl moiety forms interactions with Ile 62, Pro 63, Phe 66, Leu 69, Phe 36 and Met 33. The interactions that the biphenyl moiety makes with residues 62–69 yield the high levels of selectivity as these residues adopt a different conformation in the human enzyme. Additionally, while the structure of the DHFR from the rat pathogen, Pneumocystis carinii was first reported in 19994 several structures bound to NADPH and either pyridopyrimidine5 (PDB IDs: 3NZ6, 3NZ9, 3NZA, 3NZB, 3NZC) or trimethoprim derivatives (PDB ID: 2FZH, 2FZI)6 with carboxyalkoxy side chains were reported. These structures reveal that the ligands with carboxyalkoxy and pentynyl side chains in the trimethoprim series were potent and selective, as they formed ionic interactions between the carboxylate and Arg 75 in the active site. The carboxyalkoxy ligands in the pyridopyrimidine series were disordered, did not form the same interaction with Arg 75 and were less potent. The form of Pneumocystis that infects humans, Pneumocystis jirovecii, produces a DHFR that is similar but not identical to that found in P. carinii; a crystal structure for P. jirovecii has not been reported.
Figure 4.
Crystal structures of DHFR (cyan) bound to NADPH (orange) and antifolates (magenta) are shown. In panel a) DHFR from C. glabrata is bound to a propargyl-linked antifolate, b) S. pneumoniae DHFR is bound to methotrexate, c) S1 DHFR is bound to trimethoprim and d) T. cruzi DHFR is bound to C-448.
Structures of five new species of bacterial DHFR were determined in 2006–2010. The structure of a ternary complex of NADPH, MTX and mutant Sp9 form of Streptococcus pneumoniae DHFR, in which eight surface and one L100V active site amino acid residues were mutated relative to wild-type S. pneumonia was reported (Figure 4b, PDB ID: 3IX9). The structure shows that the bulkier leucine residue at position 100 confers TMP resistance7. Structures of DHFR from Bacillus anthracis, the bioterrorism target, were reported bound to propargyl-linked antifolates (PDB ID: 3E0B, 3JVX, 3JWM)8; in the binary state bound to NADPH (PDB ID: 2QK8)9; bound to RAB1, which has a phthalazine moiety and acryloyl linker(PDB ID: 3FL8); and trimethoprim (PDB ID: 3FL9)10. Additionally, structures of mutant B. anthracis DHFR were determined in order to investigate the mechanism of trimethoprim resistance11 and the solution NMR structure of B. anthracis DHFR (PDB ID: 2KGK) was compared to the crystal structure12. Overall, in addition to revealing paths for the design of inhibitors with increased potency and selectivity, the structures of B. anthracis DHFR show that the wild-type residue Tyr 102 confers trimethoprim resistance and that the flexibility of residues Phe 96 and Leu 55 at the active site may affect ligand binding. The structure of Mycobacterium avium DHFR was deposited bound to NADPH and TMP (PDB ID: 2W3V) or a pyridopyrimidine ligand (PDB ID: 2W3W). The structure of exogenous S1 DHFR bound to TMP (PDB ID: 2W9S, 2W9T, Figure 4c) reveals that residues Ala 43 and Tyr 98 were found to confer high levels of trimethoprim resistance in MRSA13. Additionally, the structure of DHFR from Moritella profunda, a piezophilic organism, was determined in the apo (PDB ID: 3IA5) and ternary complex bound to NADPH and MTX (PDB ID: 3IA4)14. Finally, several structures of S. aureus DHFR bound to a number of propargyl-linked antifolates (PDB IDs: 3LG4, 3I8A, 3F0Q, 3F0S, 3F0V, 3F0X, 3F0B, 3F0U, 3FQ0, 3FQC, 3FQF, 3FQO, 3FQV, 3FQZ)15–17; iclaprim (PDB ID: 3FRA, 3FRF), AR-101 (PDB ID: 3FY8, 3FYW) and AR-102 (3FY9, 3FYV)18, 19, RAB1(PDB ID: 3M08, 3M09)20 and trimethoprim (PDB ID: 3FRB, 3FRE) were reported18. These structures show that the propargyl-linked antifolates and iclaprim achieve high levels of potency by forming interactions with Leu 28, Ile 50, Leu 54, and Phe 92.
Furthering the development of antifolates as anticancer agents, several structures of human DHFR bound to diaminofuropyrimidines (PDB IDs: 3NXX, 3NXY, 3NXO, 3NXR, 3NXT, 3NXV)21, dual DHFR and thymidylate synthase inhibitors (PDB ID: 3GHC, 3GHW)22, boron-containing nonclassical antifolates(PDB ID: 2C2S, 2C2T)23 and lipophilic antifolates (PDB ID: 3W3B) were reported. One of the compounds, 2-amino-4-oxo-6-ethylthieno[2,3-d]pyrimidine, a classical antifolate, is a highly potent dual inhibitor of both TS (IC50= 54 nM) and DHFR (IC50 = 19 nM) and exhibits nanomolar GI50 values against tumor cells22; structures show that the thienopyrimidine antifolates bind in the same orientation as the substrate, folate, perhaps accounting for its activity.
2.2 Structures of TS
Several crystal structures of Lactobacillus casei thymidylate synthase bound to dUMP and phthalimidic derivatives were deposited in the Protein Database (PDB IDs: 3IJZ, 3IK0, 3IK1, 3BYX, 3BZ0, 3C06, 3C0A). The phthalimidic analogs were originally developed to achieve selectivity for pathogenic species of TS24, 25. Four compounds are potent inhibitors of L. casei TS and show no activity against human TS; one of these is also a potent inhibitor of Cryptococcus neoformans TS. Additionally, the Seattle Structural Genomics Center for Infectious Disease deposited a structure of apo TS from Encephalitozoon cuniculi (PDB ID: 3KGB), a protozoan parasite. This structure is the first from the pathogenic Microsporidia phylum.
2.3 Structures of bifunctional DHFR-TS
In protozoa, the DHFR enzyme is genetically and functionally linked to thymidylate synthase to form a bifunctional DHFR-TS protein. Structures of three different bifunctional DHFR-TS protozoal enzymes were determined during 2006–2010 with the goal of ligand design for new antiprotozoal agents. The structure of DHFR-TS from Trypanosoma cruzi was first determined by Chattopadhyay in 200826 bound to NADPH and MTX (PDB ID: 3CL9) as well as NADPH and trimetrexate (PDB ID: 3CLB). Work in 2010 describes a series of diaminoquinazoline inhibitors that have IC50 values between 27 and 57 nM for the T. cruzi enzyme27; three structures of these complexes are reported (PDB ID: 3KJS, 3KJU, 3KJW). Additionally, Chitnumsub et al. reported four structures of T. cruzi DHFR-TS bound to cycloguanil (PDB IDs: 3IRM, 3IRN), a dihydrotriazine derivative (C-448; PDB ID: 3INV), and a quinazoline derivative (Q-8) (PDB IDs: 3IRO)28. C-448 is a flexible ligand with a side chain that is similar to the compounds reported in [101]. The flexible side chain places the dichlorophenyl ring in a hydrophobic pocket with Met 76, Ile 84, Pro 85, Leu 91 and Phe 88 (Figure 4d). Also reported for the first time in 2010 by the Seattle Structural Genomics Center for Infectious Disease is the structure of DHFR-TS from Babesia bovis bound to NADP, dUMP and pemetrexed (PDB ID: 3K2H) or raltitrexed (PDB ID: 3NRR). The structure of wild-type, a double and quadruple mutant Plasmodium falciparum DHFR-TS was first reported in 200329. However, additional structures of the wild-type or quadruple mutant enzyme bound to hits from an in silico screen, RJF01302 and RJF670 (PDB IDs: 3DG8, 3DGA) reported in 2009 show that the small, lead-like nature of the RJF compounds avoid the steric interactions with the S108N mutant 30. A structure of the quadruple P. falciparum DHFR mutant, resistant to pyrimethamine, was reported in 2010 bound to QN254 (PDB ID: 3JSU), an antimalarial preclinical drug candidate that showed promising activity against P. berghei but was abandoned because of dose limiting toxicity31. QN254, a quinazoline inhibitor, shows an increased number of interactions relative to WR99210, a dihydrotriazine inhibitor: a water-mediated hydrogen bond with Asn 108, additional van der Waals and π-π interactions with Leu 46, Met 55 and Phe 58. As with WR99210, the flexibility of the linker is critical in avoiding steric interactions with the mutated residues.
3. Antifolates appearing in the patent literature 2006–2010
Novel antifolates disclosed in the patent literature for the period 2006–2010 can logically be divided into three groups. The first group includes the non-classical DHFR inhibitors that are expected to gain cellular entry though passive diffusion. The second group includes classical zwitterionic DHFR inhibitors that contain a carboxylate tail, along with any prodrugs of this class. Finally, a third group is presented that contains compounds designed for selective uptake through the α-folate receptor.
3.1 Non-Classical DHFR inhibitors
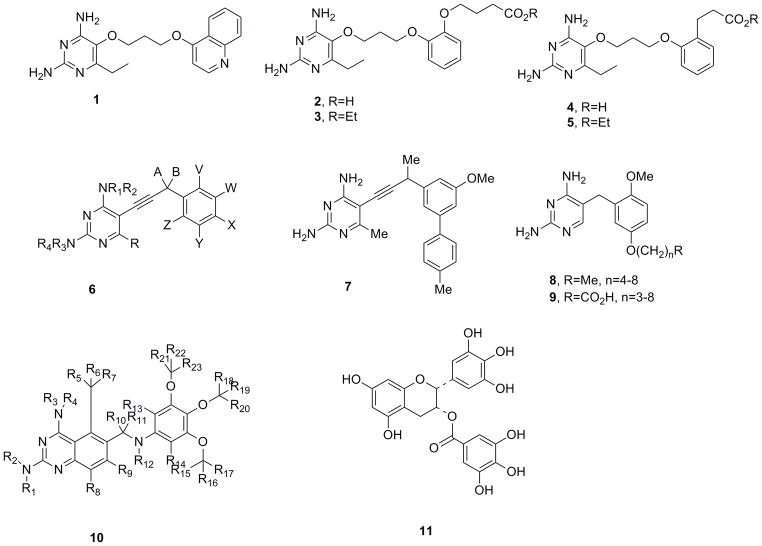
Medicines for Malaria Venture (MMV) report a series of antimalarial compounds characterized by flexible linkers32. These compounds (1–5, Figure 5) are characterized by a simple 2,4-diaminopyrimidine ring linked at C5 through an unbranched saturated linker of 4–6 atoms terminating in an aromatic or heteroaromatic residue. Using structures of P. falciparum DHFR-TS29, the compounds were designed to inhibit strains of wild-type and pyrimethamine-resistant P. falciparum DHFR possessing any or all of the resistance-conferring mutations A16V, N51I, C59R, S108N/T, I164L. The quadruple mutant enzyme (N51I, C59R, S108N, I164L) is highly resistant to pyrimethamine, exhibiting affinity losses that are approximately 500-fold33. As several of the compounds (eg. 2 and 4) are highly charged and unlikely to be well absorbed, prodrug forms such as 3 and 5 were also synthesized and evaluated. The flexible linker introduces minimal steric interactions with the mutant enzymes while maintaining a high number of enthalpic contributions. A wide range of compounds in this series are claimed and several are provided as specific examples. Table 1 illustrates the biological activity of representative members of this class.
Figure 5.
Non-classical DHFR inhibitors published in the patent literature
Table 1.
In vitro and in vivo assay results for five antimalarial compounds
| Ki WT (nM) | Ki Mut (nM)a | IC50 Wt (μM)b | IC50 W2 (μM)c | IC50 V1/S (μM)d | Cytotoxicity (μM)e | Antimalarial activity ED50 (mg/kg)f | |
|---|---|---|---|---|---|---|---|
| Pyr | 0.6 | 385 | 0.058 | 39.88 | >100 | 32 | 0.25 |
| 1 | 1.21 | 4.33 | 0.004 | 0.013 | 0.050 | 0.119 | 0.006 |
| 2 | 0.72 | 3.47 | 0.0016 | 0.005 | 0.038 | >10 | 2.0 |
| 3 | 0.69 | 3.30 | 0.004 | Ndg | Nd | 5.33 | <0.63 |
| 4 | 0.43 | 0.54 | 0.006 | Nd | Nd | >10 | <0.63 |
| 5 | 0.26 | 1.88 | 0.0025 | 0.001 | 0.068 | 1.25 | 0.21 |
against the quadruple mutant (N51I, C59R, S108N, I164L) enzyme
IC50 values are reported against the noted cell lines
W2 cells have the following mutations (N51I, C59R, S108N)
V1/S cells have the following mutations (N51I, C59R, S108N, I164L)
IC50 values against Vero cells
in a P. chabaudi AS rodent malaria model
ND: not determined
While many compounds exhibit approximately equivalent inhibition of the wild-type enzyme, they show much greater inhibition of the quadruple-mutant enzyme than pyrimethamine. Additionally, the compounds are better inhibitors of the wild-type and W2 (triple) and V1/S (quadruple) mutant strains than pyrimethamine. Many of these compounds, including 2–5 have a higher margin of safety at the cellular level than pyrimethamine. Some compounds were further evaluated and performed well in models of rodent malaria. Studies also showed that the ester prodrugs were rapidly hydrolyzed in vivo to give the carboxylic acid.
The University of Connecticut disclosed a series of antifolates characterized by the generalized structure, 6 (Figure 5), employing a propargylic linker to join a 2,4-diaminopyrimidine head group and a variably substituted hydrophobic domain34. These propargyl-linked antifolates are claimed for a wide range of indications including antibacterial, antifungal, antiprotozoal and anticancer disease states. The development of the compounds proceeded through a structure-based approach that suggested that extension of the one-carbon linker in trimethoprim would create a class of molecules that would target both TMP-sensitive and TMP-insensitive pathogenic enzymes35. Further evolution of the lead series led to the incorporation of biphenyl moieties in the hydrophobic domain, producing compounds such as 7. Some of these biphenyl compounds show very high potency (IC50 values of 0.5 nM) and selectivity (over 2300-fold) for DHFR from Candida glabrata and significant MIC values (1.5 μg/mL) that are similar to those of clinically used antifungal agents2, 3.
Several crystal structures of species of DHFR including Bacillus anthracis8, 11, Candida glabrata2, 3, and methicillin-resistant Staphylococcus aureus15–17 bound to pathogen-specific analogues from this class reveal the basis of the potency. The hydrophobic and highly directional nature of the alkyne in the propargyl linker optimally joins the 2,4-diaminopyrimidine anchor to a substituted aryl group designed to bind a hydrophobic pocket, thus making these compounds particularly well-suited DHFR inhibitors. Alterations in the hydrophobic domain joined to the propargyl linker engender selectivity over the human enzyme. Additionally, structures of TMP-sensitive and TMP-resistant S. aureus DHFR with a series of the compounds revealed that the F98Y resistance-conferring mutation induced an alternative conformation of NADPH that reduced key interactions between the inhibitor and cofactor 16, 17.
Dana-Farber Cancer Institute36 reports compounds that are hybrid forms of trimethoprim and piritrexim as inhibitors of DHFR targeting the opportunistic infections caused by Pneumocystis carinii, Toxoplasma gondii and Mycobacterium avium. The compounds maintain the basic trimethoprim scaffold linking a pyrimidine derivative to an aromatic domain through a single methylene spacer. Preferred compounds, 8 and 9 (Figure 5), maintain a 2′5′ dialkoxy pattern of substitution on the aromatic domain with the 2′ substitution being a simple methoxy unit and the 5′ group embodying an extended alkyl ether terminating in a simple alkyl or carboxylate group. Some of the compounds (specifically 9 where n=4 in Figure 5) show enzyme inhibition as low as 0.05 μM and selectivity of up to 80-fold favoring the P. carinii enzyme. The selectivity may result from an interaction of the carboxylate with a basic residue specific to the pathogenic DHFR.
Auspex Pharmaceuticals disclosed an invention focusing on diaminoquinazolines described by the general structure 10 where at least one of the hydrogen atoms is replaced by the heavier deuterium isotope37. The compound of general formula 10 that contains full protonation is trimetrexate (Figure 2) that has been shown to be a potent antifolate. The inventors discuss that trimetrexate is metabolized by oxidation of the aromatic methoxy groups leading to overall O-demethylation products as well as benzylic oxidation that leads to subsequent deamination. Their hypothesis is that the replacement of an active hydrogen by deuterium (double the atomic weight) will produce a significant kinetic isotope effect (KIE) with respect to cytochrome P450 oxidation, thus producing compounds with substantially longer half-lives and diminished side effects that are caused by metabolites. Although no specific compounds were prepared or evaluated, the inventors discuss various standard protocols that can be used to incorporate deuterium at selected positions in the diaminoquinazoline scaffold.
Rodriguez-Lopez, et al. disclose that the green tea catechin, epigallocatechin gallate (EGCG; compound 11, Figure 5), is an inhibitor of dihydrofolate reductase and that compounds in this catechin series may be useful in the treatment of cancer, infection and inflammatory conditions38. Of the green tea catechins, EGCG is the most abundant and possesses known antioxidant, antibiotic and antiviral activities, although the site of action of EGCG is not understood. The inventors discovered that EGCG inhibits chicken liver DHFR with a Ki value of 10.3 μM and inhibits the growth of L1210 cells with an IC50 of 20 μM. Models of the compound bound to the structure of human DHFR elucidate a predicted binding mode. Polyphenols lacking the ester bonded gallate moiety did not inhibit bovine DHFR. Claims include the use of EGCG as a DHFR inhibitor to treat cancer, modeling EGCG in DHFR and modifying EGCG to create new DHFR inhibitors.
3.2 Classical DHFR inhibitors
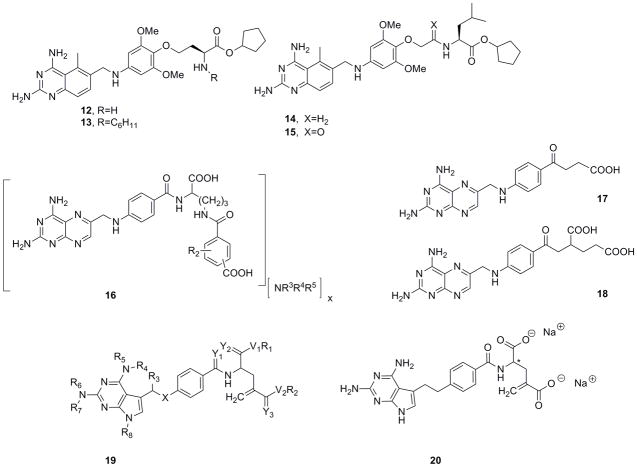
In separate patents, Chroma Therapeutics disclose two related series of diaminoquinazolines or diaminopyridopyrimidines that contain a linked aromatic region substituted with a highly variable linker terminating in a carboalkoxy group39, 40. According to the inventors, the overall design is predicated on intracellular hydrolysis of the carboalkoxy group to the corresponding carboxylic acid. This prodrug design allows for the penetration of the cell membrane by the ester and subsequent retention of the carboxylic acid form once it is liberated by esterase activity. In preferred compounds such as 12–15 (Figure 6), the terminal carboxylate is derived from an amino acid such that this domain is a substrate for a carboxyesterase. Achieving selectivity for cleavage by intracellular esterases over extracellular esterases present in the blood and liver microsomes may present an additional challenge. The major difference between the two series of compounds lies in the position of attachment to the amino ester head group. In one series, exemplified by compounds 12 and 13, the drug is appended through functionality at the Cα side chain while in the other series, for example compounds 14 and 15, the drug is linked through the amino group. Several compounds such as 12–15 were shown to be good inhibitors of DHFR and to demonstrate appreciable cytotoxicity against malignant cell lines.
Figure 6.
Classical DHFR inhibitors published in the patent literature
Dana-Farber Cancer Institute report N-acyl derivatives of N-(4-amino-4-deoxypteroyl)-L-ornithine compounds that are chemically stable and inhibit the growth of methotrexate-resistant cells41. These compounds are derivatives of methotrexate whereby the glutamate moiety has been replaced by an ornithine with varying acyl groups, typically containing a carboxylic acid, on the Nδ group(compound 16, Figure 6). This class of compounds had been shown to demonstrate superior activity against methotrexate-resistant cancer cell lines such as SCC 15/R1 and SCC 25/R1. It was found that formulation of the compounds as the ammonium salts greatly improved chemical stability and increased aqueous solubility. Preferred compounds embodied several different ammonium salts such as ammonia, piperazinium and 2-hydroxyethylammonium.
In two patents, Stoicescu disclosed a novel family of potential antifolates that is reminiscent of the structure of methotrexate with a key alteration whereby the amide unit linking the glutamyl sidechain to the PABAring is replaced by a ketonic or thioketonic linkage42, 43. The compounds are viewed not only as potential inhibitors of DHFR but of other folate-dependent enzymes such as glycinimide ribonucleotide (GAR) formyltransferase and 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) formyltransferase, both of which are dependent on the cofactor N10-formyltetrahydrofolate. As all of these enzymes are key players in the de novo synthesis of nucleotides, the compounds are proposed as potential treatments for various cancers. It is suggested that the alteration of the amide linker to the ketonic/thioketonic variants may produce lower renal toxicity than methotrexate and may increase the half-life of the antifolate. Two representative compounds 17 and 18 in the keto-series were prepared and evaluated for DHFR inhibition and anti-cancer activity. The diacid derivative 18 was shown to be slightly more potent than methotrexate against the DHFR enzyme and to potently inhibit the growth of several cancer lines including HepG2, HeLa and A549 with the strongest effects reported against a leukemia line (L1210). The compound was further evaluated for anticancer activity in the Walker-256 rat tumor model using a range of doses. At a dose of 0.1 mg/kg administered once a day for 5 days, all 10 animals in the group survived at 30 days post-transplant with a tumor volume of 14.5 ± 3.8 cm3 as compared to 2/10 survivors (tumor volume=37.3cm3) in the saline control group.
Roberts and Pedder of Chelsea Therapeutics disclose novel classical antifolates with the generalized scaffold 19 (Figure 6) designed to be “metabolically inert”, that is, they are not hydroxylated and do not form polyglutamates in the cell44. Two examples are provided: compound 20 (Figure 6), both racemic and as the S enantiomer of the disodium salt (the asymmetric center is denoted by an asterisk in the structure shown in Figure 6). The compounds possess a key methylidene group that has been shown to prevent polyglutamylation45. The examples show IC50 values that are approximately 10- and 20-fold greater than that of methotrexate (0.45 nM) against DHFR in CCRF-CEM human leukemia cells. The compounds show minimal activity against thymidylate synthase (TS) with IC50 values in the micromolar range.
3.3 Antifolates Targeting α-FR for selective uptake
BTG International Limited disclose cyclopenta[g]quinazoline compounds as classical antifolates with anticancer activity46. These compounds are designed to function as cytotoxic agents through inhibition of thymidylate synthase. Additionally, it is intended that the compounds selectively target malignant cells owing to selective uptake relative to non-malignant cells. The origin of this selectivity lies in the relative affinity of this compound class for the α-folic acid receptor (α-FR) relative to the reduced folate carrier (RFC), the two major transporters associated with uptake of classical folates. Folic acid shows greater affinity for α-FR over the RFC, with values of 0.1 nM versus 200 μM, respectively. Importantly, the α-FR is overexpressed in certain tumor types including ovarian cancers, providing a possible mechanism to selectively target agents to malignant cells.
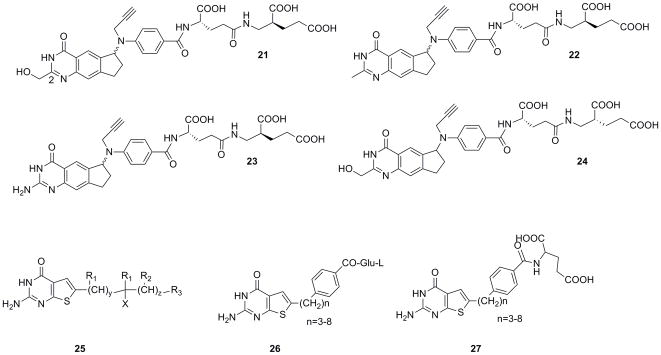
Four specific compounds, 21–24 (Figure 7), were synthesized and evaluated in A431 cells and in A431-FBP cells, which are a form of A431 cells transfected with α-FR (A431-FBP) as well as KB cells that constitutively overexpress the transporter. It was found that in the cyclopenta[g]quinazoline series, the C2 substitution did not have a significant effect on α-FR binding. Interestingly, the C2-CH2OH analogue, 21, was highly potent and selective against the A431-FBP and KB cells, with IC50 values of 0.0021 μM and 0.0034 μM, respectively, compared to the A431 cells (IC50= 9.8 μM). The difference in growth inhibition between the A431-FBP and A431 cells is 4700-fold, demonstrating that selective uptake is possible.
Figure 7.
Compounds targeting α-FR for selective uptake published in the patent literature
In a related strategy, Duquesne University of the Holy Spirit reports classical thienopyrimidine compounds that selectively target α-FR and may inhibit the GAR and AICAR formyltransferase enzymes47. A generalized scaffold, 25 and preferred embodiments(compounds 26 and 27)are shown in Figure 7. The inventors report that the distance and orientation of the major ring groups are important for determining biological activity; between 4–8 alkyl chain carbons appears to be ideal.
4. Expert Opinion
This survey of the recent patent literature shows that antifolates are a subject of considerable interest both in the pharmaceutical industry and in academic laboratories. Despite the presence of several potential enzyme targets in the folate biosynthetic pathway, focus is primarily centered on two well-known targets, dihydrofolate reductase and thymidylate synthase. Inhibitors of DHFR function as antineoplastic agents as well as antibacterial, antifungal and antiprotozoal agents. Interestingly, antifolates are one of the only classes of compounds that cross this wide range of antiproliferative therapeutics for antiproliferative applications.
Even though there have been decades of research devoted to the discovery of antifolates, there are only approximately eight compounds that are clinically used across all indications. Despite the small number of compounds, several of these such as methotrexate, trimethoprim, 5FU and pyrimethamine, continue to find wide application in the treatment of disease. Some of the challenges that face the discovery of new antifolates include the incorporation of selectivity-enhancing groups that favor the pathogenic organism over the human form, optimization of drug-like properties and resistance-conferring mutations that render drugs ineffective. Over the period from 2006–2010, researchers have continued to meet these challenges with a series of innovative designs and strategies.
The types of activities reported in these disclosures can be loosely grouped into three categories. Several compounds in the first category reveal new scaffolds. Disclosures from Medicines for Malaria Venture, University of Connecticut and Dana-Farber Cancer Institute describe three new classes of non-classical antifolates, all of which maintain a diaminopyrimidine or related heterocycle, the hallmark of all clinically used antifolates. The single exception to this conservation is a report from Rodriguez-Lopez on the inhibition of DHFR by a tea polyphenol. The new non-classical antifolates potently and selectively inhibit pathogenic species of DHFR including P. falciparum, C. glabrata, B. anthracis, S. aureus and P. carinii. The antifolates described by Medicines for Malaria Venture target pyrimethamine-resistant P. falciparum DHFR by employing a flexible linker that avoids steric interference; the compounds described by the University of Connecticut target trimethoprim-resistant species by incorporating additional interactions with the hydrophobic pocket that is occupied by the pABA ring of folate. Compounds in the second category represent attempts to make significant improvements in the properties of clinically used antifolates. Disclosures from Stoicescu, Chelsea Therapeutics and Auspex Pharmaceuticals focus on methods to increase the half-life of antifolates by blocking or slowing specific metabolic pathways. These strategies to enhance the half-life of the compounds are innovative, yet a comprehensive evaluation of the metabolic properties remains undetermined. Reports from Dana-Farber Cancer Institute and Chroma Therapeutics concentrate on improving other properties such as stability and solubility as well as prodrug strategies. The final group represents a new strategy to enhance the overall efficacy of antifolates by selectively targeting the agents to cancer cells. BTG International and Duquesne University both present compounds designed to be selectively transported by the α-folate receptor, which is overexpressed in certain cancer cell types. Targeting the α-FR is a highly innovative and potentially valuable strategy given that the compounds should be actively and selectively transported across cellular membranes. Validation of the strategy is limited to cellular systems at this time; further in vivo studies will reveal the actual therapeutic potential.
Owing to the need for high levels of potency and selectivity, especially in targeting pathogenic species, the use of high resolution crystal structures remains an important tool to guide the design of novel antifolates. Interestingly, the patents disclosing novel compounds were ones where X-ray crystallography was an integral component of the design process. Finally, a variety of new structures have been reported that may play an important role in the future development of therapeutic antifolates.
Article Highlights.
This review focuses on the discovery and development of inhibitors of targets in the folate biosynthetic pathway.
Several inhibitors of the folate biosynthetic pathway, called antifolates, have become successful drugs that inhibit the growth of proliferating malignant mammalian cells or proliferating bacterial and protozoal pathogens.
This survey of the recent patent literature shows that antifolates are a subject of considerable interest both in the pharmaceutical industry and in academic laboratories.
Some of the challenges that face the discovery of new antifolates include the incorporation of selectivity-enhancing groups that favor the pathogenic organism over the human form, optimization of drug-like properties and resistance-conferring mutations that render drugs ineffective.
Owing to the need for high levels of potency and selectivity, especially in targeting pathogenic species, the use of high resolution crystal structures remains an important tool to guide the design of novel antifolates.
Acknowledgments
The authors thank the National Institutes of Health (GM067542 and AI073375) for funding their work on DHFR inhibitors.
Footnotes
Declaration of interest
The authors are inventors of one of the included patents.
Bibliography
- 1.Bertino J. Karnofsky Memorial Lecture: Ode to methotrexate. J Clin Oncol. 1993;11:5–14. doi: 10.1200/JCO.1993.11.1.5. [DOI] [PubMed] [Google Scholar]
- 2•.Liu J, Bolstad D, Smith A, et al. Structure-guided development of efficacious antifungal agents targeting Candida glabrata dihydrofolate reductase. Chem Biol. 2008;15:990–96. doi: 10.1016/j.chembiol.2008.07.013. This paper describes the first structure of C. glabrata DHFR bound to a propargyl-linked antifolate disclosed by University of Connecticut. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu J, Bolstad D, Smith A, et al. Probing the active site of Candida glabrata dihydrofolate reductase with high resolution crystal structures and the synthesis of new inhibitors. Chem Biol Drug Des. 2009;73:62–74. doi: 10.1111/j.1747-0285.2008.00745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cody V, Galitsky N, Rak D, et al. Ligand-Induced Conformational Changes in the Crystal Structures of Pneumocystis carinii Dihydrofolate Reductase Complexes with Folate and Nadp+ Biochemistry. 1999;38:4303–12. doi: 10.1021/bi982728m. [DOI] [PubMed] [Google Scholar]
- 5.Cody V, Pace J. Structural analysis of Pneumocystis carinii and human DHFR complexes with NADPH and a series of five potent 6-[5′-(w-carboxyalkoxy)benzyl]-pyrido[2,3-d]pyrimidine derivatives. Acta Cryst. 2011;D67:1–7. doi: 10.1107/S0907444910041004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cody V, Pace J, Chisum K, et al. New insights into DHFR interactions: analysis of Pneumocystis carinii and mouse DHFR complexes with NADPH and two highly potent trimethoprim derivatives. Proteins. 2006;65:959–69. doi: 10.1002/prot.21131. [DOI] [PubMed] [Google Scholar]
- 7.Lee J, Yennawar N, Gam J, et al. Kinetic and structural characterization of dihydrofolate reductase from Streptococcus pneumoniae. Biochemistry. 2010;49:195–206. doi: 10.1021/bi901614m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beierlein J, Frey K, Bolstad D, et al. Synthetic and crystallographic studies of a new inhibitor series targeting Bacillus anthracis dihydrofolate reductase. J Med Chem. 2008;51:7532–40. doi: 10.1021/jm800776a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett B, Xu H, Simmerman R, et al. Crystal structure of the anthrax drug target, Bacillus anthracis dihydrofolate reductase. J Med Chem. 2007;50:4374–81. doi: 10.1021/jm070319v. [DOI] [PubMed] [Google Scholar]
- 10.Bourne C, Bunce R, Bourne P, et al. Crystal strucutre of Bacillus anthracis dihydrofolate reductase, with the dihydrophthalazine-based trimethoprim derivative RAB1 provides a structural explanation of potency and selectivity. Antimicrob Agents and Chem. 2009;53:3065–73. doi: 10.1128/AAC.01666-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beierlein J, Karri N, Anderson A. Targeted Mutations of Bacillus anthracis Dihydrofolate Reductase Condense Complex Structure-Activity Relationships. J Med Chem. 2010;53:7327–36. doi: 10.1021/jm100727t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beierlein J, Deshmukh L, Frey K, et al. The Solution Structure of Bacillus anthracis Dihydrofolate Reductase Yields Insight into the Analysis of Structure-Activity Relationships for Novel Inhibitors. Biochemistry. 2009;48:4100–08. doi: 10.1021/bi802319w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13•.Heaslet H, Harris M, Fahnoe K, et al. Structural comparison of chromosomal and exogenous dihydrofolate reductase from Staphylococcus aureus in complex with the potent inhibitor trimethoprim. Proteins. 2009;76:706–17. doi: 10.1002/prot.22383. This paper describes the structure of S1 DHFR, one of the major contributors to trimethoprim resistance in MRSA. [DOI] [PubMed] [Google Scholar]
- 14.Hay S, Evans R, Levy C, et al. Are the catalytic properties of enzymes from piezophilic organisms pressure adapted? Chem Bio Chem. 2009;10:2348–53. doi: 10.1002/cbic.200900367. [DOI] [PubMed] [Google Scholar]
- 15.Frey K, Georgiev I, Donald B, et al. Predicting resistance mutations using protein design algorithms. Proc Natl Acad Sci. 2010;107:13707–12. doi: 10.1073/pnas.1002162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frey K, Liu J, Lombardo M, et al. Crystal Structures of Wild-type and Mutant Methicillin-resistant Staphylococcus aureus Dihydrofolate Reductase Reveal an Alternative Conformation of NADPH that may be Linked to Trimethoprim Resistance. J Mol Biol. 2009;387:1298–308. doi: 10.1016/j.jmb.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frey K, Lombardo M, Wright D, et al. Towards the Understanding of Resistance Mechanisms in Clinically Isolated Trimethoprim-resistant, Methicillin-resistant Staphylococcus aureus Dihydrofolate Reductase. J Struc Biol. 2010;170:93–97. doi: 10.1016/j.jsb.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oefner C, Bandera M, Haldimann A, et al. Increased hydrophobic interactions of iclaprim with Staphylococcus aureus dihydrofolate reductase are responsible for the increase in affinity and antibacterial activity. J Antimicrob Chemother. 2009;63:687–98. doi: 10.1093/jac/dkp024. [DOI] [PubMed] [Google Scholar]
- 19.Oefner C, Parisi S, Schulz H, et al. Inhibitory properties and X-ray crystallographic study of the binding of AR-101, AR-102 and iclaprim in ternary complexes with NADPH and dihydrofolate reductase from Staphylococcus aureus. Acta Cryst. 2009;D65:751–57. doi: 10.1107/S0907444909013936. [DOI] [PubMed] [Google Scholar]
- 20.Bourne C, Barrow E, Bunce R, et al. Inhibition of antibiotic-resistant Staphylococcus aureus by the broad-spectrum dihydrofolate reductase inhibitor RAB1. Antimicrob Agents and Chem. 2010;54:3825–33. doi: 10.1128/AAC.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cody V, Piraino J, Pace J, et al. Preferential selection of isomer binding from chiral mixtures: alternate binding modes observed for the E and Z isomers of a series of 5-substituted 2,4-diaminofuro[2,3-d]pyrimidines as ternary complexes with NADPH and human dihydrofolate reductase. Acta Cryst. 2010;D66:1271–77. doi: 10.1107/S0907444910035808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22•.Gangjee A, Li W, Kisliuk R, et al. Design, synthesis, and X-ray crystal structure of classical and nonclassical 2-amino-4-oxo-5-substituted-6-ethylthieno[2,3-d]pyrimidines as dual thymidylate synthase and dihydrofolate reductase inhibitors and as potential antitumor agents. J Med Chem. 2009;52:4892–902. doi: 10.1021/jm900490a. This paper describes the structure and evaluation of a potent dual DHFR and TS inhibitor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reynolds R, Campbell S, Fairchild R, et al. Novel boron-containing, nonclassical antifolates: synthesis and preliminary biological and structural evaluation. J Med Chem. 2007;50:3283–89. doi: 10.1021/jm0701977. [DOI] [PubMed] [Google Scholar]
- 24.Costi M, Rinaldi M, Tondi D, et al. Phthalein Derivatives as a New Tool for Selectivity in Thymidylate Synthase Inhibition. J Med Chem. 1999;42:2112–24. doi: 10.1021/jm9900016. [DOI] [PubMed] [Google Scholar]
- 25.Stout T, Tondi D, Rinaldi M, et al. Structure-Based Design of Inhibitors Specific for Bacterial Thymidylate Synthase. Biochemistry. 1999;38:1607–17. doi: 10.1021/bi9815896. [DOI] [PubMed] [Google Scholar]
- 26.Schormann N, Senkovich O, Walker K, et al. Structure-based approach to pharmacophore identification, in silico screening, and three-dimensional quantitative structure-activity relationship studies for inhibitors of Trypanosoma cruzi dihydrofolate reductase function. Proteins. 2008;73:889–901. doi: 10.1002/prot.22115. [DOI] [PubMed] [Google Scholar]
- 27.Schormann N, Velu S, Murugesan S, et al. Synthesis and characterization of potent inhibitors of Trypanosoma cruzi dihydrofolate reductase. Bioorg Med Chem. 2010;18:4056–66. doi: 10.1016/j.bmc.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 28.Chitnumsub P, Yuvaniyama J, Chahomchuen T, et al. Crystallization and preliminary crystallographic studies of dihydrofolate reductase-thymidylate synthase from Trypanosoma cruzi, the Chagas disease pathogen. Acta Cryst. 2009;F65:1175–78. doi: 10.1107/S1744309109041979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuvaniyama J, Chitnumsub P, Kamchonwongpaisan S, et al. Insights into antifolate resistance from malarial DHFR-TS structures. Nat Struct Biol. 2003;10:357–65. doi: 10.1038/nsb921. [DOI] [PubMed] [Google Scholar]
- 30.Dasgupta T, Chitnumsub P, Kamchonwongpaisan S, et al. Exploiting structural analysis, in silico screening, and serendipity to identify novel inhibitors of drug-resistant falciparum malaria. ACS Chem Biol. 2009;4:29–40. doi: 10.1021/cb8002804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nzila A, Rottman M, Chitnumsub P, et al. Preclinical evaluation of the antifolate QN254, 5-chloro-N′6′-(2,5-dimethoxy-benzyl)-quinazoline-2,4,6-triamine, as an antimalarial drug candidate. Antimicrob Agents and Chem. 2010;54:2603–10. doi: 10.1128/AAC.01526-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Medicines for Malaria Venture. WO09048957. Antimalarial compounds with flexible side-chains. 2009 This patent describes potent and selective inhibitors of the wild-type and mutant P. falciparum DHFR.
- 33.Sirawaraporn W, Sathitkul T, Sirawaraporn R, et al. Antifolate-resistant mutants of Plasmodium falciparum dihydrofolate reductase. Proc Natl Acad Sci. 1997;94:1124–29. doi: 10.1073/pnas.94.4.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34••.University of Connecticut. US20090105287. Inhibitors of Dihydrofolate reductase with antibacterial, antiprotozoal, antifungal and anticancer properties. 2009 This patent describes novel propargyl-linked antifolates active against fungal, bacterial and protozoal species of DHFR.
- 35.Pelphrey P, Popov V, Joska T, et al. Highly efficient ligands for DHFR from Cryptosporidium hominis and Toxoplasma gondii inspired by structural analysis. J Med Chem. 2007;50:940–50. doi: 10.1021/jm061027h. [DOI] [PubMed] [Google Scholar]
- 36.Dana-Farber Cancer Institute. 7119095. Pharmaceutically active compounds and methods of use thereof. 2006
- 37.Auspex Pharmaceuticals. 20100150896. Diaminoquinazoline inhibitors of dihydrofolate reductase. 2010
- 38.US20070249545. Dihydrofolate reductase inhibition by epigallocatechin gallate compounds. 2007
- 39.Chroma Therapeutics. WO07129020. Pyrimidine derivatives useful as DHFR inhibitors. 2007
- 40.Chroma Therapeutics. WO07132146. DHFR enzyme inhibitors. 2007
- 41.Dana-Farber Cancer Institute. US6989386. Pharmaceutically active ornithine derivatives, ammonium salts thereof and methods of making same. 2006
- 42.US20100249141. Novel inhibitors of folic acid-dependent enzymes. 2010
- 43.US7718660. Inhibitors of folic acid-dependent enzymes. 2010
- 44.Chelsea Therapeutics. US20080214585. New classical antifolates. 2008
- 45.Abraham A, McGuire J, Galivan J, et al. Folate analogs. 34. Synthesis and antitumor activity of non-polyglutamylatable inhibitors of dihydrofolate reductase. J Med Chem. 1991;34:222–27. doi: 10.1021/jm00105a035. [DOI] [PubMed] [Google Scholar]
- 46••.BTG International Limited. US7705006. Anti-cancer cyclopenta[g]quinazoline compounds. 2010 This patent describes compounds that target the folate receptor for selective uptake.
- 47.Duquesne University of the Holy Spirit. US20090326224. Thieno pyrimidine compounds. 2009