Abstract
Angiotensin-converting enzyme (ACE) is an important zinc-dependent hydrolase responsible for converting the inactive angiotensin I to the vasoconstrictor angiotensin II and for inactivating the vasodilator bradykinin. However, the substrate binding mode of ACE has not been completely understood. In this work, we propose a model for an ACE Michaelis complex based on two known X-ray structures of inhibitor-enzyme complexes. Specifically, the human testis angiotensin-converting enzyme (tACE) complexed with two clinic drugs were first investigated using a combined quantum mechanical and molecular mechanical (QM/MM) approach. The structural parameters obtained from the 550 ps molecular dynamics simulations are in excellent agreement with the X-ray structures, validating the QM/MM approach. Based on these structures, a model for the Michaelis complex was proposed and simulated using the same computational protocol. Implications to ACE catalysis are discussed.
1. Introduction
The angiotensin-converting enzyme (ACE) is a zinc-dependent dipeptidase, which was discovered more than half a century ago.1 This enzyme exhibits an important biological function in regulating the conversion of the biologically inactive angiotensin I to angiotensin II, a powerful vasoconstrictor. It is also involved in the inactivation of bradykinin, a potent vasodilator. The dual functionality is now known to play a key role in the blood pressure regulating renin-angiotensin system (RAS). As a result, ACE is a prominent target for treating hypertension and cardiovascular diseases.2, 3 Although several FDA approved ACE inhibitors are already available for clinic use, the substrate binding and catalysis of ACE are still not completely understood. Interestingly, the first few ACE inhibitors were identified using carboxypeptidase A (CPA) or thermolysin (TLN) as a model,4, 5 which was believed to have a similar active-site architecture. It is only recently that the three-dimensional structure of ACE was determined via X-ray diffraction.6 While confirming the similarities of the active sites among these enzymes, ACE was shown to have a vastly different overall fold from CPA and TLN.
Two types of ACE are known. The human somatic angiotensin-converting enzyme (sACE) has two domains, namely the N domain and the C domain. They have about 55% sequence similarity,7 but both domains contain the same zinc binding motif, HEXXH, and a downstream E residue.8 This phenomenon is thought to be a result of gene duplication. The C domain was found to be the dominant angiotensin-converting site in controlling blood pressure and cardiovascular functions, based on the observation that the inhibition of the N domain has little effect on these functions.9 Another form of ACE is found in testis, which plays a role in fertilization. The testicular ACE (tACE) shows an identical active site with the C domain of sACE, but has no N domain.10 Recently, three-dimensional structures of various ACEs have been determined.6, 11–16
An interesting structural characteristic of ACE is the presence of two chloride ions outside the active site. Experiments indicated that they are essential to maintain the binding structure and catalytic activity.17–19 Based on X-ray structures of ACE, the Cl− ion at the first binding position (I) is about 21 Å away from the zinc ion. It is in hydrogen bonding distances with Arg186 and Arg489, and exhibits van der Waals interactions with a shell formed by Trp485, Trp486 side chain groups and the Asp507 backbone. These interactions are thought to be very important for the stabilization of the enzyme-substrate complex.20 The chloride ion at the second binding position (II) is about 10 Å away from the zinc ion and is in hydrogen bonding distances with Arg522, Tyr224, and a water. In addition, a hydrophobic shell formed by residues of Pro407, Pro519, and Ile521 was found to surround Cl−(II). Kinetic experiments suggested that Cl−(II) is critical for enzyme catalysis.17, 19 In our simulations, both anions are included in the model, but this work will not focus on the roles played by these two important anions.
The search for effective ACE inhibitors has a long history.3, 21, 22 It is remarkable that the first inhibitor (captopril) was discovered serendipitously using CPA as a model,23 without the structure of ACE. Subsequently, other potent inhibitors of ACE were reported,5, 24–26 again without knowledge of its structure. Very recently, several ACE structures in complex with inhibitors have been reported, as shown in Table I along with the corresponding inhibition constants. The availability of ACE structures opened the door for the rational design of new and improved ACE inhibitors.27, 28 The early stage inhibitors of ACE had less specificity to the C or N domains, thus exhibiting adverse side effects. More recently, there is a keen interest in developing new inhibitors with high specificity to one of two domains.27, 29 Some success, e.g., RXP407 to the N domain,25 and RXPA380 to the C domain,26 have been reported. The emergence of domain-selective inhibitors calls for more studies of the substrate or inhibitor binding mode to different domains, as a recent study showed that the basis of domain-dependent inhibition of ACEs might come from interactions between bulky hydrophobic side chain moieties and the domain-specific hydrophobic residues.15
Table 1.
List of X-ray structures of tACE complexed with various inhibitors
| PDB code | Inhibitors | Ki(µM) | Ref. |
|---|---|---|---|
| 2OC2 | RXPA380 | 0.003a | 14 |
| 1O86 | lisinopril | 0.00027b | 6 |
| 1UZF | captopril | 0.00111b | 11 |
| 1UZE | enalaprilat | 0.00078b | 11 |
| 3BKK | kAF | 0.83a | 15 |
| 3BKL | kAW | 0.679a | 15 |
200mM Cl−
20mM Cl−
The widespread use of ACE inhibitors in clinic settings underscores the importance for understanding the binding and catalytic modes of the enzyme at the microscopic level. Experimental studies alone are often insufficient to answer all questions and computational models can offer a complementary perspective. Despite the rapid accumulation of structural and kinetic data, however, there have been few computational studies on this important system. An earlier density functional theory study focused on a truncated active-site model,30 while more recent ones investigated docking of various small molecule inhibitors to ACE.31, 32 These studies provided no dynamical information concerning the fluctuation of the enzymatic system. A recent molecular dynamics study on an enzyme-substrate complex has been reported by Papakyriakou et al.,33 using a modified Amber force field. To avoid the well-known problems associated with a force-field description of the zinc-ligand bonds as electrostatic interactions,34, 35 a bonded approach36 was used in which an artificial force field for the zinc-ligand interactions was determined based on ab initio calculations.
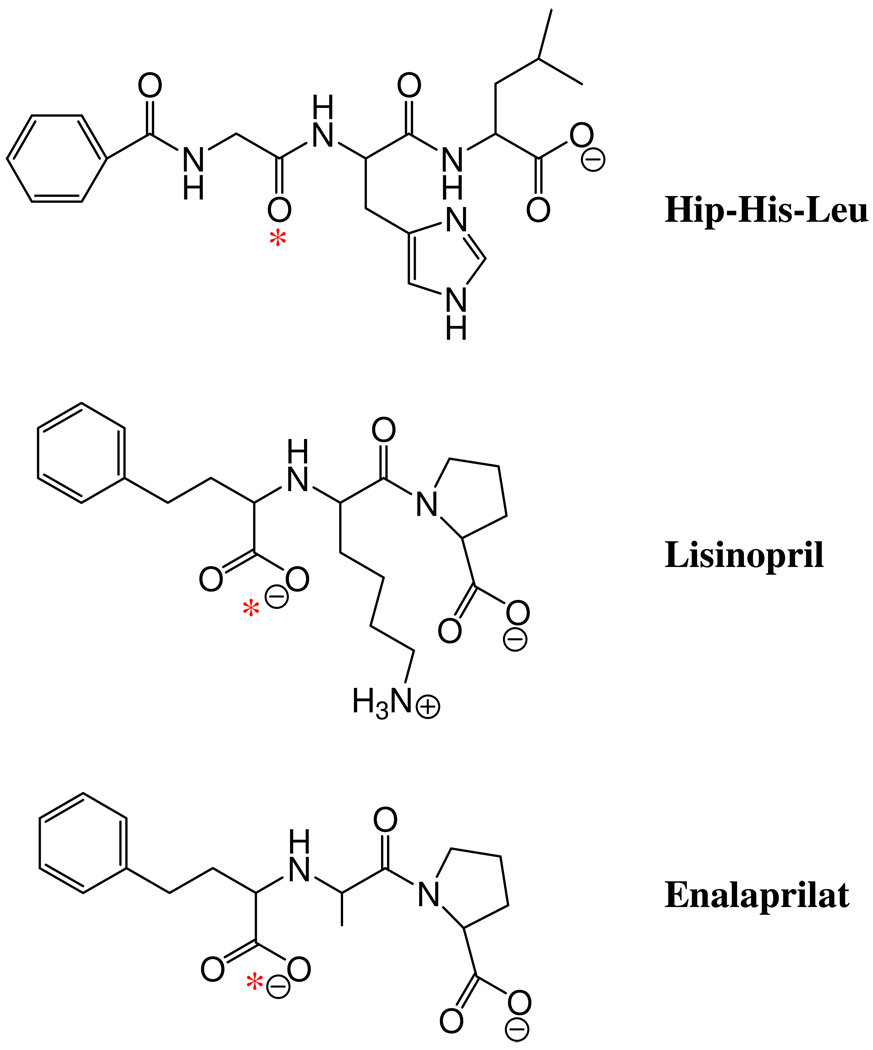
In this work, we attempt to establish the structure of the Michaelis complex for ACE using a quantum mechanical/molecular mechanical approach. Our simulations are based on the known structures of ACE in complex with two inhibitors: enalaprilat and lisinopril.6, 11 These two inhibitors are effective ACE inhibitors that have been extensively used in blood-pressure control therapy.22 The backbones of these inhibitors are very close to a natural substrate of ACE, Hip-His-Leu, as shown in Scheme 1. Such a structural similarity allowed us to propose a plausible substrate binding mode based on the simulation of inhibitor-enzyme complexes. We further constrain our model to conform with several known determinants in CPA and TLN catalysis concerned with the interaction with the zinc cofactor.
Scheme 1.
Structural comparison of Hip-His-Leu, lisinopril, and cnalaprilat, where the Zn2+ binding is indicated by red asterisks.
2. Method and Protocol
2A. Quantum mechanical/molecular mechanical method
The computational approach used to describe the active-site dynamics is a combined quantum mechanical and molecular mechanical (QM/MM) method,37–39 which divides the system into two parts. A smaller QM region consists of those residues related to reaction, while the surrounding MM region contains most of the environment residues and solvent. The MM region is represented by the all-atom CHARMM force field,40 and TIP3P water.41 The QM-MM boundary is treated with the link-atom method.38 The selection of the method to treat the QM region is critical for the computational efficiency and accuracy. In this work, we will use the self-consistent charge-density functional tight binding (SCC-DFTB) method42–44 to perform the electronic structure calculations for the QM region. The SCC-DFTB parameters for the biological zinc ion have been developed,45 and the SCC-DFTB/CHARMM approach has been successfully validated in several zinc enzymes, including carbonic anhydrase,46, 47 carboxypeptidase A,48, 49 and metallo-β-lactamases.50–55 Since ACE has a similar active-site construct, this QM/MM scheme should be able to characterize the enzyme well.
2B. Enzyme-inhibitor complex models
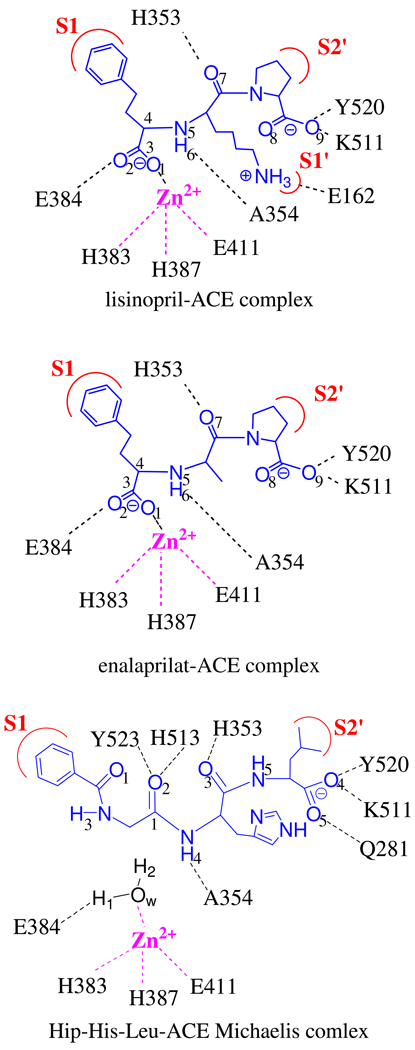
The initial structures for two enzyme-inhibitor complexes were obtained from the Protein Data Bank (PDB codes: 1O86,6 and 1UZE11). For both models, the sole zinc ion is bound with the side chains of His383, His387, and Glu411. The fourth ligand of the zinc ion is provided by the carboxylate group of the inhibitors. In our models, the disulfide bonds between Cys152 and Cys158, Cys352 and Cys370, and between Cys538 and Cys550 were enforced. The HBUILD module of CHARMM was used to assign hydrogen atoms to residues. Special attention was paid for the ionization states of key titratable groups. For example, Glu384 in the active site was assigned to be protonated, judging by the distance between its carboxylate oxygen and the inhibitor carboxylate group as shown in Figure 1. In addition, the side chain of the lysine residue of the lisinopril molecule was treated in the ionized form since the X-ray structure was obtained at pH=4.7.6
Figure 1.
Atom definition and corresponding interactions between inhibitors/substrate and active-site residues of tACE.
The systems were then solvated with a pre-equilibrated TIP3P water41 sphere of a 25 Å radius centered at the zinc ion, followed by 30 ps molecular dynamics (MD) simulation with all of enzyme residues, inhibitors, zinc and Cl− ions fixed. This process was performed several times with randomly rotated water spheres to ensure uniformly solvation. Subsequently, stochastic boundary conditions56 were applied to reduce the computational costs. In particular, those atoms that are 25 Å away from the origin were removed, while atoms in the buffer zone (22 Å < r < 25 Å) were subjected to Langevin dynamics with friction and random forces. In the inner reaction zone (r < 22 Å), the atoms follow Newtonian dynamics on the hybrid QM/MM potential energy hypersurface. A group-based switching scheme was used for non-bonded interactions.57
In this work, the QM region includes the metal ion, the inhibitor molecule, and the side chain groups of His383, His387, Glu411, and Glu384 residues. The labeling of active-site atoms is provided in Fig. 1. Since the Cl− ions are outside the active site, they were simulated in the MM region. For both models, a total 550 ps MD simulation was performed with an integration time step of 1.0 fs. The temperature was slowly heated to 300 K in 30 ps, and another 70 ps MD was allowed for further equilibration. The subsequent 450 ps MD trajectory was used for the data analysis. The SHAKE algorithm58 was applied to restrain the covalent bonds involving hydrogen atoms.
2C. Michaelis Complex
The MD simulations of the enzyme-inhibitor complexes provide a good starting point to postulate the binding mode for a bona fide substrate of ACE. In this work, a natural substrate molecule, hippuryl-L-histidyl-L-leucine (Hip-His-Leu),59 was employed in the simulation of the Michaelis complex. Our binding model differs significantly from the suggestion made by Sturrock et al.,7 with a tripeptide molecule, Phe-His-Leu. In their model, the fourth ligand of the zinc ion was postulated to be the carbonyl oxygen of the scissile bond in the substrate and a non-zinc bound water molecule is present in the active site with a hydrogen bond with Glu384. However, such an arrangement is at odds with that in the active sites of the extensively studied CPA and TLN, which are believed to have the same catalytic mechanism as ACE. In an earlier study on TLN, Hangauer et al. suggested that the fourth ligand of the zinc ion is a water molecule which serves as the nucleophile when activated by a general base (Glu143).60 Such a mechanism has recently been confirmed computationally by Blumberger et al.61 A similar binding model and mechanism were proposed for CPA,62 and recently verified with a QM/MM model.48
Our model of the Michaelis complex is based on the premise that the fundamental binding determinants of ACE should be similar to TLN and CPA. In other words, the fourth ligand of the zinc ion is the water nucleophile hydrogen bonded with the general base (Glu384) and the backbone carbonyl oxygen only interacts weakly with the metal ion. This model is reasonable, given the small (0.52 Å) RMSD between the active sites of ACE and TLN.6 It is believed that the ligand-free ACE has a water molecule bound to the zinc ion as its fourth ligand.6 Since the metal binding site of both inhibitors (labeled by red asterisks in Scheme 1) consists of a negatively charged carboxylate group, it is not difficult to image the water to be displaced by the anionic group which is known to interact strongly with the zinc ion. On the other hand, the carboxylate group is replaced by a neutral carbonyl oxygen in the substrate, which generally does not interact strongly with the zinc ion.15 Thus, when the substrate enters the binding site, it is unlikely to dislodge the zinc-bound water, as in the case of CPA and TLN.
The corresponding atom definitions and interactions between the substrate and active-site residues are given in Fig. 1. To build the model for the Michaelis complex, we first removed the lisinopril molecular from the active site of tACE (PDB code 1O86).6 The Hip-His-Leu substrate was then manually docked in the active site in the appropriate orientation. This approach is similar to the work of Papakyriakou et al.,33 but with a QM/MM description of the system. The subsequent setup protocol is essentially the same as that for the enzyme-inhibitor models. Specifically, the QM system includes the zinc ion, side chain groups of its three protein ligands, the entire substrate, the water nucleophile, and the deprotonated carboxylate of the general base (Glu384). A total of one nanosecond MD simulation was carried out for the substrate-enzyme complex. The first 300 ps were used to sufficiently relax the whole system, while the subsequent trajectory of 700 ps was employed for data analysis.
3. Results
3A. Dynamics Enzyme-inhibitor Complexes
Active site binding pattern
The binding modes for the two inhibitors based on the X-ray structures are displayed in Fig. 1. Both enalaprilat and lisinopril are bound with the enzyme through a direct contact with the metal ion via the C4-carboxylate group. The so-called carboxylate class of ACE inhibitors differs from the sulfhydryls such as captopril, which binds the enzyme via its sulfide as the fourth ligand of the zinc ion.29 No other covalent bonds between inhibitors and enzyme residues are present in the crystal structures or in our models.
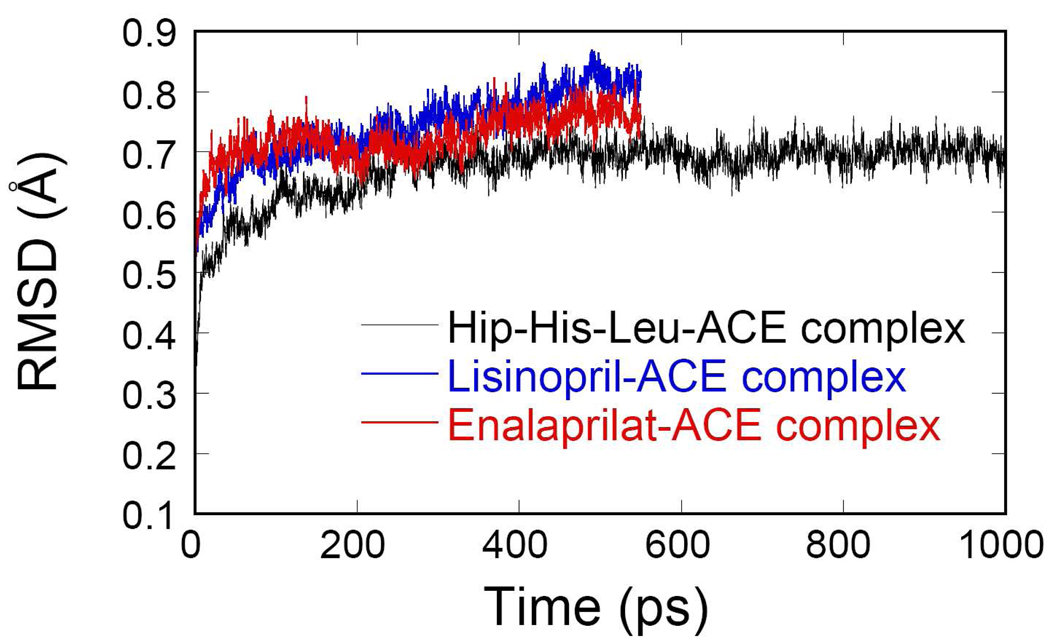
To understand the detailed dynamic features of the inhibitor binding, we performed QM/MM MD simulations for two enzyme-inhibitor complexes. Both structures are quite stable during the MD simulations. As Fig. 2 shows, the root mean square deviations (RMSDs) for the backbone atoms are 0.77±0.06Å for the lisinopril-ACE complex, and 0.76±0.04Å for the enalaprilat-ACE complex, respectively. Selected internuclear distances averaged over the trajectories are listed in Table 2 and Table 3, and two snapshots for the inhibitor-enzyme complex are displayed in Fig. 3.
Figure 2.
RMSDs for both enzyme-inhibitor complexes and enzyme-substrate complex as a function of time.
Table 2.
Key geometric parameters at the active site of the lisinopril-ACE complex
| Bond length (Å) and bond angle (deg) |
QM/MM MD | X-ray6 |
|---|---|---|
| Zn···Nε2 (H383) | 2.02±0.06 | 2.04 |
| Zn···Nε2 (H387) | 1.98±0.06 | 2.07 |
| Zn···Oε1(E411) | 2.06±0.07 | 1.99 |
| Zn···O1 | 2.09±0.14 | 2.14 |
| O2···Hε2(E384) | 1.78±0.15 | 2.69 |
| O9···Hζ1(K511) | 1.59±0.09 | 2.93a |
| O9···Hη(Y520) | 1.70±0.13 | 2.55b |
| O7···Hε2(H353) | 1.97±0.25 | 2.76a |
| O1···Hη(Y523) | 1.86±0.24 | 2.77b |
| H6···O(A354) | 2.36±0.59 | 2.92 |
| Oε2(E162)···Hζ(Lys) | 1.81±0.30e | 3.45 |
| Cl···Zn | 10.14±0.29 | 10.37 |
| Cl···Hε(R522) | 1.95±0.14 | 3.08c |
| Cl···H22(R522) | 1.93±0.15 | 3.55c |
| Cl···Hη(Y224) | 1.85±0.35 | 3.01d |
| Nε2(H383)···Zn···O1 | 110.4±8.1 | 116.8 |
| Nε2(H387)···Zn···O1 | 125.2±7.8 | 123.0 |
| Oε1(E411)···Zn···O1 | 104.2±8.3 | 98.8 |
distance between O and O atoms.
distance between O and N atoms.
distance between Cl and N atoms.
distance between Cl and O atoms.
averaged distance between Oε2 and the Hζ involved in hydrogen bonding.
Table 3.
Key geometric parameters at the active site in the enalaprilat-ACE complex
| Bond length (Å) and bond angle (deg) |
QM/MM MD | X-ray11 |
|---|---|---|
| Zn···Nε2(H383) | 1.99±0.06 | 2.10 |
| Zn···Nε2(H387) | 1.98±0.06 | 2.05 |
| Zn···Oε1(E411) | 2.04±0.07 | 1.89 |
| Zn···O1 | 2.08±0.08 | 2.01 |
| O2···Hε2(E384) | 1.82±0.16 | 2.71a |
| O9···Hζ1(K511) | 1.65±0.14 | 2.84b |
| O9···Hη(Y520) | 1.71±0.20 | 2.61a |
| O7···Hε2(H353) | 1.95±0.25 | 2.68b |
| O1···Hη(Y523) | 1.72±0.14 | 2.82a |
| H6···O(A354) | 1.94±0.24 | 2.96b |
| Cl···Zn | 10.11±0.31 | 10.40 |
| Cl···Hε(R522) | 1.89±0.12 | 3.15c |
| Cl···H22(R522) | 1.96±0.16 | 3.53c |
| Cl···Hη(Y224) | 1.85±0.31 | 3.09d |
| Nε2(H383)···Zn···O1 | 110.7±7.6 | 116.5 |
| Nε2(H387)···Zn···O1 | 118.9±7.8 | 125.2 |
| Oε1(E411)···Zn···O1 | 103.5±7.3 | 103.6 |
distance between O-O atoms.
distance between O-N atoms.
distance between Cl and N atoms.
distance between Cl and O atoms.
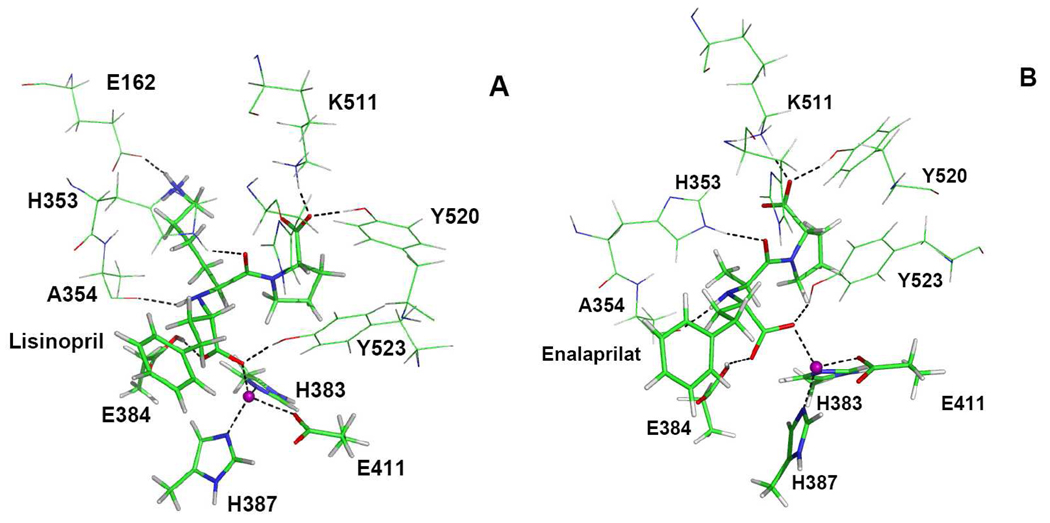
Figure 3.
Snapshots of the ACE active site bound to lisinopril (A) and enalaprilat (B), respectively. The zinc ion is color coded purple and the dash lines are for hydrogen bonds and ligand bonds to the zinc ion.
In agreement with the crystal structures, the tetra-coordination for the zinc ion was maintained for both inhibitors throughout our simulations. The zinc ligands consist of His383, His387, Glu411, and the C4 carboxylate oxygen (O1) of lisinopril or enalaprilat. The O1-Zn distance is 2.09±0.14 Å and 2.08±0.08 Å for lisinopril and enalaprilat, respectively, which can be compared with the X-ray values of 2.14 Å and 2.01 Å. As discussed below, O1 is also hydrogen bonded with the side chain of Try523.
For the two inhibitors studied in this work, the binding patterns are quite similar. As shown in Fig. 1 and Fig. 3, the lysine residue of lisinopril interacts strongly with the Glu162 residue at the S1’ subsite of the enzyme, although the three hydrogen atoms of the lysine switch their positions during the MD simulation. On the other hand, the methyl group of enalaprilat makes no such contact, which could be the reason why lisinopril has a stronger binding affinity than enalaprilat. At the S1 site, the phenyl group of the inhibitors is surrounded by two hydrophobic residues, Phe512 and Val518, and two water molecules. On the other hand, the C-terminal proline residue of the inhibitor is stabilized in the S2’ subsite of ACE by hydrogen bonds to its carboxylate oxygens. In particular, O9 of the proline terminal carboxylate group is hydrogen bonded with Lys511 and Tyr520, with the hydrogen bond distances of 1.59±0.09 Å and 1.70±0.13 Å in lisinopril, 1.65±0.14 Å and 1.71±0.20 Å in enalaprilat. The other carboxylate oxygen (O8) forms hydrogen bonds with solvent water molecules.
Both inhibitors are further stabilized by additional hydrogen bonds in the active site. For example, the central carboxylate oxygens (O1 and O2) of the inhibitor molecules form strong hydrogen bonds with the Glu384 and Tyr523 side chains. For lisinopril, the O2-Hε2(Glu384) and O1-Hη(Tyr523) distances are 1.82±0.16 Å and 1.72±0.14 Å, respectively. These distances for enalaprilat are 1.78±0.15 Å and 1.86±0.24 Å, respectively. In addition, the backbone amide NH group of the inhibitor forms a hydrogen bond with the backbone carbonyl of Ala354 with a distance of 2.36±0.59Å for lisinopril and 1.94±0.24 Å for enalaprilat. Furthermore, the backbone carbonyl oxygen (O7) is hydrogen bonded with His353, evidenced by the O7-Hε2(His353) distances of 1.97±0.25 Å for lisinopril and 1.95±0.25 Å for enalaprilat, respectively.
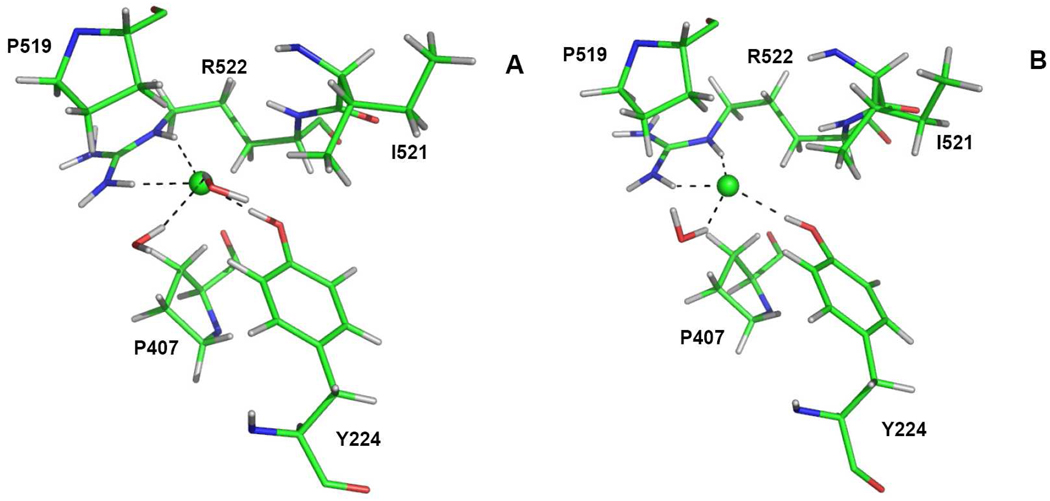
The Cl−(II) ion has strong electrostatic interactions with side chains of both Arg522 and Tyr224. The distances from Hε and H22 atom of Arg522 to Cl− are typically less than 2.0 Å, while the distance between Hη atom of Tyr224 and Cl− is around 1.85 Å for both inhibitors. Additionally, two solvent water molecules are nearby, forming two hydrogen bonds with Cl−(II). Besides hydrogen bonds from enzyme residues, a hydrophobic shell formed by Pro407, Pro519 and Ile521 may contribute as well. Snapshots of the chloride site for both inhibitor-enzyme complexes are displayed in Fig. 4.
Figure 4.
Snapshots for the Cl−(II) site for the lisinopril-tACE (A) and enalaprilat-tACE (B) complexes. The chloride ion is color coded green while the dash lines represent the hydrogen bonds to the Cl− ion.
3B. Dynamics of Michaelis Complex
The Hip-His-Leu substrate used here to construct the Michaelis complex of ACE has been used by Cushman et al. as a template to design inhibitors to ACE.23 As shown in Scheme 1, it has a backbone structure similar to those of lisinopril and enalaprilat. As a result, the inhibitor-enzyme complexes discussed above provide a good starting point for constructing the Michaelis complex of ACE. As in the inhibitor-enzyme complexes, the Michaelis complex is quite stable, evidenced by the calculated RMSD of 0.66±0.04 Å for the 1 ns MD simulation. The selected key geometric parameters are listed in Table 4.
Table 4.
Selected geometric parameters of the Michaelis complex obtained from the QM/MM MD simulations using the SCCDFTB/MM method.
| Distance (Å) Angle (Deg.) |
QM/MM MD |
|---|---|
| Zn···Ow | 2.04±0.06 |
| Zn···O2 | 4.51±0.36 |
| Zn···Nε2 (H387) | 2.01±0.06 |
| Zn···Nε2 (H383) | 2.00±0.05 |
| Zn···Oε1 (E411) | 2.05±0.07 |
| H1···Oε2 (E384) | 1.34±0.14 |
| O2···Hε2 (H513) | 2.01±0.36 |
| O2···Hη(Y523) | 1.98±0.25 |
| C1···Ow | 3.19±0.26 |
| O3···Hε2 (H353) | 1.89±0.17 |
| O4···Hη(Y520) | 1.69±0.11 |
| O4···Hζ1(K511) | 1.71±0.15 |
| O5···H21(Q281) | 2.02±0.47 |
| H4···O(A354) | 2.02±0.19 |
| Cl···Zn | 10.33±0.26 |
| Cl···Hε(R522) | 1.94±0.14 |
| Cl···Hη(Y224) | 1.83±0.11 |
| Cl···H22(R522) | 2.00±0.21 |
| Nε2(H383)···Zn···Ow | 100.9±5.4 |
| Nε2(H387)···Zn···Ow | 105.1±6.2 |
| Oε1(E411)···Zn···Ow | 110.1±7.6 |
Perhaps the most important feature in our model for the Michaelis complex is that the substrate has no direct contact with the zinc ion, in accordance with those observed in the QM/MM simulations of CPA49 and TLN.61 In particular, the O2-Zn distance is 4.51±0.36 Å. Instead, the zinc ion is tetra-coordinated by three protein ligands and a water molecule, which is 2.04±0.06Å from the zinc ion. The tetra-coordination was kept very well throughout the simulation. In addition to the zinc coordination, this water molecule is also hydrogen bonded strongly with the Glu384 carboxylate group, with an H-O distance of 1.34±0.14 Å. Like in CPA and TLN, the Glu384 residue is expected to participate in the catalysis as the general base, which activates the zinc-bound water via proton transfer. Indeed, the putative water nucleophile is ideally located in a near-attack configuration, with a Ow-C1 distance of 3.19±0.26 Å.
Similar to the lisinopril and enalaprilat inhibitors, the Hip-His-Leu substrate also has a benzene group at the S1 site, which is accommodated by a hydrophobic pocket formed by Phe512 and Val518. In addition, it also has direct contact with some solvent molecules, which suggests that the hydrolysis product can be readily released once the amide bond is cleaved. Similarly, the dimethyl group of the substrate occupies the S2’ site formed by Thr282, Phe457 and Phe527, replacing the proline residue in the inhibitors. However, the S1’ site is not occupied as the imidazole group of the substrate is much shorter than the lysyl group in lisinopril.
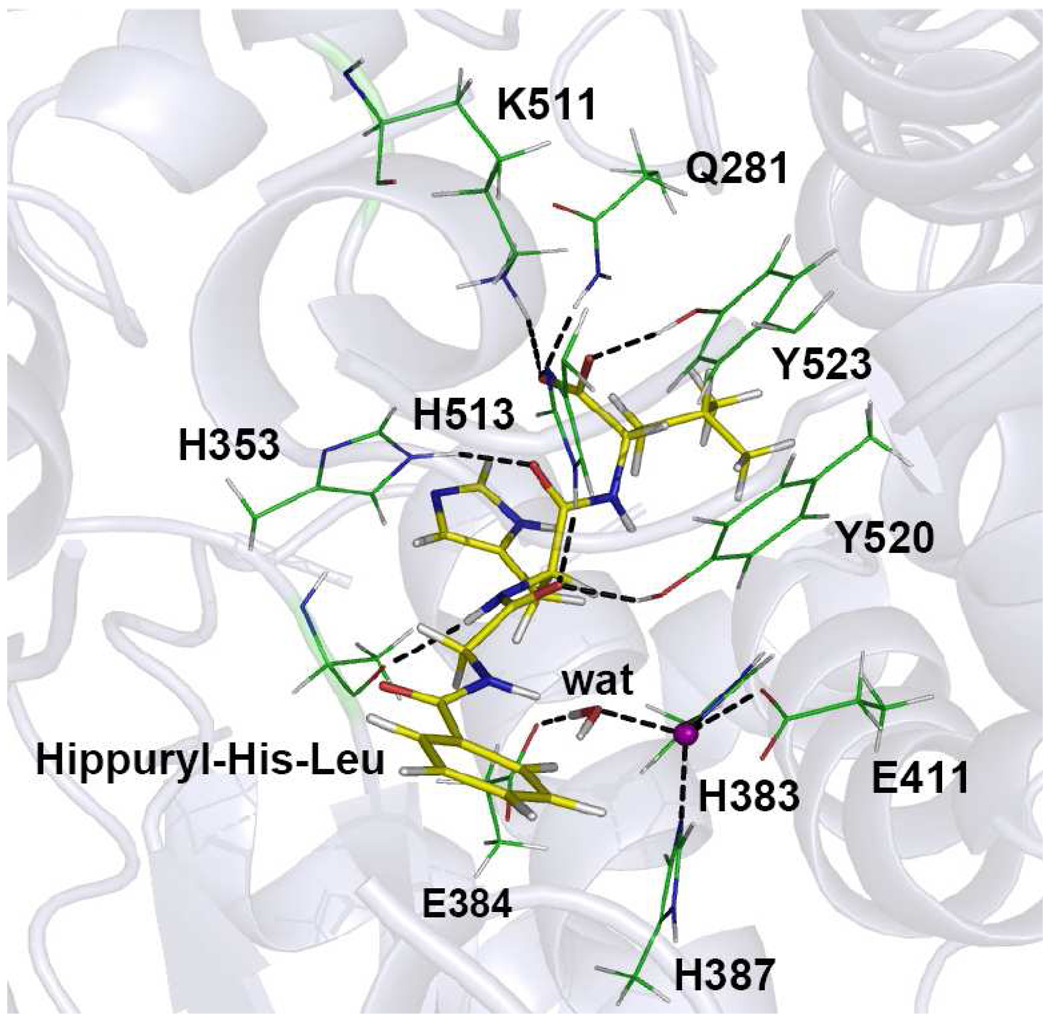
The hydrogen-bond network between the substrate and enzyme active site is quite similar to those observed in the inhibitor-enzyme complexes. For example, the C-terminal carboxylate of substrate is stabilized by hydrogen bonds with Tyr520, Lys511, and Gln281, with the O4···Hη(Tyr520), O4···Hζ1(Lys511), and O5···H21(Gln281) distances of 1.69±0.11 Å, 1.71±0.15 Å, and 2.02±0.47 Å, respectively. In addition, the Ala354 backbone oxygen is also hydrogen bonded to a peptide NH group of the substrate with an O-H4 distance of 2.02±0.19 Å. Furthermore, the substrate backbone carbonyl oxygens are hydrogen bonded with His353, His513, and Tyr523, with the hydrogen bond distances of 1.89±0.17 Å for O3-Hε2(His353), 2.01±0.36 Å for O2-Hε2(His513), and 1.98±0.25 Å for O2-Hη(Tyr523). A snapshot of the active site is displayed in the Fig. 5.
Figure 5.
Snapshot for the Michaelis complex of ACE. The Hip-His-Leu substrate is color coded yellow while the zinc ion purple.
A similar binding pattern around the Cl−(II) was found for the Michaelis complex. The chloride ion is tightly held by Tyr224 and Arg522, as well as several solvent water molecules. The distance between Cl−(II) and zinc ion is 10.33±0.26 Å, which is quite close to that in lisinopril or enalaprilat.
4. Discussion
ACE has two functional domains, the C and N domains. In this work, QM/MM MD was used to simulate the binding modes of two clinical inhibitors and a natural substrate molecule, Hip-His-Leu, of the testis ACE (tACE), an isoform of the human sACE C-domain. The structure of the substrate-enzyme complex is a pre-requisite for a better understanding of the catalysis of ACE. Due to its transient nature, it is very difficult, if not impossible, to obtain structural information of the Michaelis complex. Hence, reliable theoretical models become an important alternative to gain insight into the binding mode and catalysis. In this work, we report a plausible model for the ACE Michaelis complex using a QM/MM simulation method validated by reproducing the structural features of the inhibitor-enzyme complexes observed in X-ray diffraction experiments.
Our model suggests that the substrate in the Michaelis complex is not in direct contact with the zinc ion and the nucleophilic water is the fourth ligand of the metal ion. This model is consistent with the consensus catalytic action of both CPA62 and TLN,63 confirmed by recent QM/MM studies.49, 61 Our model assigns the zinc-bound water as the nucleophile and Glu384 as the general base. We further propose that the catalysis is initiated nucleophilic attack at the scissile carbonyl carbon by the nucleophile, assisted by proton transfer to Glu384. The resulting tetrahedral intermediate is stabilized by an oxyanion hole provide by the zinc ion and presumably Tyr523 and His513. The elimination step of the reaction cleaves the C-N bond with the protonation of the leaving group nitrogen by the general acid Glu384. This putative catalytic mechanism has been demonstrated computationally for TLN61 and CPA,49 and the resulting barriers are consistent with kinetic data for these two enzymes. Further QM/MM simulations of the catalytic mechanism of ACE will test this mechanism proposal. Work in this direction is underway in our laboratories.
In an earlier publication, Sturrock et al. has proposed a different model for the Michaelis complex of ACE,7 in which the fourth ligand of the zinc ion is the carbonyl oxygen of the scissile C-N bond, and the water is not zinc bound, but hydrogen bond with the general base (Glu384). This model was an empirical one, based on the inhibitor-enzyme structures alone. No atomistic interactions were included. Subsequently, a computational model was established for the complex between ACE and the gonadotropin-releasing hormone (GnRH), a natural substrate of ACE.33 The MD simulations employed the Amber all-atom force field.64 However, a force field description of the metal-ligand bonds as electrostatic interaction is known to be problematic,34, 35 a bonded model36 was used for the zinc ion and its ligands, in which the metal-ligand bonds were treated as covalent interactions. As can be expected, such a model has large ambiguities in devising and parameterizing the force field. Indeed, the zinc ion was modeled by Papakyriakou et al.33 as a penta-coordinated species based on gas phase density functional theory calculations, in which both the nucleophilic water and substrate carbonyl oxygen are zinc ligands. As a result, the MD simulations are restricted to a penta-coordinated zinc configuration. Although penta-coordinated zinc ions are known to exist in enzymes,65 accurate ab initio QM/MM simulations of TLN61 has clearly demonstrated that the zinc ion is tetra-coordinated in its active site that is essentially identical to ACE. It may thus be argued that the penta-coordination of the zinc ion in the truncated active-site model of Papakyriakou et al. might be an artifact, attributable to the lack of the enzyme environment. Interestingly, we note that the Amber based classical MD simulations performed by Blumberger et al.61 on TLN resulted in a stable penta-coordinated zinc cofactor, but the carbonyl oxygen was “expelled from the first coordinate shell” in the first picosecond of the QM/MM simulation.
As discussed above, the two chloride ions play an important role in the binding and catalysis of ACE. Although this study is not aimed at the elucidation of the role of the anions, we would like to comment on some differences between this work and the earlier simulation of the ACE-GnRH complex.33 In their MD simulations, Papakyriakou et al. found a direct substrate-chloride interaction, thanks to the long peptide substrate (GnRH) used in their simulations, which covers the entire substrate binding crevice of ACE. No such interaction was observed in our simulations, presumably due to the short substrate in our model. Interestingly, we note that some experimental evidence suggested that the absence of Cl−(II) in the ACE homologue from Drosophila melanogaster, AnCE, does not seem to affect the binding of small molecule inhibitors, such as lisnopril and enalaprilat.16, 66 So it is possible that its effect is more pronounced for long chain substrates.
To elucidate the structural role of chloride ions in ACE, it might be desirable to perform longer time MD simulations, with and without the chloride ions. After all, the timescale for protein conformational changes is much longer than that used in the current QM/MM MD simulations.
5. Conclusions
In this work, we investigated the binding patterns of two well established inhibitors of ACE using a combined QM/MM MD method. The calculated structures using the SCC-DFTB/CHARMM method are in good agreement with those obtained from X-ray diffraction. Based on the inhibitor-enzyme complexes, we propose a plausible model for the Michaelis complex of ACE with the Hip-His-Leu substrate, in which the nucleophilic water is a zinc ligand. QM/MM simulations of this complex yielded useful information that allowed us to propose a catalytic mechanism: The hydrolysis catalyzed by ACE is initiated by the nucleophilic attack of the zinc-bound water at the scissile carbonyl carbon of the substrate, assisted by the Glu384 general base. This is followed by the cleavage of the C-N bond via the elimination of the nitrogen leaving group, again assisted by the Glu384 as the general acid. The binding mode and catalytic mechanism of ACE are thus similar to that of thermolysin and carboxypeptidase A.
Acknowledgements
The authors would thank National Natural Science Foundation of China (Grant No. 20803048 and 21073125) to D. Xu, National Natural Science Foundation of China (Grant No.20725312 and 91021010) and Chinese Ministry of Science and Technology (2007CB815201) to D. Xie, and National Institutes of Health (R03-AI071992) to H. Guo for the financial support.
References
- 1.Skeggs LTJ, Kahn JR, Shuway NP. The preparation and function of the hypertension-converting enzyme. J. Exp. Med. 1956;103:295–299. doi: 10.1084/jem.103.3.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown NJ, Vaughan DE. Angiotensin-converting enzyme inhibitors. Circulation. 1998;97:1411–1420. doi: 10.1161/01.cir.97.14.1411. [DOI] [PubMed] [Google Scholar]
- 3.Zaman MA, Oparil S, Calhoun DA. Drugs targeting the renin-angiotensin-aldosterone system. Nat. Rev. Drug Discov. 2002;1:621–636. doi: 10.1038/nrd873. [DOI] [PubMed] [Google Scholar]
- 4.Ondetti MA, Rubin B, Cushman DW. Design of specific inhibitors of angiotensin-converting enzyme: new class of orally antihypertensive agents. Science. 1977;196:441–444. doi: 10.1126/science.191908. [DOI] [PubMed] [Google Scholar]
- 5.Patchett A, Harris E, Tristram E, Wyvratt M, Wu M, Taub D, Peterson E, Ikeler T, Ten Broeke J, Payne L, Ondeyka D, Thorsett E, Greenlee W, Lohr N, Hoffsommer R, Joshua H, Ruyle W, Rothrock J, Aster S, Maycock A, Robinson F, Hirschmann R, Sweet C, Ulm E, Gross D, Vassil T, Stone C. A new class of angiotensin-converting enzyme inhibitors. Nature. 1980;288:280–283. doi: 10.1038/288280a0. [DOI] [PubMed] [Google Scholar]
- 6.Natesh R, Schwager SLU, Sturrock ED, Acharya KR. Crystal structure of the human angiotensin-converting enzyme-lisinopril complex. Nature. 2003;421:551–554. doi: 10.1038/nature01370. [DOI] [PubMed] [Google Scholar]
- 7.Sturrock ED, Natesh R, van Rooyen JM, Acharya KR. Structure of Angiotensin I-Converting Enzyme. Cell. Mol. Life Sci. 2004;61:2677–2686. doi: 10.1007/s00018-004-4239-0. [DOI] [PubMed] [Google Scholar]
- 8.Soubrier F, Alhenc-Gelas F, Huber C, Allegrini J, John M, Tregear G, Corvol P. Two putative active centers in human angiotensin I-converting enzyme revealed by molecular cloning. Proc. Natl. Acad. Sci. USA. 1988;85:9386–9390. doi: 10.1073/pnas.85.24.9386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Junot C, Gonzales MF, Ezan E, Cotton J, Vazeux G, Michaud A, Azizi M, Vassiliou S, Yiotakis A, Corvol P, Dive V. RXP 407, a selective inhibitor of the N-domain of angiotensin I-converting enzyme, blocks in vivo the degradation of hemoregulatory peptide acetyl-Ser-Asp-Lys-Pro with no effect on angiotensin I hydrolysis. J. Pharmacol. Exp. Ther. 2001;297(2):606–611. [PubMed] [Google Scholar]
- 10.Ehlers MRW, Fox EA, Strydom DJ, Riordan JF. Molecular cloning of human testicular angiotensin-converting enzyme: the testis isozyme is identical to the C-terminal half of endothelial angiotensin-converting enzyme. Proc. Natl. Acad. Sci. U.S.A. 1989;86:7741–7745. doi: 10.1073/pnas.86.20.7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Natesh R, Schwager SLU, Evans HR, Sturrock ED, Acharya KR. Structural Details on the Binding of Antihypertensive Drugs Captopril and Enalaprilat to Human Testicular Angiotensin I-Converting Enzyme. Biochem. 2004;43:8718–8724. doi: 10.1021/bi049480n. [DOI] [PubMed] [Google Scholar]
- 12.Watermeyer JM, Sewell BT, Schwager SL, Natesh R, Corradi HR, Acharya KR, Sturrock ED. Structure of Testis ACE Glycosylation Mutants and Evidence for Conserved Domain Movement. Biochem. 2006;45:12654–12663. doi: 10.1021/bi061146z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corradi HR, Schwager SLU, Nchinda AT, Sturrock ED, Acharya KR. Crystal structure of the N domain of human somatic angiotensin I-converting enzyme provides a structural basis for domain-specific inhibitor design. J. Mol. Biol. 2006;357:964–974. doi: 10.1016/j.jmb.2006.01.048. [DOI] [PubMed] [Google Scholar]
- 14.Corradi HR, Chitapi I, Sewell T, Georgiadis D, Dive V, Sturrock ED, Acharya KR. The Structure of Testis Angiotensin-Converting Enzyme in Complex with the C Domain-Specific Inhibitor RXPA380. Biochem. 2007;46:5473–5478. doi: 10.1021/bi700275e. [DOI] [PubMed] [Google Scholar]
- 15.Watermeyer JM, Kroger WL, O'Neil HG, Sewell BT, Sturrock ED. Probing the Basis of Domain-Dependent Inhibition Using Novel ketone Inhibitors of Angiotensin-Converting Enzyme. Biochem. 2008;47:5942–5950. doi: 10.1021/bi8002605. [DOI] [PubMed] [Google Scholar]
- 16.Akif M, Georgiadis D, mahajan A, Dive V, Sturrock ED, Isaac RE, Acharya KR. High-Resolution Crystal Structures of Drosophila melanogaster Angiotensin-Converting Enzyme in Complex with Novel Inhibitors and Antihypertensive Drugs. J. Mol. Biol. 2010;400:502–517. doi: 10.1016/j.jmb.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 17.Bunning P, Riordan JF. Activation of angiotensin conveting enzyme by monovalent anions. Biochem. 1983;22:110–116. doi: 10.1021/bi00270a016. [DOI] [PubMed] [Google Scholar]
- 18.Shapiro R, Holmquist B, Riordan JF. Anion activation of angiotensin converting enzyme: dependence on nature of substrate. Biochem. 1983;22:3850–3857. doi: 10.1021/bi00285a021. [DOI] [PubMed] [Google Scholar]
- 19.Liu X, Fernandez A, Wouters MA, Heyberger S, Husain A. Arg(1097) is critical for the chloride dependence of human angiotensin I-converting C-domain catalytic activity. J. Biol. Chem. 2001;276:33518–33525. doi: 10.1074/jbc.M101495200. [DOI] [PubMed] [Google Scholar]
- 20.Tzakos AG, Galanis AS, Spyroulias GA, Cordopatis P, Manessi-Zoupa E, Gerothanassis IP. Structure-function discrimination of the N- and C- catalytic domains fo human angiotensin-converting enzyme: implications from Cl− activation and peptide hydrolysis mechanism. Protein Eng. 2003;16:993–1003. doi: 10.1093/protein/gzg122. [DOI] [PubMed] [Google Scholar]
- 21.Ondetti MA, Cushman DW. Annu. Rev. Biochem. 1982;51:283. doi: 10.1146/annurev.bi.51.070182.001435. [DOI] [PubMed] [Google Scholar]
- 22.Patchett AA, Cordes EH. The design and properties of N-carboxyalkyl-depeptide inhibitors of angiotensin-converting enzyme. Adv. Enzymol. Relat. Areas Mol. Biol. 1985;57:1–84. doi: 10.1002/9780470123034.ch1. [DOI] [PubMed] [Google Scholar]
- 23.Cushman DW, Cheung HS, Sabo EF, Ondetti MA. Design of potent competitive inhibitors of angiotensin-converting enzyme. Carboxyalkanoyl and mercaptoalkanoyl amino acids. Biochem. 1977;16:5484–5491. doi: 10.1021/bi00644a014. [DOI] [PubMed] [Google Scholar]
- 24.Almquist RG, Chao WR, Ellis ME, Johnson HL. Synthesis and biological activity of a ketomethylene analog of a tripeptide inhibitor of angiotensin converting enzyme. J. Med. Chem. 1980;23:1392–1398. doi: 10.1021/jm00186a020. [DOI] [PubMed] [Google Scholar]
- 25.Dive V, Cotton J, Yiotakis A, Michaud A, Vassiliou S, Jiracek J, Vazeux G, Chauvet MT, Cuniasse P, Corvol P. RXP 407, a phosphinic peptide, is a potent inhibitor of angiotensin I converting enzyme able to differentiate between its two active sites. Proc. Natl. Acad. Sci. U.S.A. 1999;96:4330–4335. doi: 10.1073/pnas.96.8.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Georgiadis D, Beau F, Czarny B, Cotton J, Yiotakis A, Dive V. Roles of the two active sites of somatic angiotensin-converting enzyme in the cleavage of angiotensin I and bradykinin: insights from selective inhibitors. Circ. Res. 2003;93:148–154. doi: 10.1161/01.RES.0000081593.33848.FC. [DOI] [PubMed] [Google Scholar]
- 27.Acharya KR, Sturrock ED, Riordan JF. ACE revisited: A new target for structure-based drug design. Nat. Rev. Drug Discov. 2003;2:891–902. doi: 10.1038/nrd1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daull P, Jeng AY, Battistini B. Towards Triple Vasopeptidase Inhibitors for the Treatment of Cariovascular Diseases. J. Cardiovasc. Pharmacol. 2007;50(3):247–256. doi: 10.1097/FJC.0b013e31813c6ca5. [DOI] [PubMed] [Google Scholar]
- 29.Redelinghuys P, Nchinda AT, Sturrock ED. Development of Domain-Selective Angiotensin I-Converting Enzyme Inhibitors. Ann. N.Y. Acad. Sci. 2005;1056:160–175. doi: 10.1196/annals.1352.035. [DOI] [PubMed] [Google Scholar]
- 30.Sramko M, Garaj V, Remko M. Thermodynamics of binding of angiotensin-converting enzyme inhibitors to enzyme active site model. J. Mol. Strut. (Theochem) 2008;869:19–28. [Google Scholar]
- 31.Pina AS, Roque ACA. Studies on the molecular recognition between bioactive peptides and angiotensin-converting enzyme. J. Mol. Recognit. 2009;22:162–168. doi: 10.1002/jmr.905. [DOI] [PubMed] [Google Scholar]
- 32.Dimitropoulos N, Papakyriakou A, Dalkas GA, Sturrock ED, Spyroulias GA. A Computational Approach to the Study of the Binding Mode of Dual ACE/NEP Inhibitors. J. Chem. Inf. Model. 2010;50:388–396. doi: 10.1021/ci9005047. [DOI] [PubMed] [Google Scholar]
- 33.Papakyriakou A, Spyroulias GA, Sturrock ED, Manessi-Zoupa E, Cordopatis P. Simulated Interactions between Angiotensin-Converting Enzyme and Substrate Gonadotropin-Releasing Hormone: Novel Insights into Domain Selectivity. Biochem. 2007;46:8753–8765. doi: 10.1021/bi700253q. [DOI] [PubMed] [Google Scholar]
- 34.Li X, Hayik SA, Merz KM., Jr QM/MM X-ray refinement of zinc metalloenzymes. J. Inorg. Biochem. 2010;104:512–522. doi: 10.1016/j.jinorgbio.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu R, Hu P, Wang S, Cao Z, Zhang Y. Flexibility of catalytic zinc coordination in thermolysin and HDAC8: A Born-Oppenheimer ab initio QM/MM molecular dynamics study. J. Chem. Theo. Comput. doi: 10.1021/ct9005322. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoops SC, Anderson KW, Merz JKM. Force-field design for metalloproteins. J. Am. Chem. Soc. 1991;113:8262–8270. [Google Scholar]
- 37.Warshel A, Levitt M. Theoretical studies of enzymatic reactions: Dielectric, electrostatic and steric stabilization of carbonium ion in the reaction of lysozyme. J. Mol. Biol. 1976;103:227–249. doi: 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]
- 38.Field MJ, Bash PA, Karplus M. A combined quantum mechanical and molecular mechanical potential for molecular dynamics simulations. J. Comput. Chem. 1990;11:700–733. [Google Scholar]
- 39.Gao J. Methods and applications of combined quantum mechanical and molecular mechanical potentials. In: Lipkowitz KB, Boyd DB, editors. Rev. Comput. Chem. Vol. 7. New York: VCH; 1996. pp. 119–185. [Google Scholar]
- 40.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher III WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 41.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 42.Elstner M, Frauenheim T, Kaxiras E, Seifert G, Suhai S. A self-consistent charge density-functional based tight-binding scheme for large biomolecules. Phys. Stat. Sol. 2000;B217:357–376. [Google Scholar]
- 43.Cui Q, Elstner M, Kaxiras E, Frauenheim T, Karplus M. A QM/MM implementation of the self consistent charge density functional tight binding (SCC-DFTB) method. J. Phys. Chem. B. 2001;105:569–585. [Google Scholar]
- 44.Riccardi D, Schaefer P, Yang Y, Yu H, Ghosh N, Prat-Resina X, Konig P, Li G, Xu D, Guo H, Elstener M, Cui Q. Development of effective quantum mechanical/molecular mechanical (QM/MM) methods for complex biological processes. J. Phys. Chem. B. 2006;110:6458–6469. doi: 10.1021/jp056361o. [DOI] [PubMed] [Google Scholar]
- 45.Elstner M, Cui Q, Munih P, Kaxiras E, Frauenheim T, Karplus M. Modeling zinc in biomolecules with the self consistent charge density functional tight binding (SCC-DFTB) method: Applications to structure and energetic analysis. J. Comput. Chem. 2003;24:565–581. doi: 10.1002/jcc.10201. [DOI] [PubMed] [Google Scholar]
- 46.Riccardi D, Cui Q. pKa analysis for the zinc-bound water in human carbonic anhydrase II: benchmark for "multi-scale" QM/MM simulations and mechanistic implications. J. Phys. Chem. A. 2007;111:5703–5711. doi: 10.1021/jp070699w. [DOI] [PubMed] [Google Scholar]
- 47.Riccardi D, Konig P, Guo H, Cui Q. Proton transfer in carbonic anhydrase is controlled by electrostatics rather than the orientation of the acceptor. Biochem. 2008;47:2369–2378. doi: 10.1021/bi701950j. [DOI] [PubMed] [Google Scholar]
- 48.Xu D, Guo H. Quantum mechanical/molecular mechanical and density functional theory studies of a prototypical zinc peptidase (carboxypeptidase A) suggest a general acid-general base mechanism. J. Am. Chem. Soc. 2009;131:9780–9788. doi: 10.1021/ja9027988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu SS, Zhang CC, Xu DG, Guo H. Catalysis of Carboxypeptidase A: Promoted-Water versus Nucleophilic Pathways. J. Phys. Chem. B. 2010;114(28):9259–9267. doi: 10.1021/jp101448j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu D, Zhou Y, Xie D, Guo H. Antibiotic binding to monozinc CphA β-lactamase from Aeromonas hydropila: quantum mechanical/molecular mechanical and density functional theory studies. J. Med. Chem. 2005;48:6679–6689. doi: 10.1021/jm0505112. [DOI] [PubMed] [Google Scholar]
- 51.Xu D, Xie D, Guo H. Catalytic mechanism of class B2 metallo-β-lactamase. J. Biol. Chem. 2006;281:8740–8747. doi: 10.1074/jbc.M512517200. [DOI] [PubMed] [Google Scholar]
- 52.Wang C, Guo H. Inhibitor binding by metallo-β-lactamase IMP-1 from Pseudomonas aeruginosa: Quantum mechanical/molecular mechanical simulations. J. Phys. Chem. B. 2007;111:9986–9992. doi: 10.1021/jp073864g. [DOI] [PubMed] [Google Scholar]
- 53.Xu D, Guo H, Cui Q. Antibiotic deactivation by dizinc β-lactamase: mechanistic insights from QM/MM and DFT studies. J. Am. Chem. Soc. 2007;129:10814. doi: 10.1021/ja072532m. [DOI] [PubMed] [Google Scholar]
- 54.Xu D, Guo H, Cui Q. Antibiotic binding to dizinc β-lactamase L1 from Stenotrophomonas maltophilia: SCC-DFTB/CHARMM and DFT studies. J. Phys. Chem. A. 2007;111:5630–5636. doi: 10.1021/jp068746s. [DOI] [PubMed] [Google Scholar]
- 55.Wu S, Xu D, Guo H. QM/MM studies of mono-zinc beta-lactamase CphA suggest that the crystal structure of an enzyme-intermediate complex represents a minor pathway. J. Am. Chem. Soc. 2010;132:17986–17988. doi: 10.1021/ja104241g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brooks CL, III, Karplus M. Solvent effects on protein motion and protein effects on solvent motion. J. Mol. Biol. 1989;208:159–181. doi: 10.1016/0022-2836(89)90093-4. [DOI] [PubMed] [Google Scholar]
- 57.Steinbach PJ, Brooks BR. New spherical-cutoff methods for long-range forces in macromolecular simulations. J. Comput. Chem. 1994;15:667. [Google Scholar]
- 58.Ryckaert JP, Ciccotti G, Berendsen HJ. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 59.Vrielink A, Obel-Jorgensen A, Codding PW. Hippuryl-L-histidyl-L-leucine, a Substrate for Angiotensin Converting Enzyme. Acta Cryst. 1996;C52:1300–1302. doi: 10.1107/s0108270195016180. [DOI] [PubMed] [Google Scholar]
- 60.Hangauer DG, Mozingo AF, Matthews BW. An interactive computer graphics study of thermolysin-catalyzed peptide cleavage and inhibition by N-carboxymethyl dipeptides. Biochem. 1984;23:5730–5741. doi: 10.1021/bi00319a011. [DOI] [PubMed] [Google Scholar]
- 61.Blumberger J, Lamoureux G, Klein ML. Peptide hydrolysis in thermolysin: Ab initio QM/MM investigation of the Glu143-assisted water addition mechanism. J. Chem. Theo. Comput. 2007;3:1837–1850. doi: 10.1021/ct7000792. [DOI] [PubMed] [Google Scholar]
- 62.Christianson DW, Lipscomb WN. Carboxypeptidase A. Acc. Chem. Res. 1989;22:62–69. [Google Scholar]
- 63.Matthews BW. Structural basis of the action of thermolysin and related zinc peptidases. Acc. Chem. Res. 1988;21:333–340. [Google Scholar]
- 64.Cornell WD, Cieplak P, Bayly CI, Gould IR, K. M. Merz J, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of protein, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179. [Google Scholar]
- 65.Lipscomb WM, Strater N. Recent advances in zinc enzymology. Chem. Rev. 1996;96:2375–2433. doi: 10.1021/cr950042j. [DOI] [PubMed] [Google Scholar]
- 66.Kim HM, Shin DR, Yoo OJ, Lee H, Lee JO. Crystal structure of Drosophila angiotensin I-converting enzyme bound to captopril and lisinopril. FEBS Lett. 2003;538:65–70. doi: 10.1016/s0014-5793(03)00128-5. [DOI] [PubMed] [Google Scholar]