Abstract
Human islet amyloid polypeptide is a hormone coexpressed with insulin by pancreatic beta-cells. For reasons not clearly understood, hIAPP aggregates in type II diabetics to form oligomers that interfere with beta-cell function, eventually leading to the loss of insulin production. The cellular membrane catalyzes the formation of amyloid deposits and is a target of amyloid toxicity through disruption of the membrane’s structural integrity. Therefore, there is considerable current interest in solving the 3D structure of this peptide in a membrane environment. NMR experiments could not be directly utilized in lipid bilayers due to the rapid aggregation of the peptide. To overcome this difficulty, we have solved the structure of the naturally occurring peptide in detergent micelles at a neutral pH. The structure has an overall kinked helix motif, with residues 7–17 and 21–28 in a helical conformation, and with a 310 helix from Gly 33 – Asn 35. In addition, the angle between the N- and C-terminal helices is constrained to 85º. The greater helical content of human IAPP in the amidated versus free acid form is likely to play a role in its aggregation and membrane disruptive activity.
Keywords: Islet amyloid polypeptide, amyloid, aggregation, peptide, membrane, structure
Human Islet Amyloid Polypeptide (hIAPP1, also known as amylin) is a 37 residue peptide hormone secreted from pancreatic β-cells (Fig. 1). In its normal physiological role, hIAPP is associated with appetite suppression and, in conjunction with insulin, in maintaining proper glycemic levels.1 However, a change in the cellular environment in the early stages of type II diabetes, poorly understood at present, causes it to aggregate into dense, insoluble fibrillar deposits that accumulate in the pancreas.2 These proteinaceous deposits, known as amyloid, have a characteristic β-sheet secondary structure similar to those found in Alzheimer’s, Parkinson’s, Huntington’s, and a variety of other degenerative disorders.3 Like the amyloid deposits found in these diseases, hIAPP aggregates of various forms have been linked to cellular death and impairment of normal tissue functioning.4, 5
Fig. 1.
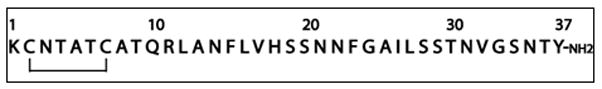
Primary sequence of human-IAPP including the disulfide bridge between Cys2 – Cys7 and amidated C-terminus.
One of the primary mechanisms by which hIAPP and amyloidogenic peptides in general cause cellular death is the disruption of the integrity of the cellular membrane.6 Human islet amyloid polypeptide, but not the non-amyloidogenic rat variant, has been shown to cause significant impairment of the integrity of the phospholipid membrane in both model membranes and in cells. 7–12 The exact mechanism of membrane disruption is unknown but has been linked to peptide aggregation on the membrane surface.7, 10, 11 Binding of hIAPP to lipid membranes also markedly accelerates the aggregation to fibril formation, a factor that is likely to be important in determining the final amount of amyloid deposition.13–15
Structural studies report that monomeric hIAPP is primarily, but not completely, unstructured before it aggregates to form β-sheet fibers.16–18 While a structural model for hIAPP amyloid-fibers has been created from solid state NMR measurements,19 solving the high-resolution structure of hIAPP in a membrane environment has been a major challenge due to the rapid aggregation of hIAPP in the presence of membranes.13 Low-resolution structural studies have shown that human-IAPP initially binds to the membrane in a helical state,20–23 and a recent NMR study reported the structure of the free-acid form of hIAPP at a low pH in SDS micelles.24 Once bound to the membrane, aggregation of the peptide causes a cooperative conformational change from the helical conformation to the β-sheet amyloid form.20, 21, 23, 25
While these studies have provided evidence for a helical conformation of the peptide, the high-resolution 3D structure of the membrane-associated hIAPP peptide in its natural form at physiological pH has not been reported. In this study, we have solved the high-resolution structure of C-terminus-amidated hIAPP (similar to in the native form) in SDS micelles and determined the membrane-binding topology with respect to the micelle at physiological pH. Our results indicate a substantial structural difference in the C-terminus of the peptide between the two versions of the peptide, which is disordered in the previous hIAPP NMR structure but is ordered under the conditions employed in this study, which likely has implications for the mechanism of membrane-mediated aggregation and membrane disruption by hIAPP.
Materials & Methods
Sample Preparation
Human-IAPP (hIAPP) was synthesized and purified by SynBioSci (Toronto, ON) with a disulfide bridge from residues Cys 2 – Cys 7 and an amidated C-terminus.
The peptide was dissolved in hexafluoroisopropanol to monomerize the preformed aggregates of hIAPP, and then lyophilized to remove the solvent. For NMR experiments, the sample was prepared by dissolving a 3 mg of lyophilized peptide in 20 mM sodium phosphate buffer at pH ~ 7.3 containing 10% D2O, 120 mM NaCl and 200 mM perdeuterated SDS (Cambridge Isotopes Laboratory) to a final peptide concentration of 2.5 mM.
NMR Data Collection and Processing
All NMR experiments on SDS micelles containing hIAPP were performed at 25 °C using a 900 MHz Bruker Avance NMR spectrometer equipped with a triple-resonance cryogenic probe. After optimizing the experimental parameters using 1D 1H NMR spectrum of the sample, a 2D 1H-1H TOCSY spectrum was recorded with an 80 ms mixing time and 2D 1H-1H NOESY spectra were recorded at 100 ms and 300 ms mixing times in order to assign backbone and side-chain resonances. Complex data points were acquired for quadrature detection in both frequency dimensions of the 2D experiments. For all the spectra, zero-filling was performed in both dimensions to yield matrices of 2048 × 2048 points. Proton chemical shifts were referenced by setting the water peak at 4.7 ppm, as DSS has been found to bind some amyloidogenic proteins and may affect the conformation.26 All 2D spectra were processed using TopSpin 2.1 software (from Bruker) and analyzed using SPARKY.27 Resonance assignments were done using a standard approach as reported elsewhere.28
Structure Calculations
The final structure calculations were carried out with the CYANA 2.1 program package using simulated annealing in combination with molecular dynamics in torsion angle space.29 NOE connectivities were used for the calculation of dihedral angle restraints.30 Unambiguous long-range NOE constraints were used during the first round of structure calculations to generate an initial low-resolution structure. The remaining ambiguous NOE cross-peaks were assigned in an iterative fashion by applying a structure-aided filtering strategy in repeated rounds of structure calculations.31 A total of 500 conformers were calculated using 8000 annealing steps for each conformer after complete assignment of resonances. The lowest 20 energy conformers were selected and visualized using MOLMOL.32 The values of Wishart et al were used for the random coil chemical shifts.33
Paramagnetic Quenching
2D 1H-1H TOCSY spectra of hIAPP embedded in SDS micelles were recorded in the absence and in the presence of 0.8 mM MnCl2. All the other experimental conditions were the same as mentioned above.
Results
3D Structure of hIAPP in SDS micelles
2D 1H-1H TOCSY and NOESY spectra of hIAPP embedded in SDS micelles were used to assign the backbone and side chain resonances of the peptide. The deviation of the Hα chemical shifts from the corresponding values for a random coil structure (Hα chemical shift index) was used as an indicator of the secondary structure of the peptide. In an α-helical structure, Hα protons experience a shift to higher field corresponding to a negative deviation in the CSI plot. A negative deviation spanning 3 or more residues indicates the propensity for an α-helical conformation. The CSI plot shows the propensity for a helix formation in two regions of the hIAPP, a longer helix in the region Ala 5 – Ser 28 and a shorter helix from Ser 34 to Tyr 37 as shown in Fig. 2. The chemical shift value for the Hα proton of Cys 2 was not observed in the spectra, most likely due to a chemical exchange process.
Fig. 2.
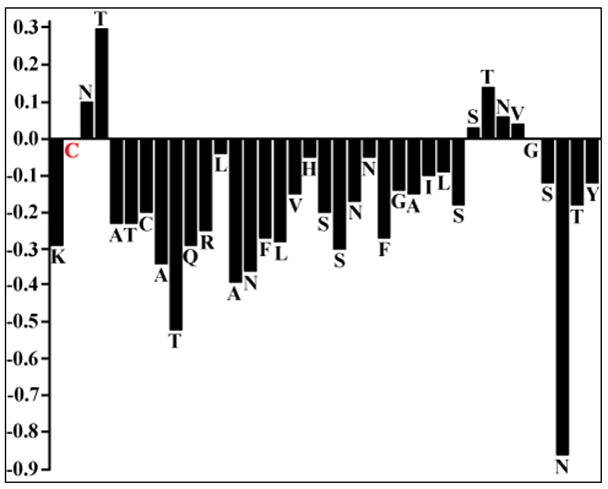
Alpha-proton chemical shift index for hIAPP embedded in SDS micelles. The CSI was calculated by subtracting the appropriate random coil chemical shifts reported in the literature. A CSI ≤ −0.1 is considered indicative of a helical conformation.
The fingerprint region of the NOESY spectra obtained at a 300 ms mixing time is shown in Fig. 3 with sequential assignment of residues. From the analysis of 2D NOESY spectra, we have assigned a total of 568 (446 short, 119 medium and 3 long range) NOEs for hIAPP (Fig. 4). The final 3D structure of the peptide is presented in Fig. 5 (pdb id 2l86). The superposition of backbone atoms from residues 7 to 37 gives an RMSD of 0.46 ± 0.15 Å as shown in Fig. 5, while the superposition of all heavy atoms gives an RMSD of 1.09 ± 0.23 Å (see Table 1 for more structure statistics). Three helical domains are evident from the numerous dNN(i,i+1), dNN(i,i+2), dαβ(i,i+3), dαN(i,i+3) and dαN (i,i+4) NOE connectivities that are diagnostic of an α-helix as shown in Fig. 5. Two of the helical domains extending from residues Cys 7 - Val 17 and Asn 21- Ser 28 are α-helices, while the third is a short 310-helix extending from Gly 33 to Asn 35. The two α-helices are separated by a turn from residues His 18 – Ser 20. The break in the N-terminal α-helix at His 18, also present in the previous NMR structure of the recombinant hIAPP 24, is confirmed by the absence of dαβ(i,i+3), dαN(i,i+3) NOEs from Leu 16 - Ser 19 and His 18 - Ser 20 that would be present if Leu 16 - Ser 19 were in a helical conformation (Fig. 4). Unlike the structure of the recombinant hIAPP, the second helix does not rotate freely about the first helix but is constrained in a bent conformation with an inter-helical angle determined by MOLMOL to be 85º ± 5º. This constraint between the two helices can be attributed to the presence of dαN(i,i+2) and dαN(i,i+3) NOEs from V17 to S19 and V17 to S20 which limit the conformational space to the bent conformation observed in the structure (Fig. 4).
Fig. 3.
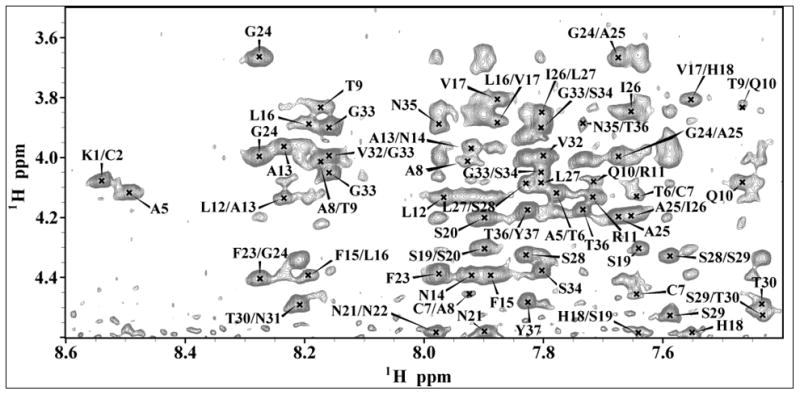
The fingerprint region of 2D 1H-1H NOESY spectrum of hIAPP in SDS micelles showing sequential Hα - HN NOE connectivities.
Fig. 4.
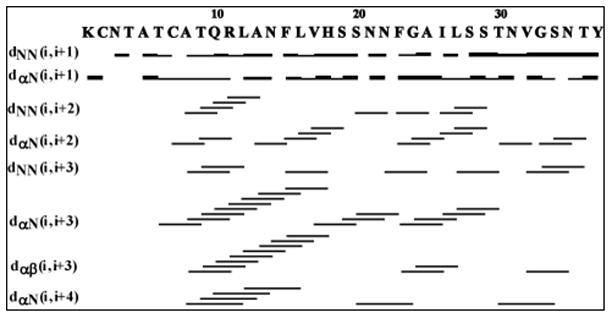
A summary of the sequential and medium range NOE connectivities for hIAPP in SDS micelles. The intensities of the observed NOEs are represented by the thickness of lines and are classified as strong, medium, and weak.
Fig. 5.
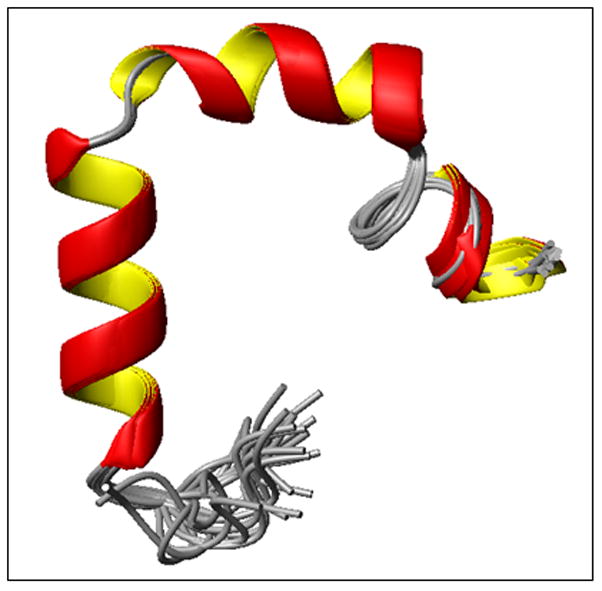
High-resolution NMR structures of hIAPP in SDS micelles at physiological pH. Note that the N-terminal helix from C7-V17 is separated from the other helix from N21-S28 by a turn comprising of residues H18-S20 and the C-terminus has a short 310-helix comprising of residues G33-N35.
Table 1.
Statistical information for the hIAPP structural ensemble
| Distance Constraints | |
|---|---|
| Total | 568 |
| Short (i − j ≤ 1) | 446 |
| Medium (i − j = 2, 3, 4) | 119 |
| Long (i − j ≥ 5) | 3 |
| Structural Statistics | |
| Violated distance constraints | 0 |
| Violated angle constraints | 0 |
| RMSD of all backbone atoms (Å) (Cys 7 - Tyr 37) | 0.49 ± 0.17 |
| RMSD of all heavy atoms (Å) (Cys 7 - Tyr 37) | 1.32 ± 0.27 |
| Ramachandran Plot | |
| Residues in most favored region (%) | 86.6 |
| Residues in additionally allowed region (%) | 10.8 |
| Residues in generously allowed region (%) | 2.6 |
The 310-helix from Gly 33 to Asn 35 found in this study was not observed in the structure of the recombinant hIAPP. The absence of dNN(i,i+3), dαβ(i,i+3), and dαN (i,i+4) NOEs in the spectra of recombinant hIAPP in this region,24 as well as their presence in the current study, support this difference between the two structures. In particular, dNN(i,i+3) NOEs from Val32 – Asn35 and Gly 33 – Thr 36, a dαβ(i,i+3) NOE from Val 32 – Asn 35, and a dαN (i,i+4) NOE from Thr30 – Ser 34 (Fig. 4) indicate the existence of a helical conformation from Gly 33 to Asn 35, none of which were observed in the spectra of recombinant hIAPP. 24 The presence of dαN(i,i+2) NOEs spanning Thr 30-Val 32, Gly 33-Asn 35, and Ser 34 -Thr 36 indicates the formation of a 310-, rather than α-, helix (Fig. 4). This contention is further supported by the observation of a hydrogen bonds in the final structure between the backbone of Gly 33 to Thr 36 and Ser 34 and the C-terminal amide, an i to to i+3 pattern typical for 310 helices.
Localization of hIAPP in SDS micelles
In order to find the orientation of the peptide with respect to the SDS micelle, we have performed paramagnetic quenching experiments using MnCl2 as a paramagnetic ion. Since Mn2+ ions are less likely to penetrate into the hydrophobic interior of the micelle, only residues exposed to solvent experience the paramagnetic induced relaxation effect that results in broadening of the corresponding peaks. In the 2D TOCSY spectrum recorded in the presence of 0.8 mM MnCl2, most of the resonances in the finger print region disappeared as shown in Fig. 6, indicating most of the residues of hIAPP are exposed to the solvent and the peptide is located close to the surface of the micelle. However, the Hα protons of T9, R11 and L12, the side chain peaks of K1, N3, Q10, R11, N14, and the amide protons of C2, A5-A13, F15, L16, S20, A25, S29, V32, T36 and Y37 are still observable but with a reduced intensity. These results indicate that the side chains of residues in the α-helical region Cys 7 – Val 17, as well as some of the residues in the C-terminus, are most likely embedded into the head group region of the detergent.
Fig. 6.
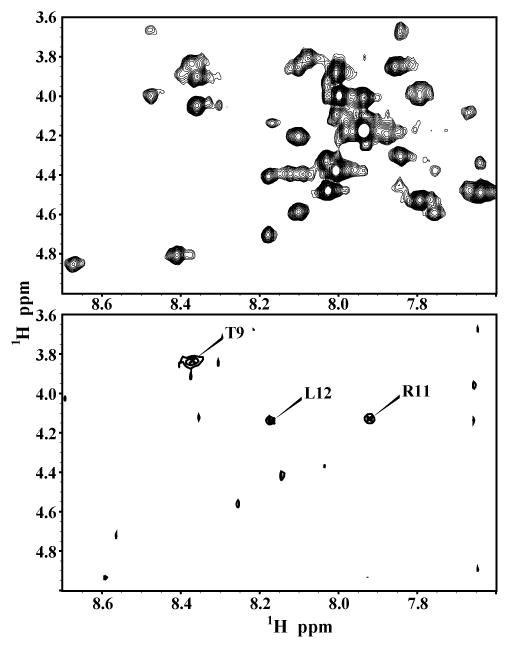
2D 1H-1H TOCSY spectra of hIAPP in SDS micelles at physiological pH in the absence (top) and presence (bottom) of 0.8 mM MnCl2.
Discussion
The misfolding and aggregation of hIAPP and related structural changes depends on a variety of experimental conditions such as pH, ionic concentration, peptide concentration and temperature in solution. Several biophysical studies have reported the structural changes accompanying the changes during aggregation in solution from the random coil monomeric peptide to the β-sheet amyloid aggregate.34–41 The interaction of the peptide with the lipid membrane is of significant interest as it increases the rate of peptide aggregation which can in turn result in membrane disruption.13, 23 While solving the high-resolution structure of the peptide in a membrane environment would provide insights into this process, it has been difficult to apply NMR techniques in lipid vesicles due to the fast aggregating nature of the sample. To overcome this limitation, in this study we have utilized SDS micelles to stabilize the intermediate structure of hIAPP at pH 7.3 and solved the 3D structure and its membrane orientation.
As shown in Figure 5, our studies reveal that the structure of amidated hIAPP in SDS has an overall kinked helix motif, with residues 7–17 and 21–28 in a helical conformation, and with a 310 helix from Gly 33 – Asn 35. In addition, the angle between the N- and C-terminal helices is constrained to 85º. The helix-turn-helix structure of hIAPP is a common motif found in many of the high-resolution structures of amyloids bound to the detergent micelles, where an amphipathic helix is separated from a more polar helix by a short flexible region. In all these high-resolution structures there is a variable degree of conformational flexibility, as the location of the linker as well as length of the helical regions vary depending upon the experimental conditions.24, 42–45 The location of the kinks in the structure in detergent micelles for amyloidogenic peptides largely correlates with the location of the turns in the structure of the amyloid fiber.19, 46–49
The overall structure of hIAPP without amidation in the C-terminal obtained at pH 4.6 (21) resembles the structure presented here of the naturally-occurring C-terminal amidated form of hIAPP obtained at neutral pH as compared in Figure 7. However, two important differences exist between the conditions used to solve the structure of the free acid form of IAPP at an acidic pH and those likely to be encountered by hIAPP in the physiological setting. First, since pH plays an important role in the membrane binding properties of hIAPP, the structure and membrane orientation obtained at an acidic pH of 4.6 may not be physiologically relevant. For example, a recent NMR study on a truncated version of hIAPP (hIAPP1-19) showed that protonation of His 18 causes a change in its membrane binding topology from a buried to a surface associated state.50 This observed change in membrane topology with pH was linked with a greatly reduced potential of hIAPP1-19 to disrupt phospholipid vesicles, a phenomenon that was also observed for β-cell membranes as demonstrated by a H18R hIAPP1-19 mutant.8 Further, the aggregation of hIAPP is strongly pH dependent,34, 51 with protonation of H18 slowing the aggregation by a factor of ~4.51 The decrease in the aggregation potential of hIAPP at acidic pHs is one factor that allows hIAPP to be safely stored in the secretory granule (pH ~5.5) in a presumably non-aggregated form.52
Fig. 7.
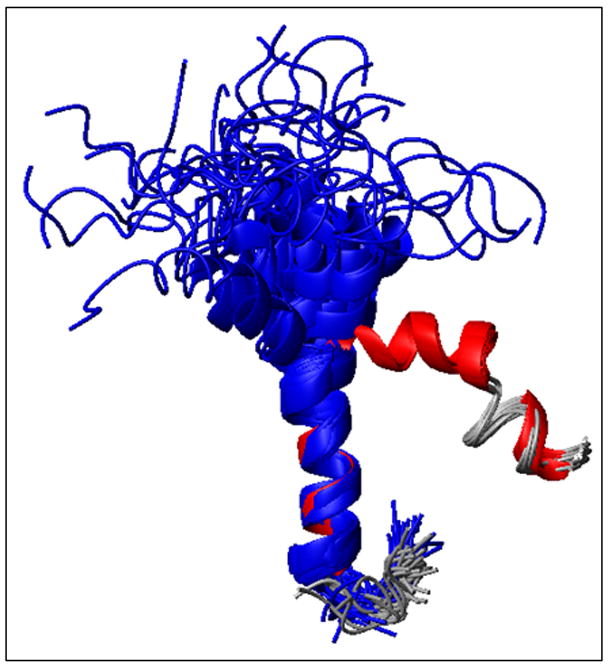
Overlay of the ensemble of NMR structures of hIAPP in SDS micelles solved at acidic pH (blue) and physiological pH (red). Note that the N-terminal helix (residues to 7 to 17) overlays quite well, while a substantial deviation is observed for the second helix from 21 to 28.
Second, hIAPP is normally expressed with an amidated C-terminus that is essential for proper biological function.53 The NMR study used a recombinant form of the peptide with a non-amidated C-terminus, altering the electrostatic interactions at the C-terminal end of the peptide.24 This small change is significant for hIAPP as studies have shown that the free acid of hIAPP is significantly less amyloidogenic than the amidated version.17 In addition, a recent study has shown relatively strong interactions between protonated His 18 and Tyr 37,40 which may partly be attributable to a salt bridge formation between His 18 and the C-terminus, an interaction that will not occur in the amidated peptide. Since environmental conditions can have a strong impact on the structure and membrane perturbing activity of the peptide, it is important to determine the structure of membrane-bound hIAPP under conditions more close to the physiological state.
Although the overall helix-turn-helix motif of the previous structure is preserved (see Figure 7), the kink is much more pronounced in the new structure of hIAPP determined in this study. While the N-terminal α-helix of both the structures overlays well from Cys 7 – Val 17 with a backbone RMSD of 0.48 ± 0.25 Å as shown in Fig. 7, the second helix deviates significantly from each other. At acidic pH, the helix from Asn 22 – Ser 28 in the the hIAPP free acid wobbles about the N-terminal helix with an inter-helical angle of 30°C.24 By contrast, the Asn21-Ser28 helix of C-amidated-hIAPP at neutral pH is constrained to lie at an interhelical angle of 85°. The strongly bent aspect of the structure resembles those of hIAPP and the related pramlintide construct determined at neutral pH in fluorinated organic solvents,51, 54 suggesting the kink in the helix is not enforced by membrane binding. In both structures the region of the peptide constrained by the disulfide ring is partially unstructured and pointing away from the hydrophobic side of the N-terminal helix.24
In addition to the differences observed in the kink region of the peptide, a substantial difference exists at the C-terminal end. In our structure residues Gly33 to Asn35 at the C-terminal end of the peptide are in a 310 helical conformation, in comparison to the conformationally unconstrained C-terminus found in the structure of Patil et al.24 The CD spectra of hIAPP at pH 4.6 and 10.8 is similar, suggesting that the change in the protonation state of His18 is not responsible for the observed differences.24 The formation of the 310 helix from Gly33 to Asn35 is therefore most likely due to the change from an unprotected, negatively charged C-terminus to the amidated, uncharged variant present in the physiologically expressed peptide. The negatively charged free acid will unfavorably interact with the headgroup of SDS and negatively charged lipids, while the amidated version not only lacks this unfavorable interaction but also has the potential for favorable hydrogen bonding interactions with the detergent or lipid headgroup.
Several lines of evidence point to the importance of the C-terminal end of hIAPP in amyloid formation. First, as a peptide fragment, residues 30-37 independently form amyloid deposits.55 Second, mutations at the C-terminal can adversely affect the kinetics of amyloid formation. For instance, mutations of Asn31 to Leu or Asn35 to Leu is sufficient for a three-fold decrease in the fibrillogenesis rate,56 while mutations of Asn31 to Ser or Val32 to Ala abolish amyloid formation.57 Third, the hIAPP free acid aggregates significantly slower (1/9 the rate) than the amidated variant, suggesting electrostatics at the C-terminal end plays a key role in aggregation.17 Fourth, the entire C-terminus is incorporated into the amyloid fiber in the current structural model of hIAPP amyloid fibers, indicating interactions of the C-terminal with the N-terminal stabilize the final structure.19 Finally, the N-terminal regions of hIAPP and the nonamyloidogenic and nontoxic rat-IAPP variant are similar, with most of the differences in structure concentrated at the C-terminal end.50, 58 Taken together, these findings raise the possibility that, while the C-terminus is not strictly essential for amyloid formation by hIAPP,59 its conformation can significantly modulate the kinetics of amyloid formation (see Fig. 8 for a possible model).
Fig. 8.
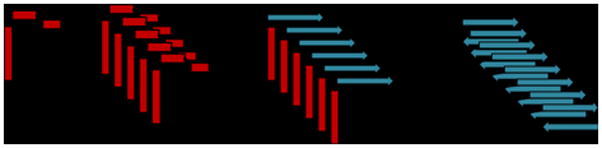
A possible schematic model for the aggregation of hIAPP in the presence of lipid membranes. (A) hIAPP initially binds to the membrane in a helical conformation. (B) Once bound to the membrane, hIAPP aggregates on the membrane surface to form helical bundles (C) Aggregation of the peptide causes a conformational change in the less structured C-terminus to the β-sheet conformation of the amyloid form. (D) Formation of β-sheets at the C-terminus triggers a corresponding conformational change at the N-terminus, producing the final amyloid fiber.
The existence of a structured C-terminus is significant for a structural interpretation of both membrane-mediated and membrane-free aggregation of hIAPP. The transition from a conformationally unconstrained monomeric peptide to the highly ordered amyloid supermolecular complex is entropically disfavored. For this reason, amyloid formation from conformationally unconstrained monomers is frequently kinetically inaccessible, despite the overall favorable free energy change associated with amyloid formation for hydrophobic sequences.60 Partially structured intermediates can reduce the entropic cost of the highly disfavored initial step of amyloid formation by favorably positioning aggregation prone regions to interact with each other. In particular, there is evidence for a mechanistic role for helical intermediates in amyloid aggregation.3, 61 Helical intermediates have been directly observed for some amyloidogenic proteins and have been indirectly inferred for others from mutational analysis and solvent perturbation studies.62, 63 Inhibitors have been constructed that utilize this fact to stop amyloid formation by either excessively destabilizing or overstabilizing the helical state.64, 65 310 helices are particularly suited for undergoing a helix to beta sheet transition due to the relative similarities in the phi/psi torsion angles between the beta-sheet and 310 helix conformations and the relative ease of partially unfolding a 310 helix compared to the α-helix conformation.3
Acknowledgments
This study was supported by research funds from NIH (DK078885 to A. R.). We thank the 900 MHz NMR facility at the Michigan State University.
Footnotes
The atomic coordinates of the best 20 conformers are deposited in the Protein Data Bank (accession code 2L86).
Abbreviations: IAPP: Islet Amyloid Polypeptide, hIAPP: Human Islet Amyloid Polypeptide, SDS: Sodium Dodecyl Sulfate, NMR: Nuclear Magnetic Resonance, CSI :Chemical Shift Index, NOE: Nuclear Overhauser Effect,
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Scherbaum WA. The role of amylin in the physiology of glycemic control. Exp Clin Endocrinol Diabet. 1998;106:97–102. doi: 10.1055/s-0029-1211958. [DOI] [PubMed] [Google Scholar]
- 2.Haataja L, Gurlo T, Huang CJ, Butler PC. Islet amyloid in type 2 diabetes, and the toxic oligomer hypothesis. Endocr Rev. 2008;29:302–316. doi: 10.1210/er.2007-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison RS, Sharpe PC, Singh Y, Fairlie DP. Amyloid peptides and proteins in review. Rev Physiol Biochem P. 2007;159:1–77. doi: 10.1007/112_2007_0701. [DOI] [PubMed] [Google Scholar]
- 4.Henson MS, Buman BL, Jordan K, Rahrmann EP, Hardy RM, Johnson KH, O'Brien TD. An in vitro model of early islet amyloid polypeptide (IAPP) fibrillogenesis using human IAPP-transgenic mouse islets. Amyloid. 2006;13:250–259. doi: 10.1080/13506120600960734. [DOI] [PubMed] [Google Scholar]
- 5.Ritzel RA, Meier JJ, Lin CY, Veldhuis JD, Butler PC. Human islet amyloid polypeptide oligomers disrupt cell coupling, induce apoptosis, and impair insulin secretion in isolated human islets. Diabetes. 2007;56:65–71. doi: 10.2337/db06-0734. [DOI] [PubMed] [Google Scholar]
- 6.Butterfield SM, Lashuel HA. Amyloidogenic protein membrane interactions: Mechanistic insight from model systems. Angew Chem Int Ed Engl. 2010;49:5628–5654. doi: 10.1002/anie.200906670. [DOI] [PubMed] [Google Scholar]
- 7.Brender JR, Durr UHN, Heyl D, Budarapu MB, Ramamoorthy A. Membrane fragmentation by an amyloidogenic fragment of human islet amyloid polypeptide detected by solid-state NMR spectroscopy of membrane nanotubes. Biochim Biophys Acta. 2007;1768:2026–2029. doi: 10.1016/j.bbamem.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brender JR, Hartman K, Reid KR, Kennedy RT, Ramamoorthy A. A single mutation in the nonamyloidogenic region of islet amyloid polypeptide greatly reduces toxicity. Biochemistry. 2008;47:12680–12688. doi: 10.1021/bi801427c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith PES, Brender JR, Ramamoorthy A. Induction of negative curvature as a mechanism of cell toxicity by amyloidogenic peptides: The case of islet amyloid polypeptide. J Am Chem Soc. 2009;131:4470–4478. doi: 10.1021/ja809002a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sparr E, Engel MFM, Sakharov DV, Sprong M, Jacobs J, de Kruijff B, Hoppener JWM, Killian JA. Islet amyloid polypeptide-induced membrane leakage involves uptake of lipids by forming amyloid fibers. FEBS Lett. 2004;577:117–120. doi: 10.1016/j.febslet.2004.09.075. [DOI] [PubMed] [Google Scholar]
- 11.Engel MF, Khemtemourian L, Kleijer CC, Meeldijk HJ, Jacobs J, Verkleij AJ, de Kruijff B, Killian JA, Hoppener JW. Membrane damage by human islet amyloid polypeptide through fibril growth at the membrane. Proc Natl Acad Sci U S A. 2008;105:6033–6038. doi: 10.1073/pnas.0708354105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janson J, Ashley RH, Harrison D, McIntyre S, Butler PC. The mechanism of islet amyloid polypeptide toxicity is membrane disruption by intermediate-sized toxic amyloid particles. Diabetes. 1999;48:491–498. doi: 10.2337/diabetes.48.3.491. [DOI] [PubMed] [Google Scholar]
- 13.Knight JD, Miranker AD. Phospholipid catalysis of diabetic amyloid assembly. J Mol Biol. 2004;341:1175–1187. doi: 10.1016/j.jmb.2004.06.086. [DOI] [PubMed] [Google Scholar]
- 14.Brender JR, Lee EL, Cavitt MA, Gafni A, Steel DG, Ramamoorthy A. Amyloid fiber formation and membrane disruption are separate processes localized in two distinct regions of IAPP, the type-2-diabetes-related peptide. J Am Chem Soc. 2008;130:6424–6429. doi: 10.1021/ja710484d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jayasinghe SA, Langen R. Membrane interaction of islet amyloid polypeptide. Biochim Biophys Acta. 2007;1768:2002–2009. doi: 10.1016/j.bbamem.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 16.Soong R, Brender JR, Macdonald PM, Ramamoorthy A. Association of highly compact type II diabetes related islet amyloid polypeptide intermediate species at physiological temperature revealed by diffusion NMR spectroscopy. J Am Chem Soc. 2009;131:7079–7085. doi: 10.1021/ja900285z. [DOI] [PubMed] [Google Scholar]
- 17.Yonemoto IT, Kroon GJ, Dyson HJ, Balch WE, Kelly JW. Amylin proprotein processing generates progressively more amyloidogenic peptides that initially sample the helical state. Biochemistry. 2008;47:9900–9910. doi: 10.1021/bi800828u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaiana SM, Best RB, Yau WM, Eaton WA, Hofrichter J. Evidence for a Partially Structured State of the Amylin Monomer. Biophys J. 2009;97:2948–2957. doi: 10.1016/j.bpj.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luca S, Yau WM, Leapman R, Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: Constraints from solid-state NMR. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Apostolidou M, Jayasinghe SA, Langen R. Structure of alpha-helical membrane-bound hIAPP and its implications for membrane-mediated misfolding. J Biol Chem. 2008;283:17205–17210. doi: 10.1074/jbc.M801383200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williamson JA, Loria JP, Miranker AD. Helix stabilization precedes aqueous and bilayer-catalyzed fiber formation in islet amyloid polypeptide. J Mol Biol. 2009;393:383–396. doi: 10.1016/j.jmb.2009.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ling YL, Strasfeld DB, Shim SH, Raleigh DP, Zanni MT. Two-dimensional infrared spectroscopy provides evidence of an intermediate in the membrane-catalyzed assembly of diabetic amyloid. J Phys Chem B. 2009;113:2498–2505. doi: 10.1021/jp810261x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jayasinghe SA, Langen R. Lipid membranes modulate the structure of islet amyloid polypeptide. Biochemistry. 2005;44:12113–12119. doi: 10.1021/bi050840w. [DOI] [PubMed] [Google Scholar]
- 24.Patil SM, Xu SH, Sheftic SR, Alexandrescu AT. Dynamic alpha-helix structure of micelle-bound human amylin. J Biol Chem. 2009;284:11982–11991. doi: 10.1074/jbc.M809085200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knight JD, Hebda JA, Miranker AD. Conserved and cooperative assembly of membrane-bound alpha-helical states of islet amyloid polypeptide. Biochemistry. 2006;45:9496–9508. doi: 10.1021/bi060579z. [DOI] [PubMed] [Google Scholar]
- 26.Laurents DV, Gorman PM, Guo M, Rico M, Chakrabartty A, Bruix M. Alzheimer's Aβ40 studied by NMR at low pH reveals that sodium 4,4-dimethyl-4-silapentane-1-sulfonate (DSS) binds and promotes β-ball oligomerization. J Biol Chem. 2005;280:3675–3685. doi: 10.1074/jbc.M409507200. [DOI] [PubMed] [Google Scholar]
- 27.Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: 1999. [Google Scholar]
- 28.Wuthrich K, editor. NMR of proteins and nucleic acids. John Wiley and Sons; New York: 1986. [Google Scholar]
- 29.Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 30.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 31.Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 32.Koradi R, Billeter M, Wuthrich K. MOMOL: A program for display and analysis of macromolecular structures. J Mol Graph Model. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 33.Wishart DS, Sykes BD, Richards FM. The chemical shift index - a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 34.Mishra R, Geyer M, Winter R. NMR spectroscopic investigation of early events in IAPP amyloid fibril formation. ChemBioChem. 2009;10:1769–1772. doi: 10.1002/cbic.200900237. [DOI] [PubMed] [Google Scholar]
- 35.Dupuis NF, Wu C, Shea JE, Bowers MT. Human islet amyloid polypeptide monomers form ordered beta-hairpins: a possible direct amyloidogenic precursor. J Am Chem Soc. 2009;131:18283–18292. doi: 10.1021/ja903814q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Padrick SB, Miranker AD. Islet amyloid polypeptide: Identification of long-range contacts and local order on the fibrillogenesis pathway. J Mol Biol. 2001;308:783–794. doi: 10.1006/jmbi.2001.4608. [DOI] [PubMed] [Google Scholar]
- 37.Green JD, Goldsbury C, Kistler J, Cooper GJS, Aebi U. Human amylin oligomer growth and fibril elongation define two distinct phases in amyloid formation. J Biol Chem. 2004;279:12206–12212. doi: 10.1074/jbc.M312452200. [DOI] [PubMed] [Google Scholar]
- 38.Zanni MT, Marek P, Mukherjee S, Raleigh DP. Residue-specific, real-time characterization of lag-phase species and fibril growth during amyloid formation: A combined fluorescence and IR study of p-cyanophenylalanine analogs of islet amyloid polypeptide. J Mol Biol. 2010;400:878–888. doi: 10.1016/j.jmb.2010.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reddy AS, Wang L, Singh S, Ling YL, Buchanan L, Zanni MT, Skinner JL, de Pablo JJ. Stable and metastable states of human amylin in solution. Biophys J. 2010;99:2208–2216. doi: 10.1016/j.bpj.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mu YG, Wei L, Jiang P, Xu WX, Li H, Zhang H, Yan LY, Chan-Park MB, Liu XW, Tang K, Pervushin K. The molecular basis of distinct aggregation pathways of islet amyloid polypeptide. J Biol Chem. 2011;286:6291–6300. doi: 10.1074/jbc.M110.166678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raleigh DP, Shim SH, Gupta R, Ling YL, Strasfeld DB, Zanni MT. Two-dimensional IR spectroscopy and isotope labeling defines the pathway of amyloid formation with residue-specific resolution. Proc Natl Acad Sci U S A. 2009;106:6614–6619. doi: 10.1073/pnas.0805957106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coles M, Bicknell W, Watson AA, Fairlie DP, Craik DJ. Solution structure of amyloid beta-peptide(1-40) in a water-micelle environment. Is the membrane-spanning domain where we think it is? Biochemistry. 1998;37:11064–11077. doi: 10.1021/bi972979f. [DOI] [PubMed] [Google Scholar]
- 43.Motta A, Andreotti G, Amodeo P, Strazzullo G, Morelli MAC. Solution structure of human calcitonin in membrane-mimetic environment: The role of the amphipathic helix. Proteins. 1998;32:314–323. [PubMed] [Google Scholar]
- 44.Motta A, Pastore A, Goud NA, Morelli MAC. Solution conformation of salmon-calcitonin in sodium dodecyl-sulfate micelles as determined by 2-dimensional NMR and distance geometry calculations. Biochemistry. 1991;30:10444–10450. doi: 10.1021/bi00107a012. [DOI] [PubMed] [Google Scholar]
- 45.Ulmer TS, Bax A, Cole NB, Nussbaum RL. Structure and dynamics of micelle-bound human alpha-synuclein. J Biol Chem. 2005;280:9595–9603. doi: 10.1074/jbc.M411805200. [DOI] [PubMed] [Google Scholar]
- 46.Tompa P. Structural disorder in amyloid fibrils: its implication in dynamic interactions of proteins. FEBS J. 2009;276:5406–5415. doi: 10.1111/j.1742-4658.2009.07250.x. [DOI] [PubMed] [Google Scholar]
- 47.Luhrs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Doeli H, Schubert D, Riek R. 3D structure of Alzheimer's amyloid-beta(1-42) fibrils. Proc Natl Acad Sci U S A. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M. Molecular-level secondary structure, polymorphism, and dynamics of full-length alpha-synuclein fibrils studied by solid-state NMR. Proc Natl Acad Sci U S A. 2005;102:15871–15876. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vilar M, Chou HT, Luhrs T, Maji SK, Riek-Loher D, Verel R, Manning G, Stahlberg H, Riek R. The fold of alpha-synuclein fibrils. Proc Natl Acad Sci U S A. 2008;105:8637–8642. doi: 10.1073/pnas.0712179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nanga RPR, Brender JR, Xu JD, Veglia G, Ramamoorthy A. Structures of rat and human islet amyloid polypeptide IAPP(1–19) in micelles by NMR spectroscopy. Biochemistry. 2008;47:12689–12697. doi: 10.1021/bi8014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brender JR, Hartman K, Nanga RP, Popovych N, de la Salud Bea R, Vivekanandan S, Marsh EN, Ramamoorthy A. Role of zinc in human islet amyloid polypeptide aggregation. J Am Chem Soc. 2010;132:8973–8983. doi: 10.1021/ja1007867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hutton JC. The internal pH and membrane-potential of the insulin-secretory granule. Biochem J. 1982;204:171–178. doi: 10.1042/bj2040171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roberts AN, Leighton B, Todd JA, Cockburn D, Schofield PN, Sutton R, Holt S, Boyd Y, Day AJ, Foot EA, et al. Molecular and functional characterization of amylin, a peptide associated with type 2 diabetes mellitus. Proc Natl Acad Sci U S A. 1989;86:9662–9666. doi: 10.1073/pnas.86.24.9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cort JR, Liu Z, Lee GM, Huggins KN, Janes S, Prickett K, Andersen NH. Solution state structures of human pancreatic amylin and pramlintide. Protein Eng Des Sel. 2009;22:497–513. doi: 10.1093/protein/gzp029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nilsson MR, Raleigh DP. Analysis of amylin cleavage products provides new insights into the amyloidogenic region of human amylin. J Mol Biol. 1999;294:1375–1385. doi: 10.1006/jmbi.1999.3286. [DOI] [PubMed] [Google Scholar]
- 56.Koo BW, Hebda JA, Miranker AD. Amide inequivalence in the fibrillar assembly of islet amyloid polypeptide. Protein Eng Des Sel. 2008;21:147–154. doi: 10.1093/protein/gzm076. [DOI] [PubMed] [Google Scholar]
- 57.Fox A, Snollaerts T, Casanova CE, Calciano A, Nogaj LA, Moffet DA. Selection for nonamyloidogenic mutants of islet amyloid polypeptide (IAPP) identifies an extended region for amyloidogenicity. Biochemistry. 2010;49:7783–7789. doi: 10.1021/bi100337p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nanga RPR, Brender JR, Xu JD, Hartman K, Subramanian V, Ramamoorthy A. Three-dimensional structure and orientation of rat islet amyloid polypeptide protein in a membrane environment by solution NMR spectroscopy. J Am Chem Soc. 2009;131:8252–8261. doi: 10.1021/ja9010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Radovan D, Smirnovas V, Winter R. Effect of pressure on islet amyloid polypeptide aggregation: revealing the polymorphic nature of the fibrillation process. Biochemistry. 2008;47:6352–6360. doi: 10.1021/bi800503j. [DOI] [PubMed] [Google Scholar]
- 60.Hall D, Hirota N, Dobson CM. A toy model for predicting the rate of amyloid formation from unfolded protein. J Mol Biol. 2005;351:195–205. doi: 10.1016/j.jmb.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 61.Abedini A, Raleigh DP. A role for helical intermediates in amyloid formation by natively unfolded polypeptides? Phys Biol. 2009;6:15005. doi: 10.1088/1478-3975/6/1/015005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kirkitadze MD, Li H, Condron MM, Zagorski MG, Teplow DB. Formation of an alpha-helix-containing, oligomeric intermediate is an obligatory step in amyloid beta-protein fibrillogenesis. Biophys J. 2001;80:174a–174a. [Google Scholar]
- 63.Fezoui Y, Teplow DB. Kinetic studies of amyloid beta-protein fibril assembly - Differential effects of alpha-helix stabilization. J Biol Chem. 2002;277:36948–36954. doi: 10.1074/jbc.M204168200. [DOI] [PubMed] [Google Scholar]
- 64.Nerelius C, Sandegren A, Sargsyan H, Raunak R, Leijonmarck H, Chatterjee U, Fisahn A, Imarisio S, Lomas DA, Crowther DC, Stromberg R, Johansson J. alpha-Helix targeting reduces amyloid-beta peptide toxicity. Proc Natl Acad Sci U S A. 2009;106:9191–9196. doi: 10.1073/pnas.0810364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saraogi I, Hebda JA, Becerril J, Estroff LA, Miranker AD, Hamilton AD. Synthetic alpha-helix mimetics as agonists and antagonists of islet amyloid polypeptide aggregation. Angew Chem Int Ed Engl. 2009;49:736–739. doi: 10.1002/anie.200901694. [DOI] [PMC free article] [PubMed] [Google Scholar]


