Abstract
Purpose
Alterations in Smad4 signaling and its loss causes genomic instability and head and neck squamous cell carcinoma (HNSCC), suggesting that agents which target both Smad4-dependent and -independent pathways could control HNSCC.
Experimental Design
Resveratrol efficacy was evaluated against HNSCC cells, FaDu, Cal27, Det562, and Cal27-Smad4 for viability, DNA damage, cell cycle progression, and apoptosis, as well as DNA damage, γH2AX expression and foci formation (γH2AX and Brca1). Resveratrol efficacy was also examined in nude mice for FaDu xenograft growth. Xenografts were analyzed for γH2AX and cleaved caspase-3.
Results
Resveratrol (5-50 μM) suppressed viability and induced DNA damage in FaDu and Cal27 cells, but not in NHEK and HFF, showing its selectivity towards HNSCC cells; however, Det562 cells were resistant to resveratrol even at 100 μM. Cal27 cells stably transfected with Smad4 showed similar resveratrol effects as parental Cal27 indicating that a lack of resveratrol effect in Det562 cells was independent of Smad4 status in these cells. Furthermore, resveratrol caused S phase arrest and apoptotic death of FaDu and Cal27 cells together with induction of Brca1 and γH2AX foci. Resveratrol (50 mg/kg bw) treatment also inhibited FaDu tumor growth in nude mice, and γH2AX and cleaved caspase-3 were strongly increased in xenografts from resveratrol-treated mice compared to controls.
Conclusion
Our findings for the first time showed anti-proliferative, DNA damaging and apoptotic effects of resveratrol in HNSCC cells independent of Smad4 status, both in vitro and in vivo, suggesting that more studies are needed to establish its potential usefulness against HNSCC.
Keywords: Head and neck cancer, resveratrol, chemoprevention, DNA damage, TGF-β, Smad4, γH2AX, Brca1
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most frequent cancer worldwide and the five-year survival rates are among the lowest (< 60%) of the major cancers. HNSCC are mainly caused by alcohol consumption, tobacco intake (smoking and smokeless products such as betel quid), poor oral hygiene and infection to high-risk types of human papilloma virus (1). According to the statistical estimates given by the American Cancer Society, ∼73,080 new cases of HNSCC would have been diagnosed in the United States in the year 2010 (2). In spite of advancements in surgical procedures, chemotherapy, radiation therapy, as well as combination of these, the patient survival rates have not improved in the last several decades. Moreover, it has been shown that the high rate of morbidity is due to both locoregional recurrence and distant metastasis (1, 3).
Transforming growth factor- β receptor II (TGF-βRII) is the main pathways that is deregulated in HNSCC (4) where aberrant signaling downstream of this receptor contributes to tumor growth. TGF-β regulates its pleiotropic biological activities through a complex formation between TGF-β receptor type I (TGF-βRI) and type II (TGF-βRII). TGF-β receptors then transmit signal through the activation of the downstream Smad signaling pathways which consists of the Smad family of proteins (Smad1-5) (4). Receptor activated Smads (Smad2 and -3 or Smad1 and -5) associate with Smad4 and form a hetero-oligomeric complex, which translocates to the nucleus where it activates transcription of various target genes namely p53, cyclin dependent kinase inhibitors, c-Myc and those regulating apoptosis (5-7). Mutation in TGF-βRII causes deregulation in growth inhibitory control and tumorigenesis (4). Mechanistic studies have established that alteration in Smad4 signaling is due to frequent mutations in TGF-βRII and this alteration is common in HNSCC. Further importance of Smad4 in HNSCC is recently established by the study where Smad4 loss caused defects in Fanconi anemia/Brca (Fanc/Brca) pathway leading to genomic instability in mice and spontaneous HNSCC (8). These observations suggest that agents are needed which could target TGF-β/Smad4 pathway for HNSCC treatment
Resveratrol, a nontoxic polyphenolic phytoalexin found in grapes skin, red wine, peanuts, berries, etc, has shown strong efficacy against various cancers in both in vitro and in vivo studies (9, 10). Recently, a number of interventional clinical trials in humans have been initiated using resveratrol as a therapeutic, oral compound (11). The voluminous literature regarding anti-cancer efficacy of resveratrol is based on an initial study by Pezzuto and colleagues showing that resveratrol inhibits preneoplastic lesions in carcinogen-treated mouse mammary glands in culture and DMBA-induced skin tumorigenesis in a mouse model (12). Despite extensive studies with resveratrol in various models and organ sites of malignancies, to date nothing is known about the anti-cancer efficacy of resveratrol against HNSCC. Because of these issues, we assessed whether resveratrol is effective against HNSCC and if resveratrol's efficacy is dependent on Smad4, a key signaling pathway deregulated in HNSCC.
Materials and Methods
Cell culture and reagents
Human HNSCC FaDu, Cal27 and Det562 cells were from ATCC (Manassas, VA) in 2006 and were confirmed by DNA fingerprinting using the ABI Identifiler kit using the following identifiler loci: D3S158, vWA, FGA, Amelogenin, D8S1179, D21S11, D18S51, D13S317, D7S820, CSF1PO, D16S539, THO1, TPOX, D2S1338 and D19S433. All results matched the fingerprint profiles in the ATCC database. Normal human epidermal keratinocytes (NHEK) and human foreskin fibroblasts (HFF) were from Lonza Walkersville, Inc (Walkersville, MD). DMEM and other cell culture materials were from Invitrogen Corporation (Gaithersburg, MD). Antibodies for γH2AX and cleaved caspase-3, and anti-rabbit peroxidase-conjugated secondary antibody were from Cell Signaling (Danvers, MA). Smad4 and Brca1 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Annexin V-Vybrant apoptosis kit was from Molecular Probes (Eugene, Oregon). Resveratrol and β-actin were from Sigma-Aldrich (St.Louis, MO). FaDu, Cal27, Det562 and HFF cells were cultured in DMEM containing 10% fetal bovine serum and 1% penicillin-streptomycin. NHEK cells were cultured in KGM-Gold bullet kit (Lonza; Cat # 00192060) under standard culture conditions.
Generation of Cal27-vector control (pcDNA3) and Smad4 over expressing stable cell lines
Following similar procedures (8), Cal27 cells were transfected with pcDNA Flag-Smad4M (Plasmid 14959) or vector control pcDNA3 (Addgene, Cambridge, MA) using Lipofectamine 2000. Stable transfectants were selected using G418 at 0.2 mg/ml in DMEM (10% FBS) for 4 weeks, and cells were pooled and maintained in the same selective medium.
MTT assay
Cells were plated in 96 well plate under standard culture conditions, next day treated with resveratrol, and at the end of desired treatment time, 20 μl MTT stock solution (5 mg/ml) was added to each well and incubated for another 5 h. Thereafter, media was aspirated, 200 μl of DMSO added to each well, and absorbance was measured at 570 nm.
Comet assay
Following desired treatments, cells were harvested, washed with PBS, 50 μl cell suspension containing 5000 cells mixed with 500 μl of 0.5% agarose at 37°C, and 75 μl of this mixture instantly added to Comet slides (OxiSelect Comet Assay Kit, Cell Biolabs, Inc). Embedded cells were immersed in lysis buffer at 4°C for 1 h, and then replaced with cold alkaline buffer and placed at 4°C for 30 min in dark followed by electrophoresis (35 V for 15 min at 4°C) in a horizontal electrophoresis chamber filled with neutral TBE electrophoresis buffer. Thereafter, slides were transferred to a container filled with cold distilled-water for 2-4 min and then placed in 70% ethanol for 5 min, and air dried overnight at room temperature. After staining with vista green for 15 min at room temperature, comets (both double and single strand DNA breaks) were observed by fluorescent microscopy (Olympus BH2) at 100× total magnification.
Cell cycle distribution and apoptosis analyses
Cells were treated with resveratrol, trypsinized and washed twice with ice-cold PBS, and pellets were incubated in 0.5 ml of saponin/propidium iodide (PI) solution at 4°C for 24 h in dark as reported earlier (13). Alternatively, cells were subjected to Annexin V and PI staining using Vybrant Apoptosis Assay Kit 2 following vendor' protocol. Stained cells were analyzed by flow cytometry using the FACS Analysis Core of the UCCC, for cell cycle distribution and apoptotic cell populations, respectively.
Immunoblotting
At 60-65% confluency, cells were treated with resveratrol, cell lysates prepared, 30-60 μg protein per sample denatured with 2× sample buffer and subjected to SDS-PAGE on 12 or 16% gel, and separated proteins were transferred onto membrane by Western blotting as described (13). Membranes were blocked with blocking buffer for 1 h at room temperature and probed with primary antibody over night at 4°C followed by incubation with peroxidase-conjugated appropriate secondary antibody and ECL detection.
Immunofluorescence staining and confocal microscopy
Cells were plated on cover slips overnight and following desired treatment, washed gently with PBS, fixed in 4% formaldehyde for 15 min, incubated with ice cold 100% methanol for 10 min and rinsed twice with PBS, and blocked in 5% BSA in PBS for 60 min. Thereafter, cells were incubated with γH2AX or Brca1 antibody overnight at 4°C, washed with PBS, incubated for 45 min with Texas Red goat anti-rabbit and Alexa Flour goat anti-mouse secondary antibodies, and counterstained with DAPI for 5 min. Cell images were captured at 1000× magnification on a Nikon D Eclipse C1 confocal microscope, and analyzed by EZ-C1 Free viewer software.
Tumor xenograft study
Exponentially growing FaDu cells were trypsinized, washed and re-suspended in serum-free and antibiotic-free DMEM. Athymic nu/nu male mice (6-week old) were maintained under standard conditions with free access to water and AIN-76 diet, randomly divided into 5 groups (n = 8/group), and injected s.c. with 3 × 106 cells mixed with Matrigel (1:1) in the right flank to initiate tumor xenograft growth. Next day, mice in groups 2 and 3 were gavaged orally with 10 and 50 mg/kg body weight doses of resveratrol in 0.2 ml of 0.5% carboxy methyl cellulose (CMC) for 5 days/week for 30 days, respectively, and those in group 1 (control) with CMC alone. For groups 4 and 5, xenograft was allowed to grow for 15 days, and then mice were gavaged orally with 0.5% CMC and 50 mg/kg resveratrol dose in CMC for 5 days/week for 15 days, respectively. Xenograft growth was measured twice weekly in all groups of animals after 7th day of cells implantation with a digital caliper, and tumor volume calculated as reported recently (14). Body weight and diet consumption were recorded twice weekly throughout study. At experiment terminations, tumors were excised, weighed and stored at -80°C. Animal care and experiments were in accordance with UCD-approved IACUC protocol.
Immunohistochemical staining
Tumor tissues, fixed in 10% phosphate-buffered formalin for 12 h at 4°C, were dehydrated in ascending concentrations of ethanol, cleared with xylene, embedded in paraffin, and tissue blocks were cut with a rotary microtome into 4-μm sections and processed for immunohistochemical (IHC) staining. Briefly, sections after deparaffinization and re-hydration were treated with 0.01M citrate buffer (pH 6.0) in a microwave for 30 min for antigen-retrieval followed by quenching of endogenous peroxidase activity with 3% H2O2 in methanol (v/v) for 5 min. Next, sections were incubated with specific antibodies, γH2AX and cleaved caspase-3, in PBS for 2 h at room temperature followed by overnight incubation at 4°C in a humidified chamber. Negative controls were incubated only with universal negative control antibodies under identical conditions. Sections were then incubated with appropriate biotinylated secondary antibody (1:200–400 dilutions) followed with conjugated horseradish peroxidase–streptavidin (Dako, Carpinteria, CA) and 3,3′-diaminobenzidine (Sigma Chemical Co., St Louis, MO) working solution and counterstained with hematoxylin (15). Microscopic images were taken by AxioCam MrC5 camera at 400× magnification and processed by Axiovision 4.6 (Carl Zeiss microimaging). IHC staining was quantified as number of positive cells (brown cells) × 100/total number of cells in 5 arbitrary selected 400× fields from each tumor.
Statistical analysis
Statistical significance of differences between control and treated samples was determined using Student's t-test (SigmaStat 3.5). P values of <0.05 were considered significant. The data in all cases are representative of at least 2–4 independent studies with reproducible results.
Results
Resveratrol inhibits cell viability and induces DNA damage selectively in HNSCC cells
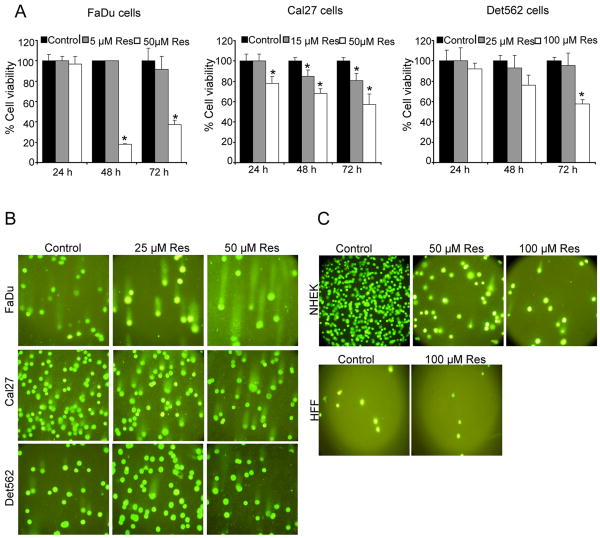
First we evaluated resveratrol efficacy in a panel of human HNSCC cell lines, namely FaDu (homologous deletion of Smad4), Cal27 (nonsense mutation in Smad4) and Det562 (wild type Smad4) cells. Resveratrol at 50 μM dose caused 83% (p<0.001) and 63% (p<0.001) decrease in viability of FaDu cells after 48 and 72 h, respectively (Fig.1A). Cal27 cells at 50 μM resveratrol dose also showed up to 43% (p<0.001) decrease in viability (Fig. 1A); however, such resveratrol treatments were not effective in Det562 cells, until a higher dose (100 μM) and longer treatment of the agent (Fig.1A). Several recent studies have shown that natural chemopreventive agents cause DNA damage in tumorigenic cells leading to apoptotic cell death (9), and accordingly, next we assessed resveratrol effect on DNA damage in HNSCC cells employing the alkaline comet assay. Compared to controls where DNA remained within the nucleus, in resveratrol-treated cells, damaged DNA exited nucleus and stained as a tail (Fig. 1B). Overall, Comet assay results clearly showed DNA-damaged FaDu and Cal27 cells following resveratrol treatment; however, Det562 cells had less DNA damage induced by resveratrol (Fig.1B) which is consistent with a lack resveratrol effect on Det562 cell viability (Fig. 1A), suggesting that the observed decrease in cell viability by resveratrol in FaDu and Cal27 cells is possibly due to its DNA damage inducing effect. Importantly, DNA damage was not observed in normal cells (NHEK and HFF) even at higher dose of resveratrol (Fig. 1C), showing it's selectivity towards HNSCC cells. In Fig. 1C, NHEK cells in the treatment groups are added less in number purposely to see the comet tail. Thus, cells are more in the control group as compared to resveratrol treated group. Further, we have found that resveratrol had no effect on the cell growth and cell number (data not shown) of NHEK cells.
Figure 1. Resveratrol inhibits cell viability and induces DNA damage selectively in HNSCC cells.
A) Resveratrol effect on cell viability of FaDu, Cal27 and Det562 cells employing MTT assay as detailed in Methods. *; p<0.001 between control and resveratrol; each bar represents mean±SE of three samples from each group. B) and C) Cells were treated for 24 h with indicated concentrations of resveratrol and then comet assay was performed as detailed in Methods. Representative pictures of damaged and undamaged nuclei in HNSCC FaDu, Cal27, Det562 cells (B), and NHEK and HFF cells (C) were captured as detailed in Methods. Res, resveratrol.
Resveratrol selectively causes cell cycle arrest and apoptosis in HNSCC cells independent of Smad4
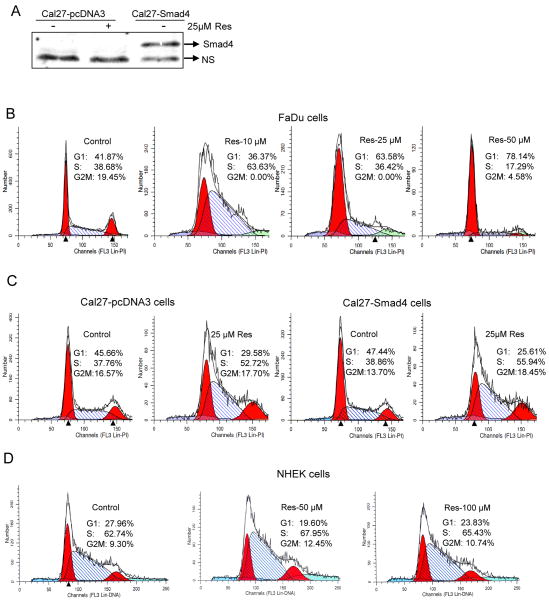
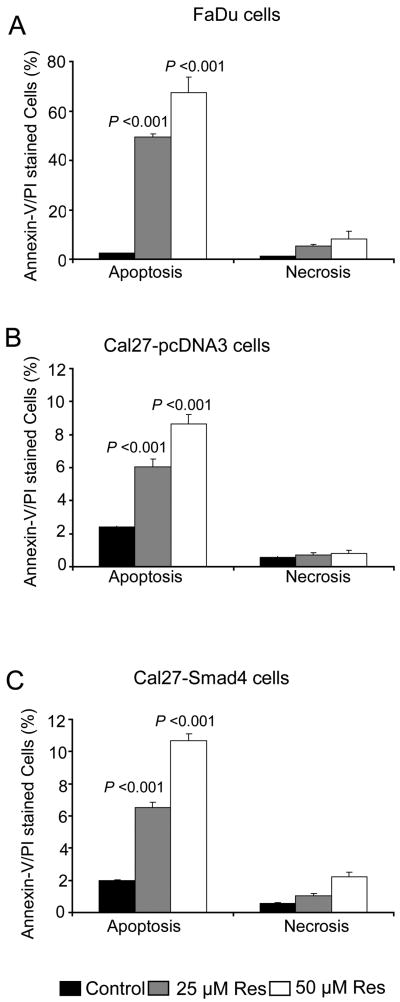
Based on results in Fig. 1 showing resveratrol-caused decreased cell viability and DNA damage selectively in HNSCC cells carrying non-functional Smad4 (FaDu and Cal27), next we assessed its effect on cell cycle progression and apoptosis in FaDu cells (deletion Smad4) and Cal27-Smad4 cells over expressing Smad4 (Fig. 2A) in order to address whether a lack of resveratrol effect in Det562 cells is due to their wild type Smad4 status. Resveratrol (10 μM) arrested FaDu cells in S phase (66%, p<0.001) compared to controls with 34% S-phase population (Fig. 2B); however, higher resveratrol concentrations (25-50 μM) for 24 h caused G1 arrest (63-78%) compared to control (41%; Fig. 2B). Regarding Cal27, compared to controls with 38-39% cells in S phase, resveratrol (25 μM) significantly induced S-phase arrest (53 and 56%; p<0.001) in both vector control Cal27-pcDNA3 and Smad4 over expressing (based on over expressed Smad4 protein, Fig.2A) Cal27-Smad4 cells, respectively (Fig. 2C). These findings revealed that resveratrol efficacy in Cal27 and a lack of efficacy in Det562 cells are independent of Smad4 expression. In other studies, resveratrol (50-100 μM) treatments did not alter cell cycle progression in NHEK cells (Fig. 2D), further supporting its selectivity towards HNSCC cells. Resveratrol (25-50 μM) treatment also induced apoptosis in FaDu, Cal27-pcDNA3 and Cal27-Smad4 cells by 20-27, 2.5-4 and 3-5 fold (p<0.001), respectively (Fig. 3). Similar to S phase arrest results, Cal27 cells with overexpression of Smad4 retained sensitivity to resveratrol-induced apoptosis (Fig. 3C), suggesting that the effect on apoptosis is also independent of Smad4.
Figure 2. Resveratrol selectively causes cell cycle arrest in HNSCC cells independent of Smad4.
A) Cal27 cells were transfected with pcDNA Flag-Smad4M (Plasmid 14959) or vector control pcDNA3 (Addgene, Cambridge, MA) using Lipofectamine 2000. Stable transfectants were selected using G418 at 0.2 mg/ml in DMEM (10% FBS) for 4 weeks, and cells were pooled and maintained in the same selective medium. Cells lysates were prepared and subjected to SDS-PAGE followed by immunoblotting, and membranes probed with Smad4 antibody as detailed in Methods. Percentage cell cycle distribution in B) FaDu, C) Cal27-pcDNA3 and Cal27-Smad4, and D) NHEK cells following resveratrol treatment for 24 h at indicated concentrations and FACS analysis as detailed in Methods. Results shown are representative of triplicate samples from each treatment, and were reproducible in two independent experiments. NS, nonspecific; Res, resveratrol.
Figure 3. Resveratrol causes apoptotic death in HNSCC cells independent of Smad4.
Percentage Annexin-V/PI stained apoptotic A) FaDu, B) Cal27-pcDNA3, and C) Cal27-Smad4 cells following resveratrol treatment for 48 h at indicated concentrations and flow cytometry analysis as detailed in Methods. Each bar represents the mean ± SE of the percentage of apoptotic cells from triplicate samples from each treatment, and these results were reproducible in three independent experiments. Res, resveratrol
Resveratrol induces γH2AX and Brca1 foci formation in HNSCC cells
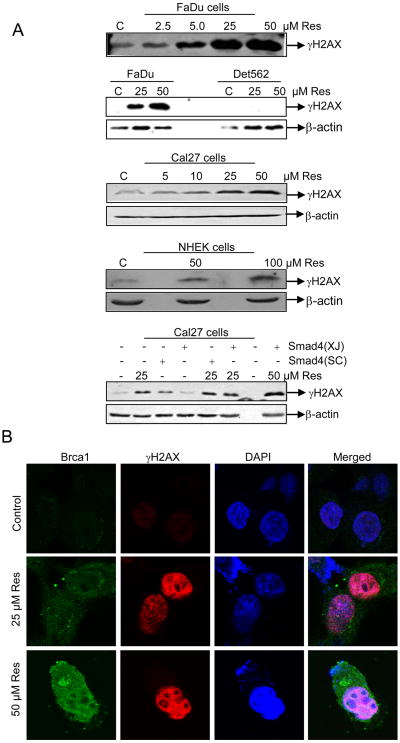
H2AX ser-139 phosphorylation (so-called γH2AX) is essential for recruiting and localizing DNA repair proteins (Brca1 and Rad51) at the sites containing damaged chromatin and for checkpoint activation which arrest cell cycle progression (16). Brca1 is a tumor suppressor protein involved in maintaining genome integrity, and following DNA damage, it accumulates in nucleus, where it concentrates in nuclear foci with other DNA double strand breaks (DSBs) repair factors including homologous recombination protein, RAD51, and end-joining RAD50-MRE11-NBS1 protein complex (17). Accordingly, to further dissect resveratrol mechanism of action, we assessed γH2AX and Brca1 foci formation. Resveratrol enhanced γH2AX levels in both FaDu and Cal27, but had no effect in Det562 cells. A higher resveratrol dose (100 μM) also induced γH2AX in NHEK but to a lesser extent in FaDu and Cal27 cells (Fig. 4A). We also compared γH2AX levels in parental Cal27 versus two independently derived Cal27-Smad4 cells (generated by us and the Wang lab (8) and found them to be comparable among these lines. Together, these results clearly indicated that resveratrol triggered DNA damage in HNSCC cells independent of Smad4 expression. Consistently, compared to controls, resveratrol also induced discrete nuclear γH2AX and Brca1 foci formation in FaDu cells where Brca1 co-localized with γH2AX at the sites of DNA damage (Fig. 4B).
Figure 4. Resveratrol induces γH2AX and Brca1 foci formation in HNSCC cells.
A) Cells were treated with DMSO alone or resveratrol for 24 h, total cell lysates prepared and subjected to SDS-PAGE followed by immunoblotting, and membranes probed with γH2AX and β-actin antibodies as detailed in Methods. B) Resveratrol induces the γH2AX and Brca1 foci formation. FaDu cells were treated with DMSO (control) or resveratrol (25-50 μM) for 24 h followed by immunocytochemical staining for γH2AX and Brca1 using specific antibody, cell images were captured at 1000× magnification on a Nikon D Eclipse C1 confocal microscope, and images analyzed by EZ-C1 Free viewer software. Red and green fluorescence represents staining for γH2AX and Brca1, respectively. Res, resveratrol; C, control.
Another important observation related to DNA damaging effect of resveratrol in FaDu cells is an S phase arrest at 10 μM but a G1 phase arrest at higher concentration (25-50 μM) (Fig. 2A), and a higher γH2AX levels with increasing resveratrol concentration indicative of increasing levels of DNA damage (Fig. 4A), which is consistent with G1 or G1/S arrest at higher concentrations. Accordingly, we also determined cyclin levels in FaDu cells after resveratrol treatment and found that whereas cyclin D1 levels decreased, cyclin E levels increased (data not shown) simultaneously, a situation representing inhibition of DNA replication and a concomitant G1/S arrest and blockade to S phase progression by resveratrol as we had seen in ovarian cancer cells (9).
Resveratrol-induced γH2AX and Brca1 foci formation is independent of Smad4 in Cal27 cells
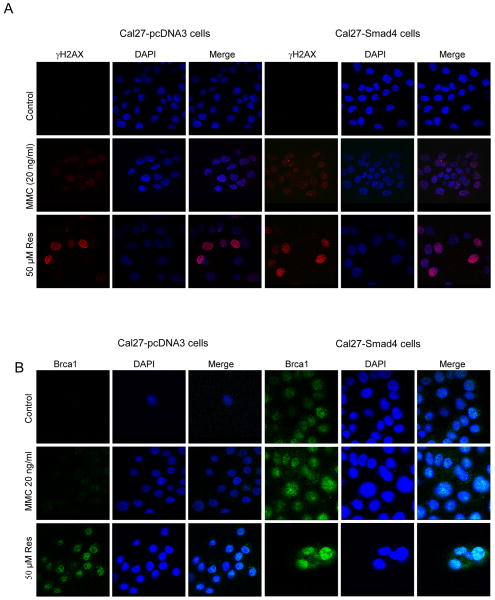
Recently, Smad4 loss was reported to cause genomic instability in mice, which correlated with less expression and function of genes encoding proteins in the Brca/Fanc in DNA repair pathway together with HNSCC development (8). Thus, we measured γH2AX and Brca1 foci formation in Cal27-pcDNA3 and Cal27- Smad4 cells. Cal27- pcDNA3 and Cal27- Smad4 cells showed almost parallel γH2AX foci formation after resveratrol (50 μM) treatment for 24 h (Fig. 5A). Mitomycin (20 ng/ml; MMC) treatment also induced the γH2AX foci formation in both the cell lines (Fig. 5A). However, MMC treated cells displayed less intense staining as compared to resveratrol treated cells (Fig. 5A). Brca1 staining was also examined in both cell lines after resveratrol treatment. Resveratrol (50 μM) treatment for 24 h induced strong nuclear localization of Brca1 as compared to untreated control (Fig. 5B). However, Cal27-Smad4 control cells showed similar Brca1 foci formation as compared to Cal27- pcDNA3 control cells (Fig. 5B). As shown previously (8), Smad4 was required for the formation of Brca1 foci by MMC (Fig. 5B). Thus, unlike MMC, resveratrol can produce Brca1 foci independently of Smad4.
Figure 5. Resveratrol-induced γH2AX and Brca1 foci formation is independent of Smad4 in Cal27 cells.
Resveratrol induces A) γH2AX and B) Brca1 foci formation in Cal27-pcDNA3 and Cal27-Smad4 cells. Following indicated treatments of the cells, immunocytochemical staining for γH2AX and Brca1 was performed using specific antibody as detailed in Methods. Cell images were captured at 1000× magnification on a Nikon D Eclipse C1 confocal microscope, and analyzed by EZ-C1 Free viewer software. Red and green fluorescence represents staining for γH2AX and Brca1, respectively. Res, resveratrol.
Resveratrol inhibits FaDu tumor xenograft growth together with in vivo DNA-damage and apoptotic effects
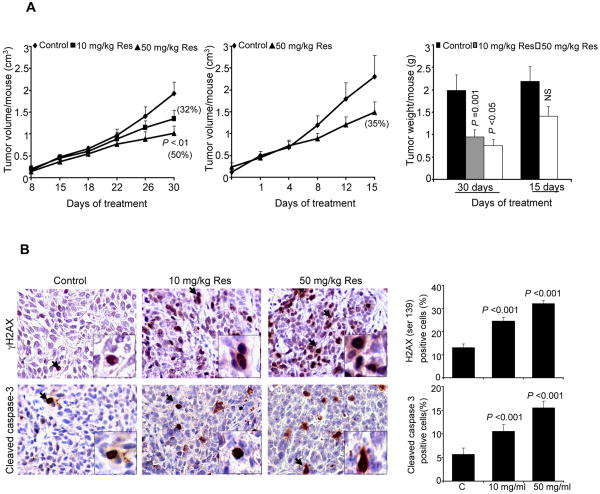
We also extended studies to an in vivo xenograft model to establish biological significance of all our in vitro findings. Resveratrol feeding by oral gavage resulted in a dose- and time-dependent inhibition in tumor growth, and at the end of the study (30 days), tumor volume/mouse decreased from 2.00 ± 0.25 in controls to 1.35 ± 0.12 and 1.01 ± 0.17 cm3 in 10 and 50 mg/kg body weight resveratrol-fed groups, respectively, accounting for 32 and 50% (p<0.01) inhibition in tumor growth (Fig. 6A). A reduction in tumor weight/mouse further supported these results where resveratrol-treated groups showed 0.95 ± 0.15 and 0.76 ± 0.13 g/mouse tumor weight, respectively, as compared to controls with 2.00 ± 0.35 g/mouse tumor weight (Fig. 6A). In contrast, resveratrol (50 mg/kg body weight) feeding for 15 days had less of an effect that was not statistically significant (Fig. 6A). In both efficacy studies, compared to controls, we did not observe any significant changes in body weight and diet consumption in resveratrol-fed groups of mice throughout the experiments (data not shown), suggesting that the resveratrol doses and treatment regimens employed in this study are non-toxic.
Figure 6. Resveratrol inhibits FaDu tumor xenograft growth together with in vivo DNA-damage and apoptotic effects.
A) Growth inhibitory effects of resveratrol in terms of tumor volume as a function of treatment and tumor weight at study end following the experiment detailed in Methods. In each case, data shown are mean±SE of eight mice/group. B) IHC analyses for γH2AX and cleaved caspase-3 in FaDu tumor xenografts from the 30 days of resveratrol treatment study as detailed in Methods. Representative images are shown for both staining in all three groups' samples. In each case quantitative data are mean ± SE of five tumor samples from five individual mouse in each group from five randomly selected microscopic (400×) fields from each tumor. NS, not statistically significant; Res, resveratrol.
The qualitative γH2AX and cleaved caspase-3 IHC analyses of tumor xenografts for DNA damage and apoptosis markers, respectively, clearly showed a strong increase in both staining in resveratrol-treated tumors compared to controls (Fig. 6B), Quantitative nuclear γH2AX immunostaining showed 25% (p<0.001) and 32% (p<0.001) induction in FaDu tumor xenografts from 10 and 50 mg/kg body weight resveratrol-treated mice, respectively, compared to 13% in controls (Fig. 6B, upper panel). Similarly, cleaved caspase-3 staining also increased 11-16% (p<0.001) in FaDu tumor xenografts from resveratrol-treated mice compared to 5 % cells in controls (Fig. 6B, lower panel).
Discussion
Following DNA damage, normal cells either repair the damage or undergo apoptotic death mostly in p53-dependent manner. However, DNA repair processes are defective in many cancer cells (18) which offer avenues for therapeutic intervention. Consequently, the present study is highly significant with major therapeutic implications in that the anti-cancer effect of resveratrol is predominately due to its ability to induce DNA damage selectively in HNSCC cells without affecting normal, repair-proficient cells.
Our detailed mechanistic studies revealed that resveratrol-induced DNA damage in HNSCC cells results in DNA strand breaks as evident by the production of Comet tails and ‘defective’ γH2AX and Brca1 repair foci resulting in irreversible arrest of HNSCC cells either at the G1/S phase boundary or in S phase eventually leading to apoptotic death. Similar results have been seen in breast cancer cells lacking Brca2 (19) and in colon cancer cells lacking Cdc14 (20) in which foci form after DNA damage but are repaired inefficiently. Since p53 is important for DNA repair mechanisms, the novelty of our study is that resveratrol effects were independent of p53 because all three HNSCC cell lines used harbor mutant p53 mutations (21). Furthermore, sensitivity to resveratrol was directly proportional to extent of basal DNA damage. For example, FaDu cells having higher level of basal DNA damage (Fig. 1B) were more sensitive to resveratrol than Cal27 cells (Fig. 1A). Similarly, Det562 cells with no detectable γH2AX even in 50 μM resveratrol were comparatively more resistant to treatment than the other two HNSCC cell lines. Because conventional cancer therapy damages DNA in normal as well as tumor cells (22), it is of considerable importance that resveratrol does not cause DNA damage and checkpoint activation in normal cells with efficient DNA repair pathways, thereby establishing the selectivity of resveratrol in affecting HNSCC cells.
The TGF-β-Smad signaling pathway is altered by genetic mutation that contributes to the carcinogenesis of HNSCC (23). Down regulation of the Smad4 tumor suppressor, the central mediator of TGF-β-Smad signaling pathway is common in malignant HNSCC and correlates with down regulated expression and an inefficient Brca/Fanc DNA repair pathway (8). Thus, DNA repair defects occur in HNSCC in which Smad4 either is frequently mutated or poorly expressed. In this regard, our results are highly significant and show resveratrol efficacy in FaDu and Cal27 HNSCC cell lines, which both contain null mutations in Smad4. To determine if the anti-cancer effects of resveratrol by the induction of DNA damage is dependent on Smad4-regulated DNA repair mechanisms, matching Cal27 and Cal27-Smad4 stable transfectants were used. Resveratrol treatment caused comparable DNA damage, ‘defective’ repair foci, cell cycle arrest, and apoptotic death in both cell lines irrespective of their Smad4 status, establishing the fact that Smad4 is dispensable for the anti-cancer effects of resveratrol in HNSCC. Perhaps, Smad4 is required to process MMC-induced damage and form foci (8), while it is not required to form the unrepairable foci induced by resveratrol. Together, these results indicate that most of the human HNSCC tumors will be sensitive to resveratrol even though more than 80 % of them are defective in Smad4 (24), signifying strong translational potential of our findings.
An important aspect of our study is that the in vitro effects of resveratrol were reproducible in xenograft studies in both preventive and therapeutic treatment regimens showing an inhibition in FaDu tumor xenograft growth in nude mice, which was associated with high levels DNA damage) and a marked induction in apoptosis. Overall, our results clearly demonstrate strong resveratrol efficacy against the growth and proliferation of HNSCC in vitro and in vivo. Specifically, resveratrol produces these effects on HNSCC via its selective ability to induce DNA damage and apoptotic death. Importantly, the anti- cancer effects of resveratrol and all associated mechanisms are independent of Smad4 status, the mutation/absence of which is one of the primary causes of failed cellular DNA repair machinery in HNSCC. Based on these findings, more studies are needed in future to evaluate both anti-cancer and chemopreventive efficacy of resveratrol in relevant pre-clinical models to establish its potential usefulness against human HNSCC.
Statement of Translational Relevance.
DNA repair processes, which are defective in many cancer cells, offer avenues for therapeutic intervention. Thus, preclinical/clinical development of non-toxic chemo preventive agents that can selectively induce DNA damage in cancer cells is highly desirable. Supporting this rationale, results of the present study are highly significant with major implications where we report that the anti-cancer effect of resveratrol is predominately due to its ability to induce DNA damage selectively in HNSCC cells without affecting normal, repair-proficient cells. This study reports that resveratrol causes S phase arrest and apoptotic death of FaDu, Cal27, Cal27-Smad4 cells together with induction of γH2AX and Brca1 foci, which are independent of Smad4 expression. The resveratrol mediated up-regulation of γH2AX and cleaved caspase-3 protein expression is also observed in FaDu tumor xenografts in athymic nude mice. These preclinical results hereby warrant further clinical investigation to determine the efficacy of resveratrol against HNSCC.
Acknowledgments
Grant Support: CCTSI (Colorado Clinical and Translational Sciences Institute NIH/NCRR #UL1-RR025780) co-pilot grant and UCCC Aging and Cancer Seed grants (NIH/NCI P20CA103680) awarded to RAS. NIH R01 grant AT003623 to CA. UCCC Core Grant (NIH/NCI P30CA046934) supported the Core facilities for FACS analysis.
Abbreviations
- HNSCC
head and neck squamous cell carcinoma
- TGF-β
transforming growth factor-β
- TGF-βRI
TGF-β type 1 receptor
- TGF-βRII
TGF-β type II receptor
- HFF
human foreskin fibroblast
- NHEK
normal human epidermal keratinocytes
- CMC
carboxy methyl cellulose
- DSBs
double strand breaks
References
- 1.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Fedele S. Diagnostic aids in the screening of oral cancer. Head Neck Oncol. 2009;1:5. doi: 10.1186/1758-3284-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grady WM. Transforming growth factor-beta, Smads, and cancer. Clin Cancer Res. 2005;11:3151–54. doi: 10.1158/1078-0432.CCR-05-0414. [DOI] [PubMed] [Google Scholar]
- 5.Chiao PJ, Hunt KK, Grau AM, Abramian A, Fleming J, Zhang W, et al. Tumor suppressor gene Smad4/DPC4, its downstream target genes, and regulation of cell cycle. Ann N Y Acad Sci. 1999;880:31–37. doi: 10.1111/j.1749-6632.1999.tb09507.x. [DOI] [PubMed] [Google Scholar]
- 6.Peng B, Fleming JB, Breslin T, Grau AM, Fojioka S, Abbruzzese JL, et al. Suppression of tumorigenesis and induction of p15(ink4b) by Smad4/DPC4 in human pancreatic cancer cells. Clin Cancer Res. 2002;8:3628–38. [PubMed] [Google Scholar]
- 7.Zhao S, Venkatasubbarao K, Lazor JW, Sperry J, Jin C, Cao L, et al. Inhibition of STAT3 Tyr705 phosphorylation by Smad4 suppresses transforming growth factor beta-mediated invasion and metastasis in pancreatic cancer cells. Cancer Res. 2008;68:4221–28. doi: 10.1158/0008-5472.CAN-07-5123. [DOI] [PubMed] [Google Scholar]
- 8.Bornstein S, White R, Malkoski S, Oka M, Han G, Cleaver T, et al. Smad4 loss in mice causes spontaneous head and neck cancer with increased genomic instability and inflammation. J Clin Invest. 2009;119:3408–19. doi: 10.1172/JCI38854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tyagi A, Singh RP, Agarwal C, Siriwardana S, Sclafani RA, Agarwal R. Resveratrol causes Cdc2-tyr15 phosphorylation via ATM/ATR-Chk1/2-Cdc25C pathway as a central mechanism for S phase arrest in human ovarian carcinoma Ovcar-3 cells. Carcinogenesis. 2005;26:1978–87. doi: 10.1093/carcin/bgi165. [DOI] [PubMed] [Google Scholar]
- 10.Gupta SC, Kannappan R, Reuter S, Kim JH, Aggarwal BB. Chemosensitization of tumors by resveratrol. Ann N Y Acad Sci. 2010;1215:150–60. doi: 10.1111/j.1749-6632.2010.05852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bishayee A. Cancer prevention and treatment with resveratrol: from rodent studies to clinical trials. Cancer Prev Res (Phila) 2009;2:409–18. doi: 10.1158/1940-6207.CAPR-08-0160. [DOI] [PubMed] [Google Scholar]
- 12.Jang M, Cai L, Udeani GO, Slowing KV, Thomas CF, Beecher CW, et al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997;275:218–20. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- 13.Roy S, Gu M, Ramasamy K, Singh RP, Agarwal C, Siriwardana S, et al. p21/Cip1 and p27/Kip1 are essential molecular targets of inositol hexaphosphate for its antitumor efficacy against prostate cancer. Cancer Res. 2009;69:1166–73. doi: 10.1158/0008-5472.CAN-08-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu M, Roy S, Raina K, Agarwal C, Agarwal R. Inositol hexaphosphate suppresses growth and induces apoptosis in prostate carcinoma cells in culture and nude mouse xenograft: PI3K-Akt pathway as potential target. Cancer Res. 2009;69:9465–72. doi: 10.1158/0008-5472.CAN-09-2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raina K, Rajamanickam S, Singh RP, Deep G, Chittezhath M, Agarwal R. Stage-specific inhibitory effects and associated mechanisms of silibinin on tumor progression and metastasis in transgenic adenocarcinoma of the mouse prostate model. Cancer Res. 2008;68:6822–30. doi: 10.1158/0008-5472.CAN-08-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Podhorecka M, Skladanowski A, Bozko P. H2AX Phosphorylation: Its Role in DNA Damage Response and Cancer Therapy. J Nucleic Acids. 2010;2010:1–9. doi: 10.4061/2010/920161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Au WW, Henderson BR. Identification of sequences that target BRCA1 to nuclear foci following alkylative DNA damage. Cell Signal. 2007;19:1879–92. doi: 10.1016/j.cellsig.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 18.Kennedy RD, D'Andrea AD. DNA repair pathways in clinical practice: lessons from pediatric cancer susceptibility syndromes. J Clin Oncol. 2006;24:3799–08. doi: 10.1200/JCO.2005.05.4171. [DOI] [PubMed] [Google Scholar]
- 19.Issaeva N, Thomas HD, Djureinovic T, Jaspers JE, Stoimenov I, Kyle S, et al. 6-thioguanine selectively kills BRCA2-defective tumors and overcomes PARP inhibitor resistance. Cancer Res. 2010;70:6268–76. doi: 10.1158/0008-5472.CAN-09-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mocciaro A, Berdougo E, Zeng K, Black E, Vagnarelli P, Earnshaw W, et al. Vertebrate cells genetically deficient for Cdc14A or Cdc14B retain DNA damage checkpoint proficiency but are impaired in DNA repair. J Cell Biol. 2010;189:631–39. doi: 10.1083/jcb.200910057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin CJ, Grandis JR, Carey TE, Gollin SM, Whiteside TL, Koch WM, et al. Head and neck squamous cell carcinoma cell lines: established models and rationale for selection. Head Neck. 2007;29:163–88. doi: 10.1002/hed.20478. [DOI] [PubMed] [Google Scholar]
- 22.Kastan MB. DNA damage responses: mechanisms and roles in human disease. Mol Cancer Res. 2008;6:517–24. doi: 10.1158/1541-7786.MCR-08-0020. [DOI] [PubMed] [Google Scholar]
- 23.Baez A, Cantor A, Fonseca S, Marcos-Martinez M, Mathews LA, Muro-Cacho CA, et al. Differences in Smad4 expression in human papillomavirus type 16-positive and human papillomavirus type 16-negative head and neck squamous cell carcinoma. Clin Cancer Res. 2005;11:3191–97. doi: 10.1158/1078-0432.CCR-04-1299. [DOI] [PubMed] [Google Scholar]
- 24.White RA, Malkoski SP, Wang XJ. TGFbeta signaling in head and neck squamous cell carcinoma. Oncogene. 2010;29:5437–46. doi: 10.1038/onc.2010.306. [DOI] [PMC free article] [PubMed] [Google Scholar]