Abstract
Enterohemorrhagic E. coli (EHEC) O157:H7 can cause serious gastrointestinal and systemic disease in humans following direct or indirect exposure to ruminant feces containing the bacterium. The main colonization site of EHEC O157:H7 in cattle is the terminal rectum where the bacteria intimately attach to the epithelium and multiply in the intestinal mucus. This study aimed to identify genomic regions of EHEC O157:H7 that contribute to colonization and multiplication at this site. A bacterial artificial chromosome (BAC) library was generated from a derivative of the sequenced E. coli O157:H7 Sakai strain. The library contains 1152 clones averaging 150 kbp. To verify the library, clones containing a complete locus of enterocyte effacement (LEE) were identified by DNA hybridization. In line with a previous report, these did not confer a type III secretion (T3S) capacity to the K-12 host strain. However, conjugation of one of the BAC clones into a strain containing a partial LEE deletion restored T3S. Three hundred eighty-four clones from the library were subjected to two different selective screens; one involved three rounds of adherence assays to bovine primary rectal epithelial cells while the other competed the clones over three rounds of growth in bovine rectal mucus. The input strain DNA was then compared with the selected strains using comparative genomic hybridization (CGH) on an E. coli microarray. The adherence assay enriched for pO157 DNA indicating the importance of this plasmid for colonization of rectal epithelial cells. The mucus assay enriched for multiple regions involved in carbohydrate utilization, including hexuronate uptake, indicating that these regions provide a competitive growth advantage in bovine mucus. This BAC-CGH approach provides a positive selection screen that complements negative selection transposon-based screens. As demonstrated, this may be of particular use for identifying genes with redundant functions such as adhesion and carbon metabolism.
Keywords: EHEC, bacterial artificial chromosome, mucus, comparative genomic hybridization, locus of enterocyte effacement, sugar utilization
Introduction
Enterohemorrhagic E. coli (EHEC) strains are associated with serious gastrointestinal disease in humans that can lead to life threatening vascular damage due to the activity of Shiga toxins. The predominant serotypes associated with human infections are O157:H7 and O26:H11 and strains persist in ruminant hosts with humans as an incidental host. It has been demonstrated that EHEC O157:H7 predominately colonizes the terminal rectum of cattle and bacterial multiplication at this site leads to the fecal excretion that is a threat to human health through contamination of bovine food products, produce, and water supplies (Naylor et al., 2003; Chase-Topping et al., 2008). There are a number of EHEC O157:H7 factors that are known to contribute to colonization of cattle at this specific gastrointestinal niche based on both in vivo studies and through research on primary epithelial cells cultured from crypts isolated from this rectal site (Chase-Topping et al., 2008). These include the locus of enterocyte effacement (LEE)-encoded type III secretion system (T3SS), various T3-secreted effector proteins, H7 flagellin, and a number of specific adhesins, including F9 fimbriae and autotransporters.
Over the last decade signature-tagged mutagenesis has been applied to extend and confirm genes important for bacterial carriage. This included screening for EHEC O157 and O26 genes important for colonization in cattle (Dziva et al., 2004; Van Diemen et al., 2005). Now with the application of massively parallel sequencing, these studies can be quantified giving exquisite information on the relative significance of each gene containing a transposon insert that is introduced into the animal (Eckert et al., 2011). This work has highlighted the importance of many of the T3-secreted effector proteins and raised interesting questions about inserts that are an advantage in vivo. However, this insertional mutagenesis primarily examines the effects of single genes or individual disrupted operons. Many virulence related phenotypes, such as iron acquisition, carbon utilization, and adherence are encoded redundantly within the genome making them difficult to interrogate with single deletions.
Based on the sequences of multiple EHEC strains that are now available, it is evident that the EHEC pathotype has arisen multiple times by independent acquisition of virulence factors on mobile genetic elements (Ogura et al., 2009). Large horizontally acquired regions, absent from the E. coli K-12 genome sequence (designated O-islands, OI), account for a significant proportion of the EHEC genome (Perna et al., 2001; Ohnishi et al., 2002; Zhang et al., 2007) and are likely critical for its niche adaptation encoding factors for nutrient acquisition and adherence. To address what these large regions contribute to the biology of the bacterium, research has been carried out on deletions of OI demonstrating their importance for colonization and persistence (Tree et al., 2011).
The aim of the research presented here was to complement these different screening approaches by generating a bacterial artificial chromosome (BAC) library from an EHEC O157:H7 strain in an E. coli K-12 background and then use competition-based assays to select for BAC clones that provide an advantage under in vitro conditions relevant to colonization of the bovine host. Comparative genome hybridization on an oligonucleotide microarray was then used to compare the input and output libraries. We have demonstrated that this approach does select for genetic regions with growth and colonization advantages. Several regions of the EHEC genome containing sugar catabolic loci were enriched using this approach and we demonstrate that BAC clones containing hexuronic acid and galactosamine/N-acetylgalactosamine catabolism genes increase growth of E. coli in bovine terminal rectal mucus. This work raises the possibility of targeting these sugar uptake systems to limit bacterial growth in certain host environments.
Results
Generation and validation of an EHEC O157 BAC library
In order to screen large chromosomal fragments that confer virulence phenotypes, a HindIII BAC library of DNA fragments with an average size of 150 kbp from gDNA isolated from E. coli O157:H7 strain Sakai stx− (Dahan et al., 2004) was generated in pCLD04541 (Jones et al., 1992) and transformed into E. coli DH10B. The BAC library was produced by Lucigen Corporation (Middleton, WI, USA) and was screened for specific sequences by Southern hybridization of colony filters made by the same company.
Type III secretion (T3S) is of central importance for EHEC and is required for colonization of the bovine host. The T3S machine is encoded by the LEE, an approx. 45 kb island inserted into the common E. coli backbone. Using a probe designed to hybridize ecs4562 (encoded within the LEE) we identified 11 BAC clones that contain ecs4562 and then screened these for clones containing the entire LEE by using PCR primers that amplified across the junctions between the LEE and common backbone (Table 1). Clone 3/A2 was found to encode the entire LEE and sequencing of the clone ends from the plasmid indicated that it contained bases 4486393–4639841 of the Sakai chromosome, a 153.4-kbp fragment containing the LEE. To determine if this region was sufficient to confer T3S on E. coli DH10B; clone 3/A2 (pBAC3/A2) was grown under T3S permissive conditions and secreted proteins isolated and analyzed for the needle filament protein EspD by western blotting. Secretion of EspD could not be detected (data not shown) in agreement with previous observations (Elliott et al., 1999). To verify that pBAC3/A2 encodes a functional copy of the LEE, a triparental mating using pBAC3/A2 as the donor, pRK2013 (Ditta et al., 1980) as helper and E. coli O157:H7 strain TUV93-0 Δ148A Nalr as the recipient was performed. TUV93-0 Δ148A Nalr contains an 8.93-kb deletion within the LEE (from 4677931 to 4686861 as defined for the EDL933 genome) and is deleted for LEE1, LEE2, and most of the LEE3 operon (escN onwards are intact) and is unable to export translocon or effector proteins (Figure 1; Campellone et al., 2004). Introduction of pBAC3/A2 restored T3S to TUV93-0 Δ148A Nalr (Figure 1) indicating that this clone is likely to contain a functional LEE and that the LEE encoded by E. coli O157:H7 strain Sakai stx− is not sufficient to confer detectable T3S on a non-pathogenic K-12 isolate such as DH10B. The extra-LEE elements that confer T3S in Sakai are still unclear.
Table 1.
Specific strains and plasmids associated with the study.
| Strain | Genotype | Source |
|---|---|---|
| DH10B | F− mcrA Δ(mrr-hsdRMS-mcrBC) Ф80lacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara leu) 7697 galU galK rpsL nupG λ− | Lucigen |
| E. coli O157:H7 strain Sakai | stx2A::kan Δstx1A′ | Dahan et al. (2004) |
| E. coli O157:H7 strain TUV93-0 | ΔBP933-W ΔCP933-V | Campellone et al. (2004) |
| E. coli O157:H7 strain TUV93-0 Δ148A | TUV93-0 ΔLEE1-3 (4677931–4686861 as defined for the EDL933 genome) | Campellone et al. (2004) |
| 3/A2 | DH10B pBAC3/A2 | This study |
| I12 | DH10B pV41 containing agaWEFA and uxaC fragment | This study |
Figure 1.

Complementation of T3S in TUV93-0 Δ148A. The LEE containing clone pBAC3/A2 was introduced into the TUV93-0 Δ148A containing a partial deletion of the LEE. A western blot of secreted proteins isolated from TUV93-0 (wt), TUV93-0 Δ148A, and the complemented strain TUV93-0 Δ148A pBAC3/A2 are shown.
| Plasmid | Genotype | Source |
|---|---|---|
| pV41 | Bacterial artificial chromosome | Lucigen |
| pRK2013 | Mobilization helper plasmid | Ditta et al., 1980 |
| pBAC3/A2 | pV41 containing the LEE (4486393–4639841 as defined for the Sakai genome) | This study |
Selection of clones that adhere to bovine primary cells
E. coli O157:H7 colonize the terminal rectal tissue of cattle and replication at this site and associated mucus leads to excretion in the feces and contamination of the environment and the food-chain. High levels of colonization at the terminal rectum are correlated with high numbers of organisms shed into the environment and a higher probability of causing human disease (so called “supershedders”; Matthews et al., 2006). Colonization factors that confer adherence to terminal rectal tissues are important potential targets to prevent colonization by vaccination or more direct interventions. In an effort to identify genomic regions of E. coli O157:H7 that may confer adhesion 384 BAC clones were used in repeated rounds of adhesion and culture on bovine primary terminal rectal cells. BAC clones were incubated with primary tissue culture for 4 h at 37°C after which the monolayer was washed to remove unattached bacteria and disrupted to recover adherent bacteria. This pool of adherent bacteria was further selected by another two rounds of adherence. Selected bacteria were inoculated into DMEM for a final round of growth to amplify the amount of gDNA for labeling and hybridization.
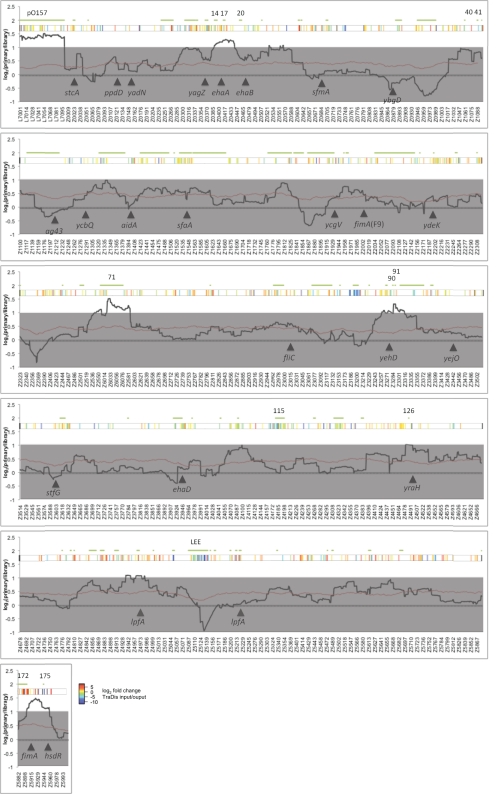
In order to determine which BAC clones increase adherence to primary tissue culture total gDNA was isolated from the input and output pools and comparative genomic hybridization (CGH) performed. CGH allows screening of the entire output pool for regions of the genome that have been enriched by selection. This data is normalized using a LOWESS regression and can be visualized as a ratio of signal within the input and output pools for each gene (Figure 2 and Data files 1,3 in Supplementary Material). To reduce variation between individual genes and to identify large regions (containing potentially overlapping BAC clones) that are uniformly increased a sliding window of 50 genes was used to average the signal for each gene. In line with our earlier observations that the LEE did not confer T3S on DH10B, LEE containing BAC clones were not selected on primary cells and in fact were negatively selected under these conditions. A number of genomic regions were enriched above two-fold in the primary tissue culture selected pools and these regions encode adhesins or other factors known to confer attachment to cultured epithelial cells. These include pO157 (Grys et al., 2005b; Dziva et al., 2007; Ho et al., 2008), the autotransporter ehaA (Wells et al., 2008), and type 1 fimbriae (Galfi et al., 1998). Encouragingly, autotransporters that have been demonstrated not to confer adhesion to primary bovine terminal rectal cells, such as ehaB, ehaC, and ehaD were not enriched in this genome-wide screen indicating a level of specificity (Wells et al., 2008). By screening the BAC library by CGH we have also identified two novel regions that are enriched by selection on bovine primary cells, a region encompassing O-island 71 (OI-71), and a region containing OI-90 and 91. We were not able to identify a known adhesin on, or surrounding OI-71 but note that the fimbrial cluster designated loc9 (indicated by yehD in Figure 2) is encoded adjacent to OI-90. Previous work on loc9 has indicated that this gene cluster can be expressed under in vitro conditions (Low et al., 2006b) and does provide a colonization advantage in cattle (Low et al., 2006a). These enriched regions provide promising targets for further study.
Figure 2.
Bacterial artificial chromosome-CGH of primary tissue culture. E. coli O157:H7 strain Sakai BAC clones with increased adherence were selected by multiple rounds of adherence to primary bovine terminal rectum cells. Total gDNA from output (adherent) and input (384 clones from the library) pools were subjected to CGH analysis. The ratio of output and input were normalized and a sliding average (window = 50 genes) applied. The black solid line indicates average ratio (output/input), the red line indicates the average p value and gray shading indicates an arbitrary two-fold cut-off for significance. Green lines and numbers above the plot indicate the location of O-islands. Solid arrowheads and designations indicate select genes implicated in adherence of E. coli.
Selection of clones that provide a growth advantage in bovine rectal mucus
The restricted colonization site of EHEC O157 in cattle means that majority of bacterial in vivo replication is limited to the rectal epithelium or mucus layer before excretion in feces. EHEC O157 should have evolved to grow efficiently in rectal mucus which is likely to require genetic regions allowing use of specific carbon sources at this site, the capacity to deal with host innate defense products and potentially the production of factors that can restrict the growth of competitive microbes. Our recent work modeling bacterial replication rates required at the terminal rectum to account for the excretion levels measured indicates doubling times between 20 and 30 min (unpublished data), toward the maximum rates established for E. coli in vitro. Doubling times measured in bovine rectal mucus diluted in Hanks buffer were between 30 and 40 min (data not shown) supporting the nutritional quality of even the diluted substrate.
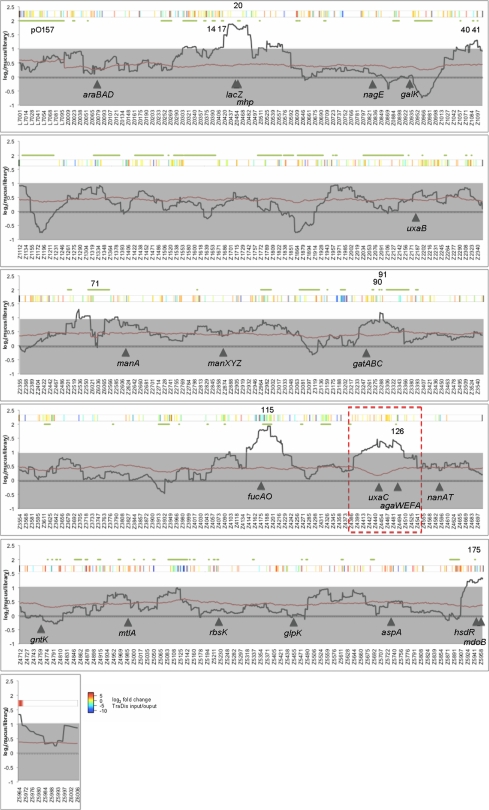
To identify genomic regions that provide a selective advantage at the terminal rectum we cultured 384 BAC clones in bovine terminal rectal mucus and screened output pools by CGH. Four regions were strongly enriched by culturing in mucus and contain OIs 20, 115, 126, and 175 (indicated in (Figure 3 and Data files 2,4 in Supplementary Material). lacZ is encoded adjacent to OI-20 and is inactivated in the E. coli background DH10B. Lactose is reported to be absent from mucus, but the metabolic pathway for metabolism of lactose is induced by culturing on murine mucus (Chang et al., 2004; Fabich et al., 2008). The region adjacent to lac also contains the mhp gene cluster allowing bacterial breakdown of 3-(3-hydroxyphenyl)propionic acid. Phenylpropanoic and phenylpropenoic acids and their derivatives are common in the environment, arising as breakdown products of lignin and other plant-derived flavonoids and phenylpropanoids (Torres et al., 2003). Adjacent to this region and central to the selected fragments, is also a two component regulatory system for sensing hexose phosphate and an associated hexose phosphate transporter (z0461–z0463).
The region containing OI-175 was also enriched and contains a lesion in the restriction–modification system hsdR in DH10B. It seems unlikely that hsdR would confer a growth advantage in mucus and this region may contain novel genes involved in mucus growth. For example at the start of the selected region is gntP, a fructuronate transporter and central to the region is phosphoglycerol transferase I (mdoB).
Two regions contained genes that are known to play an important role in EHEC catabolism of mucus sugars and colonization of cattle. fucAO are encoded adjacent to OI-115 and confer a selective advantage on wild-type E. coli in mucus when cultured in the presence of an isogenic fucAO mutant (Fabich et al., 2008). A strong colonization defect has also been observed in a fucAO mutant during cattle colonization (Snider et al., 2009). Similarly, the uronate isomerase uxaC (required for hexuronate catabolism) is encoded adjacent to OI-126 and confers a growth advantage on wild-type cells (Fabich et al., 2008). This region also encodes the agaWEFA cluster that is required for catabolism of galactosamine and N-acetylgalactosamine (GalNAc) and is inactivated in commensal E. coli K-12 strains. Both sugars are present in mucus and support growth of EHEC. The presence of the agaWEFA cluster has been reported not to confer a significant advantage on wild-type cells in vitro however an agaWEFA mutant is outcompeted during cattle colonization with significantly less CFU found in terminal rectal mucus (Snider et al., 2009). Enrichment of regions containing fucAO and uxaC/agaWEFA supports the validity of our results and the ability to BAC-CGH to identify phenotypically relevant regions of the chromosome.
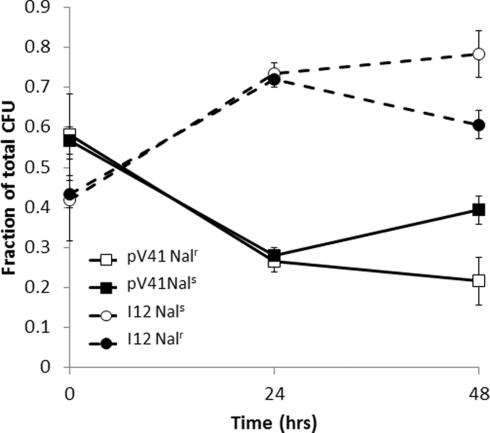
To further confirm our CGH data we have isolated BAC clones that contain ecs4487 (containing Sakai specific sequence within the uxaC/agaWEFA region) and determined their growth phenotype relative to the BAC vector in mucus. A clone containing the uxaC/agaWEFA region was identified, designated I12. Both I12 and the BAC vector only (DH10B pV41) strain were marked with Nalr to allow selection. In reciprocal experiments, the Nalr tagged I12 or pV41 strain was inoculated at equal CFU into bovine mucus with Nals pV41 or I12 respectively. After 24 h culturing in mucus the total CFU/milliliter increased from an average 4.4 × 105–6.5 × 107 (Figure 4). During these seven generations the I12 clone was able to outcompete the control vector, increasing from 42 and 43 to 74 and 72% of the total population in reciprocal experiments (p < 0.05 in both experiments using a Student’s t-test). These results confirm that the uxaC/agaWEFA region identified in our BAC-CGH screen can confer a selective advantage during growth in bovine mucus.
Figure 4.
Growth of clone I12 in bovine terminal rectal mucus. Clone I12 encodes the positively selected region containing uxaC and agaWEFA (see Figure 3). DH10B control (pV41) and I12 were tagged with Nalr and competed against reciprocal Nals strains in bovine rectal mucus. CFU/ml of total and Nalr cells were recorded over 48 h.
Discussion
Based on the sequences of multiple EHEC strains that are now available, it is evident that the EHEC pathotype has arisen multiple times by independent acquisition of virulence factors on mobile genetic elements (Ogura et al., 2009). These horizontally acquired regions add over 1 Mb of extra genetic information to the “core” genome present in non-pathogenic E. coli K-12 and encode colonization factors including the T3SS, associated secreted effector proteins, fimbriae, and Shiga toxins (Hayashi et al., 2001; Perna et al., 2001; Ogura et al., 2009). Therefore these often large, horizontally acquired regions account for a significant proportion of the EHEC genome (Ohnishi et al., 2002; Zhang et al., 2007). In an effort to define elements within the EHEC genome that confer a selective advantage when colonizing bovine terminal rectal tissue we have generated a BAC library from a derivative of E. coli O157:H7 strain Sakai. This library contains a total of 1152 clones with an average fragment size of 150 kbp and provides 10–15 times coverage of the Sakai genome. This library is likely to be of use for complementing large deletions such as those generated by deletion of entire OIs and we have demonstrated that clones from this library can be selected under in vitro colonization relevant conditions identifying regions encoding both established and novel colonization factors. By using CGH to screen output pools we were able to assay enrichment of sequences throughout the genome. This technique may also yield higher resolution of sequences encoding relevant phenotypes than the more traditional approach of isolating individual clones, as overlapping BAC clones will increase the signal recovered for the relevant region.
Using primary bovine rectal tissue culture to select for BAC clones that confer adhesion we have been able to identify regions known to aid attachment to cultured epithelial cells including pO157, ehaA, and type 1 fimbriae. The later presents in interesting situation where DH10B may complement the deficiency that has been reported in the O157 type 1 fimbriae phase switch and fimH adhesin (Roe et al., 2001; Low et al., 2006a). Additional novel regions were identified containing OI-71 and the fimbrial cluster loc9. OI-71 is known to encode T3-secreted effectors (nleG2-1′, nleA, nleH1-2, nleF, and espO1-2) although it is unclear whether these are responsible for enrichment of OI-71. While a LEE encoding BAC clone could not confer a T3S phenotype on E. coli DH10B, it is possible that these proteins could still exert a phenotype, for example by secretion through the flagellar secretion system (Lee and Galan, 2004). Further investigation is required to establish the minimal sequences that are positively selected within the OI-71 and loc9 encoding regions.
It is evident that positive selection of the BAC library is dependant on appropriate expression of virulence factors in the host strain. This is highlighted by negative selection of the LEE (Figure 2), which is required for colonization of bovine terminal rectal tissues but does not confer T3S on DH10B and is strongly negatively selected in our primary tissue culture screen.
The plasmid, pO157 was also selected on primary tissue culture. While not essential for adherence, pO157 confers increased adhesion on O157:H7 (Sheng et al., 2006; Lim et al., 2007) and has been demonstrated to encode at least four loci that are known to increase adherence to epithelial cells. The autotransporter EspP is encoded on pO157 and has been shown to increase adherence to primary bovine terminal rectal cells (Dziva et al., 2007). StcE encodes a protease that cleaves mucin 7 (MUC7), glycoprotein 340 (gp340), and C1-esterase inhibitor (Grys et al., 2005a). MUC7 and gp340 are constituents of the protective glycocalyx covering the epithelium and digestion may allow access to the underlying epithelial cell. StcE is secreted by the type 2 secretion system encoded by the etp locus on pO157 and, with YodA, are the only identified substrates. Deletion of etpC reduces adherence to HeLa cells in a stcE independent fashion indicating that this secretion system is required for function of an as yet unidentified protein(s) involved in adhesion (Ho et al., 2008). ToxB has also been shown to increase adherence, potentially by increasing T3S post-transcriptionally (Tatsuno et al., 2001; Stevens et al., 2004).
Using growth in bovine mucus as a selection for our BAC library we have also selected for multiple regions that contain genes important for uptake and utilization of different sugars present in mucus. Mucus is predominately composed of the glycoprotein, mucin, but also contains smaller fractions of other glycoproteins, proteins, sugars, glycolipids, and lipids, potentially released from epithelial cells sloughed from the intestinal lining (Conway et al., 2007). Mucin is extensively decorated with oligosaccharides that are degraded to monosaccharides by the anaerobic microflora (Conway et al., 2004). A growing body of work has demonstrated that commensal and pathogenic E. coli are able to metabolize free gluconate and monosaccharides released from mucin and these likely form a rate limiting substrate for growth (Fabich et al., 2008). Commensal E. coli metabolizes at least seven sugars present in mucus: gluconate, N-acetylglucosamine, N-acetylneuraminic acid (sialic acid), glucuronate, mannose, fucose, and ribose (Chang et al., 2004). E. coli O157 can additionally use galactosamine and N-acetylgalactosamine (Fabich et al., 2008). Of these, the hexuronates, D-glucuronate and D-galacturonate, are thought to comprise 0.6% of mouse cecal mucus by weight (Conway et al., 2007).
Using CGH to analyze the BAC library after mucus growth, two regions encoding catabolic genes (fucAO, uxaC, and agaWEFA) were selected. These have recently been shown to be important for EHEC colonization of cattle and growth within terminal rectal mucus (Fabich et al., 2008; Snider et al., 2009). Enrichment of these regions lends weight to our CGH analysis of the BAC library pools. We have been able to confirm that screening of our BAC library does produce reliable hits in regions that confer positive selection in mucus by isolating a BAC clone containing the entire uxaC/agaWEFA region, designated I12. I12 was able to outcompete DH10B with pCLD04541 when inoculated at equal CFU into mucus after approximately seven generations indicating that the uxaC/agaWEFA containing region did confer an enhanced growth phenotype on commensal E. coli in mucus.
Genome-wide studies of insertional inactivation events that confer negative selection during host colonization (signature-tagged mutagenesis, TraSH, and TraDIS) have provided a wealth of data about genes that are essential for colonization of host tissue. With the advent of massively parallel sequencing, the depth of coverage and resolution of genes essential for colonization has increased by an order of magnitude (Eckert et al., 2011). Using CGH to analyze an EHEC BAC library selected in vitro has provided reliable hits within regions that are positively selected under in vivo relevant conditions. This technique is likely to complement existing insertional inactivation screens by provide positive selection of large DNA fragments. This will be of particular use for phenotypes where there is significant redundancy engineered into the genome of pathogens such as adhesion and carbon utilization.
Materials and Methods
Bacterial strains, plasmids, oligonucleotides and media
The bacterial strains and plasmids used in the study are described in Table 1. LB broth was also used (Oxoid). Antibiotics were included when required at the following concentrations: chloramphenicol (12.5 μg/ml), kanamycin (25 μg/ml), tetracycline (12.5 μg/ml), and ampicillin (50 μg/ml).
Preparation of secreted proteins and bacterial fractions for protein analyses
Bacteria were cultured in 30 ml of MEM–HEPES at 37°C (200 rpm) to an OD600 of 0.8 unless specifically stated. The bacterial cells were pelleted by centrifugation at 4000 × g for 15 min, and supernatants were passed through filters (0.22 μm). Supernatant proteins were precipitated overnight at 4°C with 10% TCA, and separated by centrifugation at 3220 × g for 30 min at 4°C; the proteins were suspended in 100 μl of 1.5 M Tris (pH 8.8). The bacterial pellet was initially suspended in 50 μl sample buffer (Sigma#S3401) and 50 μl molecular biology grade water. Proteins were separated by SDS-PAGE using standard methods and western blotting performed as described previously (Roe et al., 2003).
Selection process with bovine primary cells and bovine rectal mucus
Bacterial artificial chromosome library mixture (384 colonies) and pV41 vector only were separately cultured overnight in triplicate in LB. One volume of the above overnight cultures was incubated in 50 volumes of DMEM until the OD600 reached 0.6, then adjusted to 0.4. Cultures were diluted 1/100 and then 100 μl/well was transferred into 6-well plates of primary cells. Tissue samples for primary cell culture and isolation of mucus were sourced from a local abattoir. Primary tissue culture was prepared as described previously (Mahajan et al., 2005). Parallel experiments using 1 ml of mucus (verified E. coli negative by plating on sorbitol-MacConkey agar) from the first scrap of cattle rectum tissues were also inoculated with 100 μl of bacterial culture. Tetracycline was added to mucus samples to select for the BAC library or vector, and samples were serially diluted and plated to determine CFU/ml on sorbitol-MacConkey agar at indicated times. For each experiment bacteria with primary cells were incubated for 4 h. Culture medium for primary cells was replaced with DMEM 1 h prior to the above treatment. After unattached bacteria were removed by washing twice with pre-warmed bacterial medium, bacteria from primary cell cultures were collected by mechanical disruption of the monolayer. Bacteria were inoculated and collected from primary tissue culture two further times. Bacterial cells were collected at the end of three rounds of adhesion and subjected to genomic DNA extraction for CGH.
Comparative genomic hybridization
Genomic DNA was extracted using an Invitrogen ChargeSwitch gDNA Mini Bacteria Kit (Invitrogen). Labeling was carried out using a Bioprime Plus Array CGH Genomic Labeling System (Invitrogen). Protocol for pre-hybridization, hybridization and washing was essentially as previously described for hybridization of cDNA (Roe et al., 2007). Briefly, array slides were washed twice in Wash Buffer II (0.1 × SSC and 0.1% × SDS), each time for 30 s. Slides were transferred in pre-warmed pre-hybridization buffer (0.1% BSA, 0.1% SDS and 5× SSC) for 120 min at 42°C. Slides were washed once in Wash Buffer II and twice in Wash buffer III (0.1 × SSC), and the denatured (95°C for 3 min) hybridization probe mixture (20 pmol of each labeled probe, 0.4 × Ultrahyb, 0.8 × SSC, 90 ng/μl polyA, 10 μg/μl BSA) was added to slides and hybridized overnight at 42°C. Following hybridization slides were washed in Wash Buffer I (0.1% SDS, 2× SSC), Wash buffer II and twice in Wash buffer III. Slides were dried by centrifugation and scanned using an Axon Genepix 4000A scanner (Axon Instruments, Union City, CA, USA). Slide Images were processed using Genepix software and data analyzed using Genespring GX 7.3 (Agilent). Data were normalized using a LOWESS regression. To visualize genome-wide data; a window of 25 genes on either side of each gene was used to average the signal in that region. Data plotted for each gene in Figures 2 and 3 represents this average. Regions with an average intensity above two-fold across >50 consecutive genes (on average ∼35 kb or ∼25% of the average BAC insert length) were considered significant.
Figure 3.
Bacterial artificial chromosome-CGH of mucus-cultured clones. E. coli O157:H7 strain Sakai BAC clones were selected cultured were selected by multiple rounds of growth in bovine terminal rectal mucus. Total gDNA from output (mucus grown) and input (384 clones from the library) pools were subjected to CGH analysis. The ratio of output and input were normalized and a sliding average (window = 50 genes) applied. The black solid line indicates average average ratio (output/input), the red line indicates the average p value and gray shading indicates an arbitrary two-fold cut-off for significance. Green lines and numbers above the plot indicate the location of O-islands. Solid arrowheads and designations indicate select genes implicated in sugar utilization of E. coli. The red dashed box indicates the region selected for further confirmation (see Figure 4).
Competition of defined BAC clones in mucus
For competition experiments of individual BAC clones in mucus, spontaneous nalidixic acid resistant (Nalr) mutants were recovered for both DH10B pCLD04541 and I12 by plating onto LB Nal plates. Competition experiments were performed between DH10B pCLD04541 Nals and I12 Nalr or DH10B pCLD04541 Nalr and I12 Nals. Equal CFU were inoculated into bovine terminal rectal mucus and incubated at 37°C for 24 h. The start and end CFU/ml of each culture was determined by plating on both LB Tet (to select for both strains carrying the BAC vector) or LB Tet/Nal to select for the tagged strain. The relative proportion each strain was determined by subtracting the number of Nalr colonies from the total Tet resistant population.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Jai J. Tree and David L. Gally were supported by DEFRA funding under VTRI 0102 and by the Wellcome Trust. Jianing Bai was funded by a scholarship from the China Scholarship Scheme. David L. Gally and Sean P. McAteer were supported by a BBSRC Institute program grant to the Roslin Institute.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/cellular_and_infection_microbiology/10.3389/fmicb.2011.00168/abstract
References
- Campellone K. G., Robbins D., Leong J. M. (2004). EspFU is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Dev. Cell 7, 217–228 10.1016/j.devcel.2004.07.004 [DOI] [PubMed] [Google Scholar]
- Chang D. E., Smalley D. J., Tucker D. L., Leatham M. P., Norris W. E., Stevenson S. J., Anderson A. B., Grissom J. E., Laux D. C., Cohen P. S., Conway T. (2004). Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. U.S.A. 101, 7427–7432 10.1073/pnas.0408439102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase-Topping M., Gally D., Low C., Matthews L., Woolhouse M. (2008). Supershedding and the link between human and livestock infection with Escherichia coli O157:H7. Nat. Rev. Microbiol. 6, 904–912 10.1038/nrmicro2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Krogfelt K. A., Cohen P. S. (2004). The Life of Commensal Escherichia coli in the Mammalian Intestine. Washington, DC: ASM Press; [DOI] [PubMed] [Google Scholar]
- Conway T., Krogfelt K. A., Cohen P. S. (2007). “Escherichia coli at the intestinal mucosal surface,” in Virulence Mechanisms of Bacterial Pathogens, 4th Edn, eds Brogden K. A., Minion F. C., Cornick N., Stanton T. B., Zhang Q., Nolan L. K., Wannemuehler M. J. (Washington, DC: ASM Press; ), 175–196 [Google Scholar]
- Dahan S., Knutton S., Shaw R. K., Crepin V. F., Dougan G., Frankel G. (2004). Transcriptome of enterohemorrhagic Escherichia coli O157 adhering to eukaryotic plasma membranes. Infect. Immun. 72, 5452–5459 10.1128/IAI.72.9.5452-5459.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. (1980). Broad host range DNA cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. U.S.A. 77, 7347–7351 10.1073/pnas.77.12.7347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziva F., Mahajan A., Cameron P., Currie C., Mckendrick I. J., Wallis T. S., Smith D. G., Stevens M. P. (2007). EspP, a Type V-secreted serine protease of enterohaemorrhagic Escherichia coli O157:H7, influences intestinal colonization of calves and adherence to bovine primary intestinal epithelial cells. FEMS Microbiol. Lett. 271, 258–264 10.1111/j.1574-6968.2007.00724.x [DOI] [PubMed] [Google Scholar]
- Dziva F., Van Diemen P. M., Stevens M. P., Smith A. J., Wallis T. S. (2004). Identification of Escherichia coli O157:H7 genes influencing colonization of the bovine gastrointestinal tract using signature-tagged mutagenesis. Microbiology 150, 3631–3645 10.1099/mic.0.27448-0 [DOI] [PubMed] [Google Scholar]
- Eckert S. E., Dziva F., Chaudhuri R. R., Langridge G. C., Turner D. J., Pickard D. J., Maskell D. J., Thomson N. R., Stevens M. P. (2011). Retrospective application of transposon-directed insertion site sequencing to a library of signature-tagged mini-Tn5Km2 mutants of Escherichia coli O157:H7 screened in cattle. J. Bacteriol. 193, 1771–1776 10.1128/JB.01292-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott S. J., Yu J., Kaper J. B. (1999). The cloned locus of enterocyte effacement from enterohemorrhagic Escherichia coli O157:H7 is unable to confer the attaching and effacing phenotype upon E. coli K-12. Infect. Immun. 67, 4260–4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabich A. J., Jones S. A., Chowdhury F. Z., Cernosek A., Anderson A., Smalley D., Mchargue J. W., Hightower G. A., Smith J. T., Autieri S. M., Leatham M. P., Lins J. J., Allen R. L., Laux D. C., Cohen P. S., Conway T. (2008). Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect. Immun. 76, 1143–1152 10.1128/IAI.01386-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galfi P., Neogrady S., Semjen G., Bardocz S., Pusztai A. (1998). Attachment of different Escherichia coli strains to cultured rumen epithelial cells. Vet. Microbiol. 61, 191–197 10.1016/S0378-1135(98)00175-8 [DOI] [PubMed] [Google Scholar]
- Grys T. E., Siege M. B., Lathem W. W., Welch R. A. (2005a). The StcE protease contributes to intimate adherence of enterohemorrhagic Escherichia coli O157:H7 to host cells. Infect. Immun. 73, 1295–1303 10.1128/IAI.73.3.1295-1303.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grys T. E., Siegel M. B., Lathem W. W., Welch R. A. (2005b). The StcE protease contributes to intimate adherence of enterohemorrhagic Escherichia coli O157:H7 to host cells. Infect. Immun. 73, 1295–1303 10.1128/IAI.73.3.1295-1303.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Makino K., Ohnishi M., Kurokawa K., Ishii K., Yokoyama K., Han C. G., Ohtsubo E., Nakayama K., Murata T., Tanaka M., Tobe T., Iida T., Takami H., Honda T., Sasakawa C., Ogasawara N., Yasunaga T., Kuhara S., Shiba T., Hattori M., Shinagawa H. (2001). Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8, 11–22 10.1093/dnares/8.1.47 [DOI] [PubMed] [Google Scholar]
- Ho T. D., Davis B. M., Ritchie J. M., Waldor M. K. (2008). Type 2 secretion promotes enterohemorrhagic Escherichia coli adherence and intestinal colonization. Infect. Immun. 76, 1858–1865 10.1128/IAI.00795-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. D., Shlumukov L., Carland F., English J., Scofield S. R., Bishop G. J., Harrison K. (1992). Effective vectors for transformation, expression of heterologous genes, and assaying transposon excision in transgenic plants. Transgenic Res. 1, 285–297 10.1007/BF02525170 [DOI] [PubMed] [Google Scholar]
- Lee S. H., Galan J. E. (2004). Salmonella type III secretion-associated chaperones confer secretion-pathway specificity. Mol. Microbiol. 51, 483–495 10.1111/j.1365-2958.2003.03946.x [DOI] [PubMed] [Google Scholar]
- Lim J. Y., Sheng H., Seo K. S., Park Y. H., Hovde C. J. (2007). Characterization of an Escherichia coli O157:H7 plasmid O157 deletion mutant and its survival and persistence in cattle. Appl. Environ. Microbiol. 73, 2037–2047 10.1128/AEM.02643-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low A. S., Dziva F., Torres A. G., Martinez J. L., Rosser T., Naylor S., Spears K., Holden N., Mahajan A., Findlay J., Sales J., Smith D. G., Low J. C., Stevens M. P., Gally D. L. (2006a). Cloning, expression, and characterization of fimbrial operon F9 from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 74, 2233–2244 10.1128/IAI.74.4.2233-2244.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low A. S., Holden N., Rosser T., Roe A. J., Constantinidou C., Hobman J. L., Smith D. G., Low J. C., Gally D. L. (2006b). Analysis of fimbrial gene clusters and their expression in enterohaemorrhagic Escherichia coli O157:H7. Environ. Microbiol. 8, 1033–1047 10.1111/j.1462-2920.2006.00995.x [DOI] [PubMed] [Google Scholar]
- Mahajan A., Naylor S., Mills A. D., Low J. C., Mackellar A., Hoey D. E. E., Currie C. G., Gally D. L., Huntley J., Smith D. G. E. (2005). Phenotypic and functional characterisation of follicle-associated epithelium of rectal lymphoid tissue. Cell Tissue Res. 321, 365–374 10.1007/s00441-005-1080-1 [DOI] [PubMed] [Google Scholar]
- Matthews L., Low J. C., Gally D. L., Pearce M. C., Mellor D. J., Heesterbeek J. A., Chase-Topping M., Naylor S. W., Shaw D. J., Reid S. W., Gunn G. J., Woolhouse M. E. (2006). Heterogeneous shedding of Escherichia coli O157 in cattle and its implications for control. Proc. Natl. Acad. Sci. U.S.A. 103, 547–552 10.1073/pnas.0503776103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor S. W., Low J. C., Besser T. E., Mahajan A., Gunn G. J., Pearce M. C., Mckendrick I. J., Smith D. G., Gally D. L. (2003). Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect. Immun. 71, 1505–1512 10.1128/IAI.71.3.1505-1512.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y., Ooka T., Iguchi A., Toh H., Asadulghani M., Oshima K., Kodama T., Abe H., Nakayama K., Kurokawa K., Tobe T., Hattori M., Hayashi T. (2009). Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 106, 17939–17944 10.1073/pnas.0903585106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi M., Terajima J., Kurokawa K., Nakayama K., Murata T., Tamura K., Ogura Y., Watanabe H., Hayashi T. (2002). Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc. Natl. Acad. Sci. U.S.A. 99, 17043–17048 10.1073/pnas.262441699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna N. T., Plunkett G., III, Burland V., Mau B., Glasner J. D., Rose D. J., Mayhew G. F., Evans P. S., Gregor J., Kirkpatrick H. A., Posfai G., Hackett J., Klink S., Boutin A., Shao Y., Miller L., Grotbeck E. J., Davis N. W., Lim A., Dimalanta E. T., Potamousis K. D., Apodaca J., Anantharaman T. S., Lin J., Yen G., Schwartz D. C., Welch R. A., Blattner F. R. (2001). Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409, 529–533 10.1038/35054089 [DOI] [PubMed] [Google Scholar]
- Roe A. J., Currie C., Smith D. G., Gally D. L. (2001). Analysis of type 1 fimbriae expression in verotoxigenic Escherichia coli: a comparison between serotypes O157 and O26. Microbiology 147, 145–152 [DOI] [PubMed] [Google Scholar]
- Roe A. J., Tysall L., Dransfield T., Wang D., Fraser-Pitt D., Mahajan A., Constandinou C., Inglis N., Downing A., Talbot R., Smith D. G. E., Gally D. L. (2007). Analysis of the expression, regulation and export of NleA-E in Escherichia coli O157:H7. Microbiology 153, 1350–1360 10.1099/mic.0.2006/003707-0 [DOI] [PubMed] [Google Scholar]
- Roe A. J., Yull H., Naylor S. W., Woodward M. J., Smith D. G. E., Gally D. L. (2003). Heterogeneous surface expression of EspA translocon filaments by Escherichia coli O157:H7 is controlled at the posttranscriptional level. Infect. Immun. 71, 5900–5909 10.1128/IAI.71.10.5900-5909.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng H., Lim J. Y., Knecht H. J., Li J., Hovde C. J. (2006). Role of Escherichia coli O157:H7 virulence factors in colonization at the bovine terminal rectal mucosa. Infect. Immun. 74, 4685–4693 10.1128/IAI.00406-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider T. A., Fabich A. J., Conway T., Clinkenbeard K. D. (2009). E. coli O157:H7 catabolism of intestinal mucin-derived carbohydrates and colonization. Vet. Microbiol. 136, 150–154 10.1016/j.vetmic.2008.10.033 [DOI] [PubMed] [Google Scholar]
- Stevens M. P., Roe A. J., Vlisidou I., Van Diemen P. M., La Ragione R. M., Best A., Woodward M. J., Gally D. L., Wallis T. S. (2004). Mutation of toxB and a truncated version of the efa-1 gene in Escherichia coli O157:H7 influences the expression and secretion of locus of enterocyte effacement-encoded proteins but not intestinal colonization in calves or sheep. Infect. Immun. 72, 5402–5411 10.1128/IAI.72.9.5402-5411.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuno I., Horie M., Abe H., Miki T., Makino K., Shinagawa H., Taguchi H., Kamiya S., Hayashi T., Sasakawa C. (2001). toxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect. Immun. 69, 6660–6669 10.1128/IAI.69.11.6660-6669.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres B., Porras G., Garcia J. L., Diaz E. (2003). Regulation of the mhp cluster responsible for 3-(3-hydroxyphenyl)propionic acid degradation in Escherichia coli. J. Biol. Chem. 278, 27575–27585 10.1074/jbc.M201926200 [DOI] [PubMed] [Google Scholar]
- Tree J. J., Roe A. J., Flockhart A., Mcateer S. P., Xu X., Shaw D., Mahajan A., Beatson S. A., Best A., Lotz S., Woodward M. J., La Ragione R., Murphy K. C., Leong J. M., Gally D. L. (2011). Transcriptional regulators of the GAD acid stress island are carried by effector protein-encoding prophages and indirectly control type III secretion in enterohemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 80, 1349–1365 10.1111/j.1365-2958.2011.07650.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Diemen P. A., Dziva F., Stevens M. P., Wallis T. S. (2005). Identification of enterohemorrhagic Escherichia coli O26:H-genes required for intestinal colonization in calves. Infect. Immun. 73, 1735–1743 10.1128/IAI.73.3.1735-1743.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells T. J., Sherlock O., Rivas L., Mahajan A., Beatson S. A., Torpdahl M., Webb R. I., Allsopp L. P., Gobius K. S., Gally D. L., Schembri M. A. (2008). EhaA is a novel autotransporter protein of enterohemorrhagic Escherichia coli O157:H7 that contributes to adhesion and biofilm formation. Environ. Microbiol. 10, 589–604 10.1111/j.1462-2920.2007.01479.x [DOI] [PubMed] [Google Scholar]
- Zhang Y., Laing C., Steele M., Ziebell K., Johnson R., Benson A. K., Taboada E., Gannon V. P. (2007). Genome evolution in major Escherichia coli O157:H7 lineages. BMC Genomics 8, 121. 10.1186/1471-2164-8-121 [DOI] [PMC free article] [PubMed] [Google Scholar]