Abstract
DNA double strand breaks (DSB) can lead to development of genomic rearrangements, which are hallmarks of cancer. TMPRSS2-ERG gene fusions in prostate cancer (PCa) are among the most common genomic rearrangements observed in human cancer. We show that androgen signaling promotes co-recruitment of androgen receptor (AR) and topoisomerase II beta (TOP2B) to sites of TMPRSS2-ERG genomic breakpoints, triggering recombinogenic TOP2B-mediated DSB. Furthermore, androgen stimulation resulted in de novo production of TMPRSS2-ERG fusion transcripts in a process requiring TOP2B and components of DSB repair machinery. Finally, unlike normal prostate epithelium, prostatic intraepithelial neoplasia (PIN) cells showed strong co-expression of AR and TOP2B. These findings implicate androgen-induced TOP2B-mediated DSB in generating TMPRSS2-ERG rearrangements.
Genomic rearrangements creating cell-type-specific fusion oncogenes are hallmarks of cancer1,2. The critical step for development of such genomic rearrangements is the generation of DNA double strand breaks (DSB) and subsequent illegitimate repair of these breaks3,4. However, the factors involved in DSB genesis and the mechanisms producing cell-type specific genomic rearrangements are not well understood for most malignancies. In human PCa, a number of recurrent rearrangements involving various AR target genes fused to oncogenic ETS family transcription factors have been described5. The most common of these rearrangements, occurring in nearly 50% of all prostate cancers, involves fusion of the AR target TMPRSS2 with the ETS transcription factor ERG, resulting in activation of the TMPRSS2-ERG fusion oncogene via subversion of androgen signaling5,6. Given the high prevalence of prostate cancer in the population7 and the high fraction of prostate cancers harboring this fusion oncogene5, the TMPRSS2-ERG gene fusion is the most common fusion gene observed in any human cancer. However, the mechanisms involved in generating the DSB allowing development of these TMPRSS2-ERG rearrangements are not completely understood.
Dysfunction of topoisomerase II (TOP2), an enzyme that catalyzes a transient DSB to resolve DNA topological constraints, can produce DSB implicated in the pathogenesis of rearrangements in treatment related acute myeloid leukemia (t-AML) and childhood leukemias8–11. Estrogen receptor (ER) signaling has been reported to recruit the TOP2 isoform TOP2B to regulatory regions of ER target genes, leading to TOP2B mediated DSB12. We hypothesized that AR signaling may lead to TOP2B mediated DSB, and that such breaks may be involved in the generation of TMPRSS2-ERG recurrent rearrangements in PCa.
We show that androgen signaling leads to co-recruitment of AR and TOP2B and TOP2B mediated double strand breaks at regulatory regions of AR target genes as well as to regions of the TMPRSS2 and ERG loci that were highly aligned with genomic breakpoints of TMPRSS2-ERG fusion genes in prostate cancer cases. These androgen-induced TOP2B mediated DSB were highly recombinogenic and could lead to de novo production of TMPRSS2-ERG fusion transcripts in a TOP2B dependent manner. In human prostate tissues, AR and TOP2B, which were rarely both present in normal TMPRSS2-ERG fusion negative prostate cells at high levels, were both robustly co-expressed in TMPRSS2-ERG fusion-positive neoplastic cells comprising prostatic intraepithelial neoplasia (PIN) lesions, known precursors to prostate cancers. These findings implicate AR signaling and TOP2B action in the generation of TMPRSS2-ERG rearrangements. TOP2B-mediated DSB occurring during regulated transcription represents a new paradigm for development of DSB involved in generating gene rearrangements in cancer.
RESULTS
TOP2B is required for efficient androgen-mediated gene expression
We used LNCaP and LAPC4 PCa cell lines, which have intact androgen signaling (Supplementary Fig. 1) but are known not to harbor the TMPRSS2-ERG rearrangements, to examine whether TOP2B was required for efficient activation of AR target genes in androgen-deprived cells stimulated with dihydrotestosterone (DHT), a prototypical androgen. By genome-wide gene expression microarray analysis, pharmacological inhibition (with etoposide (ET) or merbarone (Mer)) or RNAi mediated depletion of TOP2B (sh-TOP2B) coordinately attenuated the up-regulation of DHT-induced genes, a finding confirmed by quantitative RT-PCR of candidate genes in both LNCaP and LAPC4 cells (Fig. 1 and Supplementary Fig. 1, 2, 3). Gene set enrichment analysis13,14 revealed that androgen induced transcriptional programs were enriched for down-regulation by TOP2 inhibition (p < 10−14) or sh-TOP2B (p < 10−2) compared to all other genes (Fig. 1a,b, and Supplementary Fig. 2,3). These analyses demonstrate that there is a concordant down-regulation of AR regulated genes with both pharmacological and genetic modulations of TOP2B.
Figure 1.
TOP2B is required for efficient induction of AR target gene expression following androgen stimulation. a, Inhibition of TOP2B with ET or Mer prior to DHT stimulation of LNCaP cells attenuated expression of an androgen-induced geneset (p < 10−14), defined as the 45 genes upregulated by >2-fold in DHT vs. control treated LNCaP cells. b, Geneset enrichment analysis shows highly statistically significant enrichment for down regulation of three independently derived but overlapping androgen-responsive genesets in ET/DHT vs. DHT, Mer/DHT vs. DHT, and sh-TOP2B/DHT vs DHT (see also Supplementary Fig. 2,3). Y-axis shows the degree of confidence for geneset enrichment as −log10(p-value). Significance threshold corresponding to p = 0.05 is indicated. c–d, Real-time RT-PCR analysis of relative expression of selected genes with respect to GAPDH expression confirms down-regulation of representative androgen-responsive genes in ET/DHT, Mer/DHT, and sh-TOP2B/DHT. Data are shown as mean ± SE of two to three replicates.
Androgen-induced co-recruitment of AR/TOP2B and TOP2B catalytic activity at regulatory regions of AR target genes
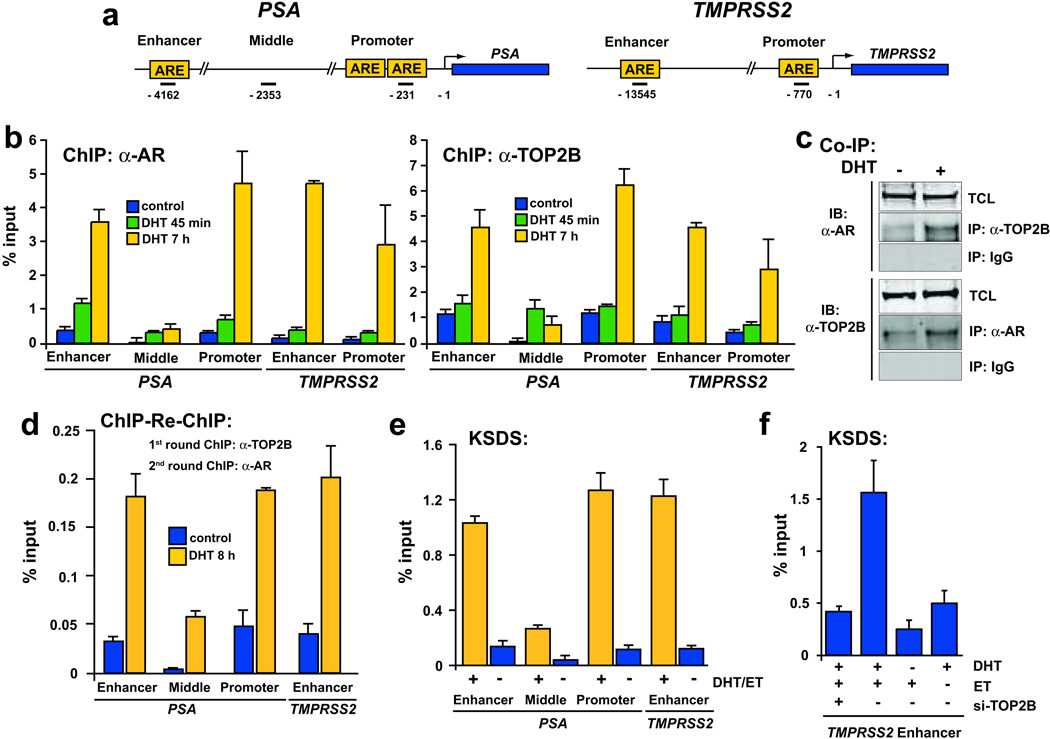
We next examined whether TOP2B was directly involved in androgen induction of AR target genes. Chromatin immunoprecipitation (ChIP) experiments revealed coordinate recruitment of both AR and TOP2B to the enhancer and promoter regions of PSA and TMPRSS2. This coordinate recruitment at regulatory regions appeared to be fairly specific since our ChIP assay did not show recruitment to the “middle” region of PSA or to regulatory regions of other arbitrarily selected genes (Fig. 2a,b, and Supplementary Fig. 4). AR and TOP2B are recruited to these regulatory regions as part of a complex, since DHT-stimulation resulted in co-immunoprecipitation of TOP2B and AR (Fig. 2c) and co-recruitment of TOP2B and AR to AREs of PSA and TMPRSS2 in ChIP-re-ChIP experiments (Fig. 2d). This AR/TOP2B complex formation was not dependent on the presence of DNA since pre-treatment of lysates with DNAseI or vast excesses of ethidium bromide did not significantly disrupt complex formation (Supplementary Fig. 4). To ascertain whether TOP2B was catalytically cleaving genomic DNA at PSA and TMPRSS2 regulatory regions, a potassium SDS (KSDS) assay15, which allows isolation of etoposide-stabilized TOP2 catalytic cleavage complexes, was conducted, demonstrating robust TOP2 cleavage at the PSA enhancer and promoter and TMPRSS2 enhancer but not at regions that did not contain AR or TOP2B binding sites (Fig. 2e, and Supplementary Fig. 5). This activity was abolished by si-TOP2B treatment prior to DHT stimulation (Fig. 2f, and Supplementary Fig. 6), suggesting that the observed catalytic activity was due to TOP2B.
Figure 2.
Androgen stimulation induced co-recruitment of AR/TOP2B and TOP2B catalytic cleavage at known regulatory regions of AR target genes. a, schematic of AREs in the promoter and enhancer region of PSA and TMPRSS2. Bars indicate positions (relative to transcriptional start site) of amplicons analyzed. b, ChIP analysis of androgen-deprived LAPC4 cells that were stimulated with 100 nM DHT for varying timepoints as indicated reveals DHT-induced recruitment of AR and TOP2B to the enhancer and promoter of PSA and TMPRSS2 but not to an intervening region (middle) between the enhancer and promoter of PSA. c, Stimulation of androgen-depleted LAPC4 cells with 100 nM DHT for 6 h allows co-immunoprecipitation of TOP2B with AR antibodies and vice versa. IP, immunoprecipitating antibody; IB, immunoblotting antibody; TCL, total cell lysate. d, ChIP-re-ChIP experiments demonstrate that stimulation of androgen-deprived LAPC4 cells with 100 nM DHT induces co-recruitment of AR and TOP2B to AREs of PSA and TMPRSS2. A first round of ChIP was performed using anti-TOP2B antibodies and the resulting immunoprecipitates were subjected to a second round of ChIP with anti-AR antibodies. Relative enrichment was determined by qPCR and is shown as percentage of input DNA. e–f, KSDS method shows TOP2 catalytic cleavage of AR target sites in LAPC4 cells in a DHT and TOP2B dependent fashion. Results are presented as mean percentage to input with ± SE of two to three replicates.
Androgen stimulation induces AR and TOP2B binding and catalytic activity at TMPRSS2 and ERG loci near genomic breakpoints observed in prostate cancer tissues
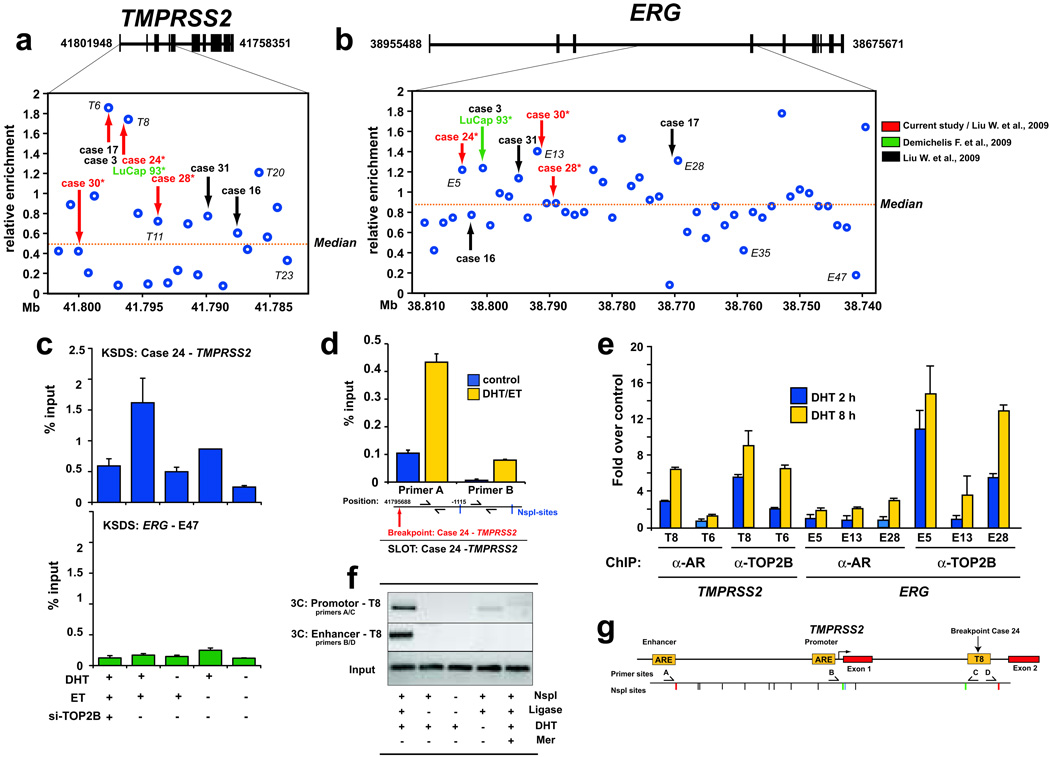
Since TOP2 cleavage can produce fragile sites for development of DSB leading to genomic rearrangements in other malignancies8–11, we hypothesized that androgen mediated TOP2 catalytic cleavage at key sites within TMPRSS2 and/or ERG might create DSB and TMPRSS2-ERG genomic rearrangements in PCa. Examination of previously published datasets revealed that genomic breakpoints in TMPRSS2 and ERG occur most frequently at the ~25 kbp region between exon 1 and exon 4 of TMPRSS2 and at a ~75 kbp stretch in intron 3 of ERG16–18 (Supplementary Table 1). We identified breakpoints at high resolution in 8 cases described in these previously published reports, and sequenced the TMPRSS2 and ERG genomic breakpoints to nucleotide resolution in 3 of these cases (Fig. 3a,b and Supplementary Fig. 7 and Supplementary Table 1). Surprisingly, using the KSDS assay and tiling 71 real-time PCR amplicons across the ~100 kbp surrounding the TMPRSS2 and ERG genomic segments showing hotspots for breakpoints, we observed regions with high levels of DHT-induced TOP2 cleavage that were significantly enriched for proximity to TMPRSS2 (p = 0.010) and ERG (p = 0.013) breakpoints (Fig. 3a,b).
Figure 3.
Androgen stimulation induces AR/TOP2B recruitment and TOP2B catalytic cleavage at genomic breakpoints of TMPRSS2 and ERG observed in human PCa. a–b, Sites closest to TMPRSS2-ERG breakpoints from 8 PCa cases determined from various studies (arrows) were significantly enriched for high KSDS enrichment of TOP2 catalytic cleavage (p = 0.010 and 0.013 respectively) in LAPC4 cells. Labeled sites (e.g., T8, T23, E5, E13, etc.) are analyzed in subsequent experiments. c, DHT and TOP2B dependent TOP2 catalytic cleavage in LAPC4 cells around the case 24 breakpoint aligning with region T8 (upper panel). The lack of KSDS enrichment at region E47 at ERG was re-confirmed in an independent KSDS experiment (lower panel). d, SLOT assay showed that DHT-induced TOP2 catalytic activity in LAPC4 cells was significantly higher at an NspI fragment most proximal to the TMPRSS2 breakpoint observed in case 24 than to the adjacent, more distal NspI fragment (see Supplementary Fig. 8, 9 for overview of SLOT). e, ChIP enrichment of AR and TOP2B at representative TOP2 catalytic cleavage sites in DHT-stimulated LAPC4 cells relative to untreated controls. f–g, 3C analysis reveals DHT-dependent spatial chromatin interaction of TMPRSS2 enhancer and promoter with region T8 (see first lane vs. fourth lane). Omission of NspI restriction enzyme and/or DNA ligase served as assay negative controls. Inhibition with Mer prevented these DHT-induced interactions. Error bars indicate ± SE of two to three experiments.
Examining the breakpoints of 2 of the 3 subjects (cases 24 and 30) for whom we sequenced rearrangement junctions, we found DHT-induced TOP2 cleavage that was abolished by si-TOP2B treatment in LAPC4 cells at the precise TMPRSS2 and ERG breakpoints seen in these cases (Fig. 3c and Supplementary Fig. 8). To verify this association further, we developed a novel assay for selective labeling of TOP2 cleavage sites (SLOT), which allows specific labeling of the etoposide-stabilized TOP2-DNA cleavage complex (Supplementary Fig. 8,9). Using SLOT, we observed a strong enrichment for DHT-dependent TOP2 cleavage at the NspI restriction fragment closest to the precise TMPRSS2-ERG genomic breakpoint observed in cases 24 and 30 (Fig. 3d and Supplementary Fig. 8).
Recent studies have demonstrated that AR binds specifically to thousands of genomic sites even if those sites are devoid of canonical AREs19. Since the sites showing high DHT-induced TOP2B cleavage within the TMPRSS2 and ERG gene bodies did not contain known AREs, we carried out ChIP experiments to understand whether TOP2 cleavage at these sites was a consequence of such non-canonical DHT-induced AR recruitment. As expected, all of the sites that showed high TOP2 cleavage (e.g., T8, T6, E5, E13, E28 shown in Fig. 3a) demonstrated DHT-induced co-recruitment of both TOP2B and AR (1.5 – 15 fold over control; Fig. 3e), suggesting direct androgen-induced AR-coupled TOP2B recruitment to these sites, while sites that did not show strong TOP2B cleavage (e.g., T23, E47, E35) showed weak or absent recruitment of both TOP2B and AR by ChIP (Supplementary Fig. 11).
In further support of a DHT-induced regulatory role for these intragenic AR/TOP2B target sites, a representative site, T8, showed a specific DHT-induced chromatin interaction with both the TMPRSS2 enhancer and promoter in 3C experiments (Fig. 3f,g, and Supplementary Fig. 12)20. Consistent with the hypothesis that TOP2B recruitment and activity at these sites as shown above is required for assembly of these enhancer- and promoter-T8 chromatin complexes, treatment of androgen deprived LAPC4 cells with Mer prior to DHT-stimulation abolished formation of these complexes. We can speculate that TOP2B activity allows resolution of topological entanglements created during formation of these transcription-associated chromatin loops.
Androgen stimulation leads to formation of TOP2B dependent DSB that are recognized by the DSB repair machinery
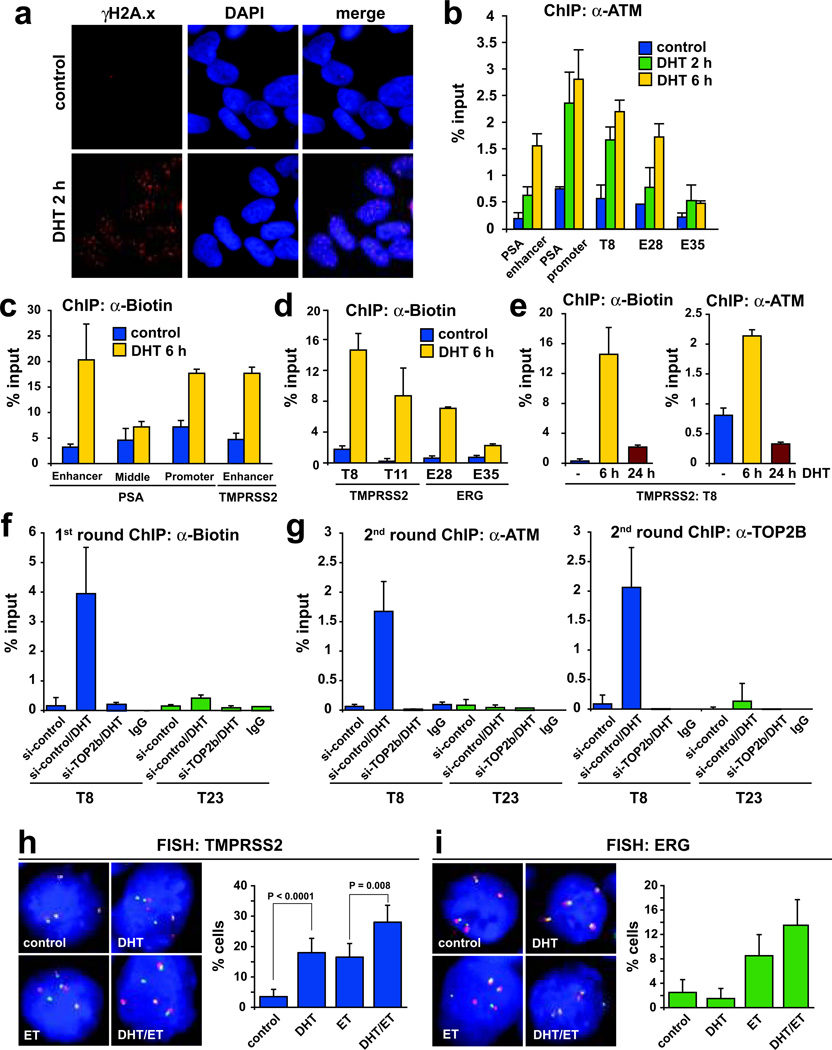
We next tested whether sites showing DHT-induced AR/TOP2B recruitment and TOP2B catalytic cleavage may be fragile sites for persistent DSB formation. We found that DHT-stimulation could induce widespread formation of γH2A.x foci (Fig. 4a), which are well known to form around DSB21. We also observed pronounced ATM recruitment, a central feature of DSB repair pathways22,23, to sites showing strong AR/TOP2B recruitment and TOP2B catalytic activity (PSA enhancer and promoter, regions T6, T8, T20, E5, E13, E28), but much less so to a region in the ERG gene (E35) that did not show significant TOP2 catalytic activity (Fig. 4b and Supplementary Fig. 13).
Figure 4.
Androgen stimulation results in TOP2B dependent DSB formation. a, Androgen stimulation induced γH2A.x foci formation in LAPC4 cells. b, Androgen stimulation led to recruitment of ATM to representative sites of high TOP2B activity but much less so to a region showing low TOP2B catalytic cleavage (E35). c–d, Stimulation of LAPC4 cells with DHT induced DSB that could be end-labeled with biotin at representative TOP2 cleavage sites, but not to regions showing low TOP2 cleavage (PSA middle, E35). e, DSB largely resolve by 24 h following DHT stimulation of LAPC4 cells as evidenced by reduced biotin labeling and ATM recruitment. The time course of DSB formation/resolution and ATM recruitment are highly parallel. f, Left panel, DHT-induced DSB at a representative TOP2B enriched site (T8) are dependent on TOP2B. A site showing low TOP2B enrichment (T23) served as a negative control. g, DHT-induced strand breaks at representative TMPRSS2 sites are enriched for TOP2B and ATM binding as seen by ChIP-re-ChIP experiments. Results are presented as mean percentage to input with ± SE of two to three replicates. h, DHT induces chromosomal breaks at TMPRSS2 in LAPC4 cells, monitored as fraction of cells (n > 200) showing “split-apart” of FISH probes 5’ (green) and 3’ (red) of TMPRSS2. i, Treatment of LAPC4 cells with ET plus/minus DHT increased split-aparts at ERG compared to control or DHT treatment. Error bars indicate 95% confidence intervals.
To detect DSB at relevant sites directly, we labeled these breaks with biotin-conjugated nucleotides in situ using terminal DNA transferase 12(Supplementary Fig. 13). In parallel with ATM recruitment, DHT induced DSB at sites of TOP2B recruitment/catalytic activity and AR binding within the TMPRSS2 and ERG genes (Fig. 4c,d). Conversely, regions at PSA and ERG that did not show significant TOP2B catalytic activity (Middle, E35) did not show significant DSB formation (Fig. 4d). DSB and ATM recruitment appeared within the first 6 hours after DHT stimulation and largely resolved after 24 hours (Fig. 4e and Supplementary Fig. 13). ChIP-re-ChIP experiments showed recruitment of ATM to the biotin-labeled strand breaks, indicating that these breaks were truly DSB and not just single strand nicks (Fig. 4f,g). A ligation-mediated PCR (LM-PCR) method allowed single nucleotide mapping of breaks occurring at a representative region showing high DHT induced TOP2Β enrichment/catalytic cleavage, ATM recruitment, and close alignment to a TMPRSS2 genomic breakpoint in a clinical PCa case (region T6; Supplementary Fig. 14). This provided further direct evidence of DSB at these sites. TOP2B was also present at these biotin-labeled DSB in ChIP-re-Chip experiments, pointing to its involvement in generating these breaks (Fig. 4g and Supplementary Fig. 13). In support of this, RNAi mediated depletion of TOP2B or AR (sh-AR) significantly reduced formation of biotin-labeled strand breaks (Fig. 4f, and Supplementary Fig. 13).
To investigate whether these DSB could result in genomic breaks and/or chromosomal rearrangements at an individual cell level, we performed dual-color fluorescence in situ hybridization (FISH) assays with a pair of probes flanking the TMPRSS2 or ERG genes (Supplementary Fig. 15). A significant separation, or “split-apart”, of these probes would indicate the presence of a genomic break or rearrangement occurring at the intervening genomic sequence. As a positive control, treatment of LAPC4 cells with ET, a known TOP2-dependent genomic-break inducing agent, resulted in generation of “split-aparts” at both the TMPRSS2 and ERG loci in ~16% and ~8% of cells respectively (Fig. 4h,i). Interestingly, stimulation of LAPC4 and LNCaP cells with DHT alone resulted in a significant increase in frequency of genomic breaks at TMPRSS2 compared to unstimulated cells (Fig. 4h and Supplementary Fig. 16). Similarly, stimulation with DHT followed by treatment with ET showed increased split-aparts compared to control, DHT alone, or ET alone treated LNCaP and LAPC4 cells at both the TMPRSS2 and ERG loci (Fig. 4h,i, and Supplementary Fig. 16). This DHT induced increase in split-aparts was significantly attenuated by si-TOP2B (Supplementary Fig. 16). These data suggest that DHT stimulation could cooperate with TOP2B action to induce genomic breaks and/or chromosomal rearrangements at these loci.
Androgen-induced TOP2B mediated breaks are highly recombinogenic and can lead to de novo production of TMPRSS2-ERG fusion genes
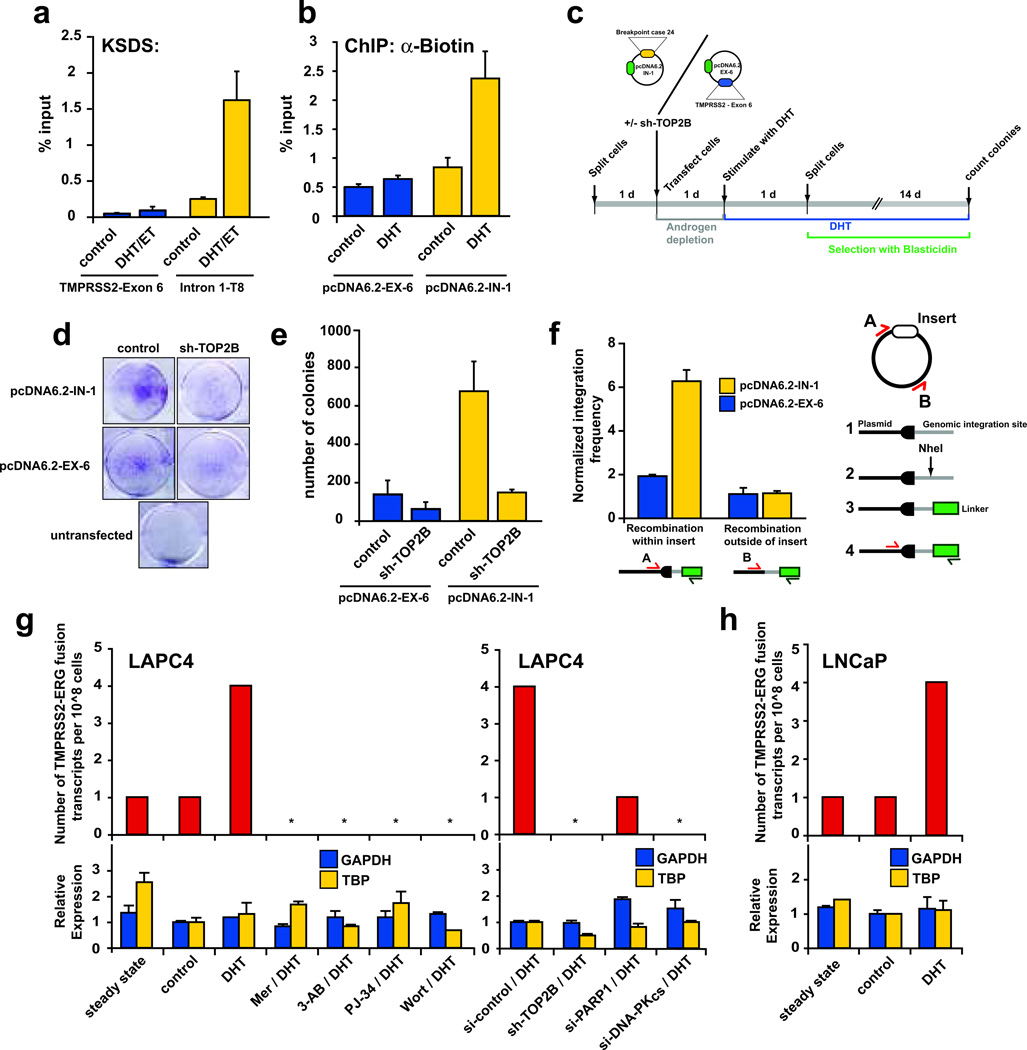
We next devised an androgen induced genomic recombination assay to assess whether these androgen-induced TOP2B mediated DSB were competent recombination foci, and whether these breaks could significantly increase genomic recombination events at these sites. For this assay, we cloned a region showing high DHT-induced TOP2B catalytic activity (T8 in Intron 1 of TMPRSS2) and one showing low TOP2B catalytic activity (TMPRSS2 exon 6; Fig. 5a), into a plasmid vector containing a blasticidin resistance gene to create the pcDNA6.2-IN-1 (IN-1) and pcDNA6.2-EX-6 (EX-6) constructs respectively. Transfection of these constructs into LAPC4 cells followed by DHT stimulation led to increased formation of biotin-labeled DSB in the IN-1 insert, but not at the EX-6 insert (Fig. 5b). To assess whether these DHT-induced breaks increased recombination efficiency, we transfected the IN-1 or EX-6 constructs along with control or sh-TOP2B constructs into LAPC4 cells, depleted media of androgens, stimulated with DHT for 24h to induce DSB formation, split cells to clonogenic density, and subjected to blasticidin selection in androgen containing media for 2 wks. The number of colonies formed after 14 days would indicate the number of successful recombination events allowing incorporation of blasticidin resistance vectors into genomic DNA (Fig. 5c). Consistent with our hypothesis, the IN-1 vector produced significantly more colonies than the EX-6 construct, and this increased frequency was abolished by sh-TOP2B (Fig. 5d,e). Transfection of LAPC4 cells with sh-TOP2B alone without blasticidin selection did not decrease clonogenic survival compared to control transfected cells (Supplementary Fig. 17). Furthermore, both plasmids were equally competent in producing recombination events if they were linearized with a restriction enzyme prior to transfection (Supplementary Fig. 18). Additionally, an LM-PCR strategy showed that the increased frequency of recombination in the IN-1 construct was due to recombination events occurring within the insert and not non-specifically at the vector backbone (Fig. 5f). These data provide evidence that DHT-induced TOP2B mediated DSB at target sites can promote genomic recombination of these target sites.
Figure 5.
Androgen-induced TOP2B mediated DSB are recombinogenic and promote de novo production of TMPRSS2-ERG fusion genes. a, Selection of TMPRSS2 regions showing high (Intron 1-T8) and low (TMPRSS2-Exon 6) KSDS enrichment in response to DHT stimulation of LAPC4 cells. b, DHT-induced breaks can be detected in plasmids containing sequences surrounding region T8 of TMPRSS2 intron 1 (pcDNA6.2-IN-1) as evidenced by increased biotin labeling in DHT stimulated LAPC4 cells transfected with this plasmid. Plasmids containing TMPRSS2 exon 6 (pcDNA6.2-EX-6) served as a negative control. c, Schematic of androgen-induced genomic recombination assay in LAPC4 cells transfected with pcDNA6.2-IN-1 or pcDNA6.2-EX-6, which contain a blasticidin resistance gene. Number of colonies represents the number of recombination events allowing integration of the blasticidin resistance vectors into the LAPC4 genome. d, Representative results of genomic recombination assays. e, pcDNA6.2-IN-1 transfected LAPC4 cells produced significantly more androgen induced recombination events than pcDNA6.2-EX-6 transfected cells. Treatment with sh-TOP2B abolished this effect. f, While both showed similar recombination frequency at the vector backbone, recombination frequency within the IN-1 insert was significantly higher than that in the EX-6 insert in pooled colonies as determined using the strategy shown on the right. Data are shown as mean ± SE of two to three replicates. g, DHT-stimulation of LAPC4 cells leads to increased TMPRSS2-ERG fusion transcripts compared to background levels in LAPC4 cells grown in androgen-containing (steady-state) or androgen-deprived (control) media. Pharmacological or genetic modulation of TOP2B (Mer, sh-TOP2B), PARP1 (3-AB, PJ-34, si-PARP1), or DNA-PKCS (Wort, si-DNA-PKCS), reduces TMPRSS2-ERG fusion transcripts without significantly altering GAPDH or TBP expression. h, DHT-stimulation leads to de novo formation of TMPRSS2-ERG fusion transcripts in LNCaP cells.
We next assessed whether DHT-induced TOP2B mediated DSB can result in production of de novo TMPRSS2-ERG fusion genes in LNCaP and LAPC4 cells. We developed a highly sensitive RT-PCR assay capable of detecting fusion transcripts juxtaposing exon 1 of TMPRSS2 with exon 4 of ERG, at a frequency of ~1 in 10^8 cells (Supplementary Fig. 19). Since LAPC4 and LNCaP cells are well known not to harbor TMPRSS2-ERG genomic rearrangements, the low-level of TMPRSS2-ERG fusion transcripts seen when these cells were maintained in their fully supplemented, androgen-containing media (steady state), or when grown in androgen-depleted media (control) represents the background level for this sensitive assay. In four independent experiments involving two different cell lines, DHT stimulation of LAPC4 or LNCaP cells led to increased de novo production of TMPRSS2-ERG fusion transcripts (Fig. 5g,h and Supplementary Fig. 19). Interestingly, pharmacological inhibition or RNAi mediated depletion of TOP2B, or DSB repair pathway components PARP1 or DNA-PKCS, prior to DHT stimulation of LAPC4 cells resulted in complete reduction of de novo TMPRSS2-ERG fusion transcript formation back to background levels (Fig. 5g).
TOP2B and AR are highly co-expressed in luminal cells of prostate cancer precursor lesions but not in normal prostate glands
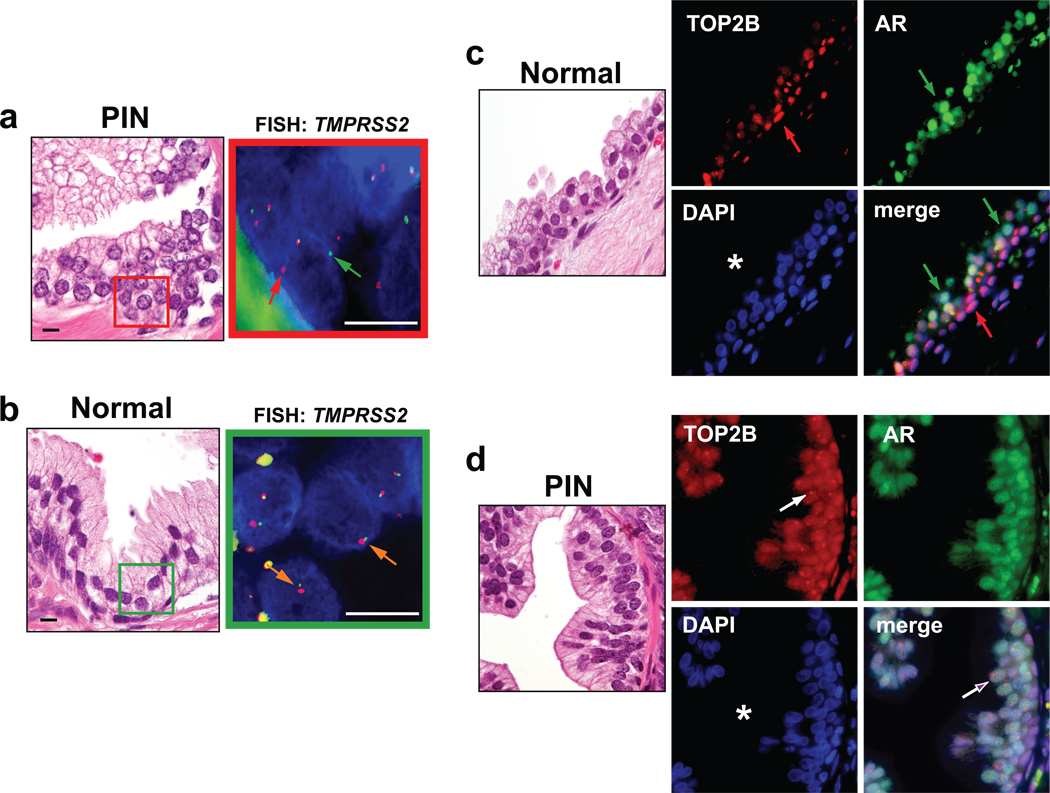
The TMPRSS2-ERG rearrangements are known to occur as early as the PIN PCa precursor lesions24 and are likely selected for during formation of invasive adenocarcinoma5,25. We carried out FISH and co-immunostaining of AR and TOP2B in PIN and matched normal prostatic tissue. In agreement with previous studies24,25, 10 of 23 subjects with PIN and none of the matched benign prostate tissues showed TMPRSS2-ERG rearrangement as determined by TMPRSS2 FISH (Fig. 6a, b and Supplementary Tables 2, 3). Basal cells in the normal prostate glands showed much stronger staining for TOP2B than the luminal cells (Fig. 6c). Inversely, the luminal cells in normal glands showed much stronger staining for AR than the basal cells (Fig. 6c). However, this compartmentalization of TOP2B staining in normal prostate glands became more promiscuous in PIN, with luminal cells expressing nearly as much TOP2B as basal cells in these lesions. AR continued to be highly expressed in these PIN luminal cells, creating a more pronounced co-expression of both TOP2B and AR in the PIN luminal cells (Fig. 6d and Supplementary Table 3). This increased co-expression of TOP2B and AR may predispose PIN luminal cells to developing the AR and TOP2B mediated DSB observed in DHT stimulated LNCaP and LAPC4 cells.
Figure 6.
TMPRSS2-ERG rearrangements are observed in PIN PCa precursor lesions and are associated with changes in the TOP2B expression pattern. a, H&E stain and corresponding TMPRSS2 FISH in a representative PIN lesion. FISH image of red boxed area shows interphase nuclei of PIN lesion with split-aparts of 5’ (green arrow) and 3’ (red arrow) TMPRSS2 FISH probes. b, H&E stain and corresponding TMPRSS2 FISH (green boxed area) of normal prostate epithelium. Orange arrows indicate normal configuration of 5’ and 3’ TMPRSS2 FISH probes. Scale bars indicate 10 µm c, Normal prostate epithelium shows strong expression of TOP2B in basal cells (red arrow) and low expression in luminal cells, whereas AR is predominantly expressed in luminal cells (green arrow). Lumen of prostate gland is marked with an asterisk. d, PIN lesion shows high TOP2B in basal and luminal cells (purple arrow). Note the accentuated nucleolar localization of TOP2B (white arrow).
DISCUSSION
These data suggest a model in which intrinsic androgen signaling in neoplastic prostate cells induces TOP2B mediated DSB at many genomic loci including breakpoints of TMPRSS2-ERG rearrangements in PCa, even without extrinsic DSB inducing genotoxic stress. These androgen-induced TOP2B-dependent DSB are competent to participate in genomic recombination events and can generate rare TMPRSS2-ERG gene rearrangements (Fig. 7). Another report has suggested that in the presence of very high levels of exogenous genotoxic stress, other enzymes such as activation induced cytidine deaminase (AID) and LINE-1 repeat-encoded ORF2 endonuclease may cooperate with AR signaling to establish DSB and, ultimately, prostate cancer gene rearrangements26. Taken together, the data from the current study and this previous report suggest that multiple different enzymatic activities, including that of TOP2B in the case of intrinsic androgen signaling (current study) and that of other nucleases in the case of high levels of exogenous genotoxic stress26, can cooperate with androgen signaling to generate recombinogenic DSB and TMPRSS2-ERG rearrangements.
Figure 7.
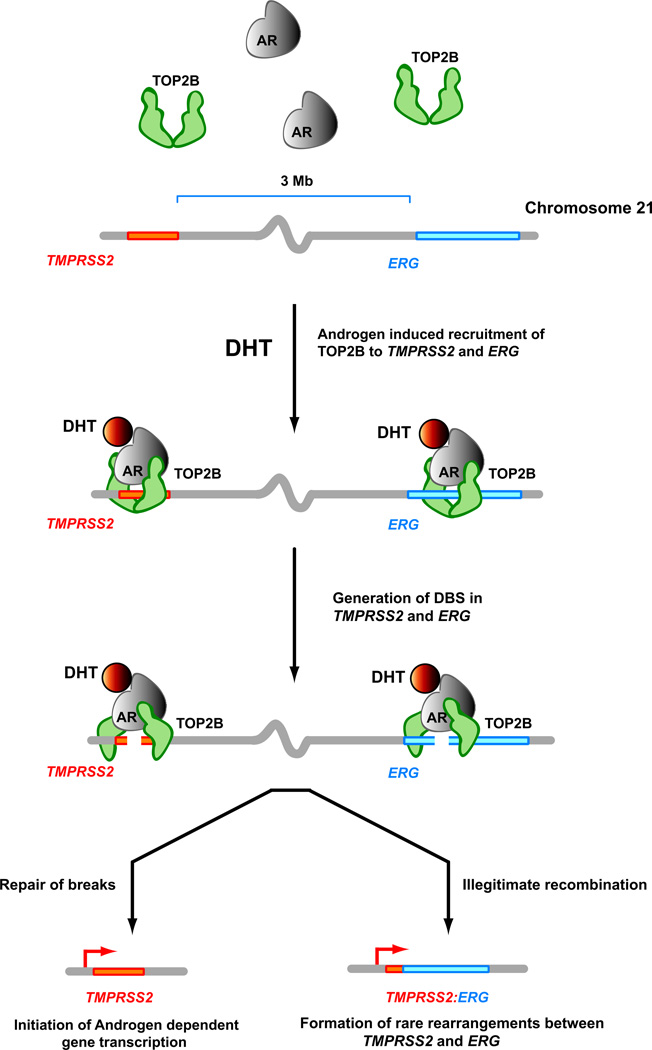
A proposed model for androgen-induced TOP2B mediated double strand breaks and TOP2B instability (TIN) for the generation of DSB and formation of TMPRSS2-ERG gene fusions. Androgen stimulation leads to co-recruitment of liganded AR and TOP2B to regulatory regions of androgen responsive genes as well as to regions of TMPRSS2 and ERG genes observed to participate in genomic rearrangements. TOP2B catalytic activity and the associated DSB formation is required for efficient activation of androgen responsive genes, and these DSB are usually resolved shortly after induction. In some circumstances, stabilization of these TOP2B-mediated DSB can lead to illegitimate recombination and rare rearrangements between TMPRSS2 or other androgen responsive genes and ERG that are then subject to selection during neoplastic outgrowth.
Although the precise biochemical mechanisms by which TOP2B can mediate generation of frank DSB are not well understood, a number of hypotheses can be drawn from previous studies. First, reactive oxygen species, such as those elaborated by androgen signaling in prostate cells in vitro27, or the inflammatory milieu often seen surrounding prostate preneoplastic lesions in vivo28, can stabilize TOP2-DNA cleavable complexes29. Dietary constituents, such as various bioflavonoids, can also stabilize TOP2-DNA cleavable complexes11. These stabilized cleavable complexes can be converted to overt DSB by proteasomal degradation of the TOP2B30 or via collision with replication or transcriptional machinery31. Of interest, these hypotheses may help to explain the well-known epidemiological evidence linking pro-oxidant stress, inflammation and dietary factors in prostate cancer etiology28,32. Indeed, the relatively high rate at which we observed genomic breaks in clinical PIN cells and in androgen-stimulated LAPC4 and LNCaP cells may be a reflection of somewhat dysregulated androgen signaling, which is known to promote both caricaturized differentiation as well as proliferation/survival pathways in these cells33,34. Due to androgen-induced increases in both proliferation and transcription in these cells, such dysregulation of androgen signaling may cause increased rates of collision between transcription or replication forks and androgen-induced TOP2B cleavable complexes with resultant DSB formation. In contrast, androgen signaling in normal cells induces terminal differentiation with suppression of proliferation, a state that may be largely resistant to generation of genomic breaks and rearrangements35. Furthermore, since DNA damage sensing pathways are likely to be functional in normal cells, even if rare breaks occur in these normal cells, they are likely to undergo senescence, cell death, or appropriate repair36. However, if a rare genomic error leading to rearrangement of the TMPRSS2 and ERG loci does occur, then this event is likely to become subject to strong selection5,37–39. That these illigetimate repair events may be extremely rare and subject to selection helps to explain the observations that fusion of TMPRSS2 with either ERG or other ETS family transcription factors are mutually exclusive5.
If our model of androgen-induced TOP2B instability (TIN) (Fig. 7) during regulated transcription holds, we would predict that several different AR target genes should participate in genomic rearrangements in PCa. In support of this, several recent studies have shown rearrangements involving different androgen-induced genes including TMPRSS2, KLK2, CANT1, SLC45A3, AX747630, and HERV-K_22q11.235,40. Interestingly, several of these previously identified androgen-induced fusion targets were modulated by TOP2B inhibition or depletion (see Fig. 1 and Supplementary Fig. 2–4). These data suggest that androgen-induced TOP2B mediated DSB contribute to the PCa specificity of TMPRSS2-ERG fusions. We can speculate that the androgen-induced TOP2B mediated DSB may cooperate with androgen-induced proximity of TMPRSS2 and ERG genes as suggested previously41,26 to confer this lineage specificity.
Additionally, given that other transcriptional processes, including ER, RAR, and AP1 mediated transcriptional programs, also seem to involve TOP2B activity and associated DSB12, the TIN model may be more generalizable to lineage specific genomic rearrangements in other malignancies. Finally, we can speculate that the TIN model may underlie some of the genomic instability and copy number alterations observed during cancer progression42.
Methods
Cell Culture, Reagents and Antibodies
LAPC4 cells were grown in ISCOVE’s Modified Dulbecco’s Medium (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum and 1 nM R1881 (PerkinElmer, Waltham, MA). LNCaP cells were propagated in RPMI medium supplemented with 10% fetal bovine serum. In order to deplete cells of basal androgens, cells were washed with serum-free medium 3 times for 1 h and incubated in 5% charcoal-stripped FBS containing medium for 48 h prior to experiments. Dihydrotestosterone (DHT), Merbarone (Mer), Etoposide (ET), PJ-34, 3-Aminobenzamide (3-AB) and Wortmannin (WORT) were purchased from Sigma-Aldrich (St. Louis, MO). Antibodies were anti-AR rabbit polyclonal (N20), anti-TOP2β rabbit polyclonal (H286) from Santa Cruz Biotechnology (Santa Cruz, CA), anti-ATM (Ab-3) from Calbiochem (Gibbstown, NJ) and anti-H2A.x p139 (clone JBW301) from Millipore (Billerica, MA) anti-PARP1 mouse monoclonal (F2) from Santa Cruz, anti-DNA-PKCS (Ab-2) from Calbiochem, anti-Biotin (clone BN-34) and anti-β-Actin (clone AC-15) from Sigma-Aldrich. Anti-TOP2B (IHC-00166) for immunohistochemistry was from Bethyl Laboratories (Montgomery, TX). Rabbit Gamma Globulin (IgG) was purchased from Jackson ImmunoResearch (West Grove, PA). Anti-AR mouse monoclonal antibody (N441) was kindly provided by Dr. Edwards43. AR (ON-TARGETplus SMARTpool L-003400), TOP2β (ON-TARGETplus SMARTpool L-004240) and control non-targeting pool siRNA oligonucleotides were purchased from Dharmacon (Lafayette, CO). siPARP-1 (cat. # 1027400) was purchased from Qiagen, siDNA-PKcs (sc-35200) was purchased from Santa Cruz Biotechnology. shRNA constructs against TOP2B in pGFP-V-RS vectors (cat. # TG308698) were purchased from Origene (Rockville, MD).
Fusion transcript detection assay
Total RNA was extracted from 108 cells using RNeasy Maxi Kits (Qiagen). First strand synthesis was carried out using Superscript III (Invitrogen) with primers specific to exons 5 to 11 of ERG (Supplementary Table 4). Reactions were cleaned up using PCR purification kit (Qiagen) and resulting cDNA libraries were split into 192 reactions and subjected to PCR amplification using primers and Taq-man probes specific to exon 1 of TMPRSS2 and exon 4 of ERG. The presence of an amplification product was monitored by real-time PCR. Products were also separated in an agarose gel and bands were sequence verified.
Chromatin Immunoprecipitation (ChIP) and ChIP-Re-ChIP
Chromatin immunoprecipitation was performed as described previously44 with some modifications.
Potassium–SDS assay (KSDS)
KSDS assay was performed as described previously15, with modifications.
Chromosome conformation Capture (3C)
3C was basically performed as described previously45.
SLOT assay
Cells were lysed in SLOT lysis buffer (2% SDS, 10 mM EDTA in 10 mM Tris-HCl pH 8) and DNA was extracted after proteinase K digestion using the Qiagen Blood and Tissue Kit (Qiagen). Tyrosine residues covalently linked to DNA were labeled using EDC and Amine-PEG3-Biotin (Thermo Scientific). Biotin-labeled DNA was precipitated using Strepavidin coated magnetic beads (NEB), digested with NspI (NEB), and unbound fragments were removed by washing with wash Buffer (10 mM Tris HCl, 1mM EDTA, 2 M NaCl). Precipitated DNA was eluted in 95% formamide, 10 mM EDTA for 5 min at 65°C, purified using the Qiagen PCR purification Kit (Qiagen) and analyzed by real-time PCR. (See Supplementary Fig. 8 for an overview of the SLOT assay). To test the specificity of the SLOT assay, the pRYG plasmid, which contains a TOP2 cleavage and recognition site46 was incubated with recombinant TOP2A (TopoGen, Inc, Port Orange, FL) for 30 min at 37°C in the absence or presence of Etoposide (Sigma). Reactions were stopped by addition of SDS and were digested by proteinase K (Invitrogen). DNA was extracted by phenol-chloroform and recovered DNAs were subjected to the SLOT assay.
DNA break labeling assay
DNA strand breaks (DSB) were labeled in intact nuclei with Biotin-11-dUTP by Terminal transferase. Biotin labeled DNA fragments were then subjected to ChIP analysis using anti-Biotin antibodies as described previously12.
Fluorescence in-situ Hybridization (FISH)
BAC clones RP11-35C4 (TMPRSS2-5’), RP11-825A8 (TMPRSS2-3’), RP1195I2I (ERG-5’), RP11-476D17 (ERG-3’) were purchased from BACPAC (http://bacpac.chori.org). Probes were labeled with SpectrumGreen (all 5’ probes) and SpectrumOrange (all 3’ probes) conjugated dUTP (Abbott, Abbott Park, IL). Slides were pretreated using Paraffin Pretreatment reagent kits (Abbott). Samples were analyzed under a 100x oil immersion objective using a Nikon E400 fluorescence microscope (Nikon Instruments, Melville, NY). Samples were scored as positive for TMPRSS2 rearrangements if >10% of cells showed either split-apart or deletion FISH configurations.
Clonogenic survival assay
PCR amplicons encompassing the breakpointsite of case 1 in intron 1 (IN-1) of TMPRSS2 and a site in exon 6 (EX-6) of TMPRSS2 were cloned into pcDNA6.2N-YFP TOPO vectors (Invitrogen). LAPC4 cells were deprived of androgens and transfected with pcDNA6.2 vectors together with sh-TOP2β or shRNA-control vectors using FuGENE6 (Roche). Twenty-four hours after transfection, cells were stimulated with 100 nM DHT and split the following day to clonogenic density in medium containing 5 µg/ml Blasticidin (Invitrogen). Cells were cultured for 14 days and medium was replaced every third day. To assess clonogenic survival, cultures were fixed in 25% methanol and stained with crystal violet as described previously47. Colonies containing at least 50 cells were counted using the Versa Doc Imaging system 3000 and the Quantity One software package (Biorad).
SNP-microarray analysis and cloning of genomic breakpoints
Primers spanning the putative TMPRSS2-ERG deletion breakpoints identified in a previously published Affymetrix SNP 6.0 metastatic prostate cancer dataset16 were used to amplify breakpoints from the corresponding cases from that dataset. Products were cloned into pCR2.1TOPO vectors (Invitrogen) and analyzed by Sanger sequencing.
Prostate Cancer and PIN tissue in Tissue Microarray (TMA)
TMAs from formalin-fixed paraffin-embedded tissue blocks containing PIN and matched normal tissue from 23 men were constructed as described previously48.
Expression array analysis
RNA from inhibitor and knockdown experiments was processed, labeled, and hybridized to HG-U133 2.0 Plus whole genome gene expression microarrays (Affymetrix) according to the manufacturer’s protocols at the Johns Hopkins Microarray facility. The microarray gene expression data was pre-processed with RMA14,49. Statistical significance was assessed by Wilcoxon rank-sum p-values after ranking genes by absolute log fold change of expression between conditions. For genes represented by more than one probe set, the one with the highest fold change was chosen. All analysis was carried out using R 2.9 (R Development Core Team, 2009) and Bioconductor 2.450.
Co-Immunoprecipitation
Co-Immunoprecipitation was performed as described previously51. To test whether the interaction between TOP2B and AR is dependent on the presence of DNA, cell lysates were incubated with 50 µg/ml Ethidium Bromide (Sigma) or 200 U/ml DNase (NEB) for 12 h at 4°C52. Lysates were then subjected to co-immunoprecipitation assays using AR specific antibodies. Precipitated complexes were separated by SDS-PAGE and immunoreactive bands were detected by western blotting.
shRNA knockdown of TOP2B and Cell sorting
LNCaP cells were transfected with pGFP-V-RS shRNA-control or pGFP-V-RS shRNA-TOP2b vectors. Sixteen hours after DHT treatment cells were harvested by trypsinization resuspended in 2 mM EDTA in PBS. Fifty-thousand GFP positive singlet cells were sorted by FACS and RNA and protein was extracted using the AllPrep™-Kit from Qiagen.
Ligation-mediated Linker PCR for identification of DHT induced DNA strand breaks at nucleotide resolution
DNA was extracted from intact cells by resuspending cells in Lysis Buffer (2% SDS, 10 mM EDTA, 10 mM TRIS-HCl ph 7.5) and incubating lysates for at least 6 h in the presence of 0.2 mg/ml Proteinase K (Invitrogen) at 37°C. Proteinase K was inactivated by heating the lysate for 10 min at 70°C and SDS was sequestered by adding Triton X-100 to a final concentraion of 1% and incubating 1 h at 37 °C. RNase (Invitrogen) was added to a final concentration of 40 µg/ml and incubated for 1 h at 37 °C. DNA was then extracted in 1 volume of Ultrapure Phenol:Chloroform:Isoamyl Alcohol (25:24:1, v/v; Invitrogen) and precipitated with ethanol. DNA ends were repaired using the End-It™ DNA End-Repair Kit (Epicenter Biotechnologies, Madison, WI). DNA was recovered by Phenol-Chloroform extraction followed by ethanol precipitation. Double stranded Linkers (Linker 1; Linker 2) were prepared by annealing 20 pmol/µl of each oligonucleotide in 250 mM Tris-HCl, [pH 7.8] by heating at 95°C for 3 min, quickly transferring to 70°C and gradually cooling to 4°C over a period of 3 h. Linkers were then ligated to 1 µg of processed DNA using T4 ligase (NEB). Gene and linker specific primers used in amplification reactions are listed in below. Products of PCR amplification reactions were separated in acrylamide Gels (Invitrogen) and sequenced by Sanger sequencing.
Determination of integration frequencies
To determine the integration frequency of the plasmid insert sequences of stable clones in the genome, DNA from pooled clones selected for blasticidin resistance was digested with NheI (New England Biolabs), which does not have cutting sites in the plasmid sequence. Blunt DNA ends were generated by using the End-It™ DNA End-Repair Kit (Epicenter Biotechnologies, Madison, WI) following manufacturer’s recommendations. A double stranded linker (Linker 3; Linker 4) was annealed and ligated to DNA molecules as described above. After DNA clean-up using the DNeasy kit (Qiagen), DNA was analyzed by quantitative real-time PCR using primers. Relative integration frequencies were calculated for pcDNA6.2-IN-1 and pcDNA6.2-EX-6 according to the formula: 2^{(ctClone_DHT_insert − ctClone_DHT_blast) − (ctClone_control_insert − ctClone_control_blast)} where ctClone_DHT_insert = the ct value of clones that were androgen deprived and then DHT treated prior to clonogenic survival using a forward primer flanking the plasmid (pcDNA6.2_insert_A) insert and reverse linker specific primers (Linker primer); ctClone DHT blast = ct value of the same DNA using forward and reverse primers specific to the blasticidin resistance gene (pcDNA6.2_blast) and ctClone_control_insert/blast = the ct value of clones that were not androgen deprived prior to clonogenic survival using a forward primer flanking the plasmid (pcDNA6.2_insert_A) insert and reverse specific to the linker sequence (Linker primer) or the pcDNA6.2_blast primers.
Supplementary Material
Acknowledgements
We would like to thank Dr. Donald Coffey for his comments and Dr. Christopher Heaphy, Dr. Hao Zhang, and Lillian Dasko-Vincent from the SKCC Cell Imaging Core Facility for technical support. We also thank the Brady Urological Research Institute Prostate Specimen Repository for providing TMA sections. This work was supported by funding from the NIH/NCI, Department of Defense PCRP, Prostate Cancer Foundation, Maryland Cigarette Restitution Fund, and the Patrick C. Walsh Prostate Cancer Research Fund/Dr. and Mrs. Peter S. Bing Scholarship.
Footnotes
Supplementary Information is available at the Nature Genetics website.
Author Contributions M.C.H. executed and analyzed all experiments and assisted in writing the manuscript. M.J.A. analyzed microarray data and assisted with statistical analysis of data. A.T., R.A., B.G., A.K.M., G.N., and A.M.D.M. assisted with execution and analysis of FISH, immunostaining, and pathology experiments. D.M.E. assisted with execution of experiments. W.B.I., G.S.B., W.L., and J.X., contributed to analysis of microarray data in determination of prostate cancer TMPRSS2-ERG genomic breakpoints. W.G.N. assisted in experimental design and analysis and contributed to writing the manuscript. S.Y. conceived of the study together with W.G.N., assisted in experimental design, execution, and analysis, and wrote the manuscript. All authors assisted in editing the manuscript.
DNA microarray data reported in the manuscript have been deposited in the Gene Expression Omnibus (accession number GSE19445).
References
- 1.Rowley JD. Chromosomal translocations: revisited yet again. Blood. 2008;112:2183–2189. doi: 10.1182/blood-2008-04-097931. [DOI] [PubMed] [Google Scholar]
- 2.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 3.Morgan WF, et al. DNA double-strand breaks, chromosomal rearrangements, and genomic instability. Mutat Res. 1998;404:125–128. doi: 10.1016/s0027-5107(98)00104-3. [DOI] [PubMed] [Google Scholar]
- 4.Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- 5.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 7.Jemal A, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 8.Pedersen-Bjergaard J, Andersen MK, Johansson B. Balanced chromosome aberrations in leukemias following chemotherapy with DNA-topoisomerase II inhibitors. J Clin Oncol. 1998;16:1897–1898. doi: 10.1200/JCO.1998.16.5.1897. [DOI] [PubMed] [Google Scholar]
- 9.Smith MA, McCaffrey RP, Karp JE. The secondary leukemias: challenges and research directions. J Natl Cancer Inst. 1996;88:407–418. doi: 10.1093/jnci/88.7.407. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Rowley JD. Chromatin structural elements and chromosomal translocations in leukemia. DNA Repair (Amst) 2006;5:1282–1297. doi: 10.1016/j.dnarep.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 11.Strick R, Strissel PL, Borgers S, Smith SL, Rowley JD. Dietary bioflavonoids induce cleavage in the MLL gene and may contribute to infant leukemia. Proc Natl Acad Sci U S A. 2000;97:4790–4795. doi: 10.1073/pnas.070061297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ju BG, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 13.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irizarry RA, Wang C, Zhou Y, Speed TP. GENE SET ENRICHMENT ANALYSIS MADE SIMPLE. The Berkeley Electronic Press; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson WG, Liu LF, Coffey DS. Newly replicated DNA is associated with DNA topoisomerase II in cultured rat prostatic adenocarcinoma cells. Nature. 1986;322:187–189. doi: 10.1038/322187a0. [DOI] [PubMed] [Google Scholar]
- 16.Liu W, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15:559–565. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demichelis F, et al. Distinct genomic aberrations associated with ERG rearranged prostate cancer. Genes Chromosomes Cancer. 2009;48:366–380. doi: 10.1002/gcc.20647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu W, et al. Multiple genomic alterations on 21q22 predict various TMPRSS2/ERG fusion transcripts in human prostate cancers. Genes Chromosomes Cancer. 2007;46:972–980. doi: 10.1002/gcc.20482. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q, et al. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 21.Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 23.Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol. 2008;9:759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- 24.Clark J, et al. Complex patterns of ETS gene alteration arise during cancer development in the human prostate. Oncogene. 2008;27:1993–2003. doi: 10.1038/sj.onc.1210843. [DOI] [PubMed] [Google Scholar]
- 25.Perner S, et al. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31:882–888. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- 26.Lin C, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ripple MO, Henry WF, Rago RP, Wilding G. Prooxidant-antioxidant shift induced by androgen treatment of human prostate carcinoma cells. J Natl Cancer Inst. 1997;89:40–48. doi: 10.1093/jnci/89.1.40. [DOI] [PubMed] [Google Scholar]
- 28.De Marzo AM, et al. Inflammation in prostate carcinogenesis. Nat Rev Cancer. 2007;7:256–269. doi: 10.1038/nrc2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li TK, et al. Activation of topoisomerase II-mediated excision of chromosomal DNA loops during oxidative stress. Genes Dev. 1999;13:1553–1560. doi: 10.1101/gad.13.12.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Azarova AM, et al. Roles of DNA topoisomerase II isozymes in chemotherapy and secondary malignancies. Proc Natl Acad Sci U S A. 2007;104:11014–11019. doi: 10.1073/pnas.0704002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, D'Arpa P, Liu LF. A model for tumor cell killing by topoisomerase poisons. Cancer Cells. 1990;2:23–27. [PubMed] [Google Scholar]
- 32.Nelson WG, De Marzo AM, Isaacs WB. Prostate cancer. N Engl J Med. 2003;349:366–381. doi: 10.1056/NEJMra021562. [DOI] [PubMed] [Google Scholar]
- 33.Denmeade SR, Lin XS, Isaacs JT. Role of programmed (apoptotic) cell death during the progression and therapy for prostate cancer. Prostate. 1996;28:251–265. doi: 10.1002/(SICI)1097-0045(199604)28:4<251::AID-PROS6>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 34.Marker PC. Does prostate cancer co-opt the developmental program? Differentiation. 2008;76:736–744. doi: 10.1111/j.1432-0436.2008.00297.x. [DOI] [PubMed] [Google Scholar]
- 35.Litvinov IV, De Marzo AM, Isaacs JT. Is the Achilles' heel for prostate cancer therapy a gain of function in androgen receptor signaling? J Clin Endocrinol Metab. 2003;88:2972–2982. doi: 10.1210/jc.2002-022038. [DOI] [PubMed] [Google Scholar]
- 36.Hallstrom TM, Laiho M. Genetic changes and DNA damage responses in the prostate. Prostate. 2008;68:902–918. doi: 10.1002/pros.20746. [DOI] [PubMed] [Google Scholar]
- 37.Carver BS, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.King JC, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–526. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klezovitch O, et al. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci U S A. 2008;105:2105–2110. doi: 10.1073/pnas.0711711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hermans KG, et al. Two unique novel prostate-specific and androgen-regulated fusion partners of ETV4 in prostate cancer. Cancer Res. 2008;68:3094–3098. doi: 10.1158/0008-5472.CAN-08-0198. [DOI] [PubMed] [Google Scholar]
- 41.Mani RS, et al. Induced chromosomal proximity and gene fusions in prostate cancer. Science Express. 2009:1–2. doi: 10.1126/science.1178124. 10.1126/science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
References for Methods
- 43.Boonyaratanakornkit V, et al. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol Cell Biol. 1998;18:4471–4487. doi: 10.1128/mcb.18.8.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 45.Splinter E, Grosveld F, de Laat W. 3C technology: analyzing the spatial organization of genomic loci in vivo. Methods Enzymol. 2004;375:493–507. doi: 10.1016/s0076-6879(03)75030-7. [DOI] [PubMed] [Google Scholar]
- 46.Spitzner JR, Chung IK, Muller MT. Eukaryotic topoisomerase II preferentially cleaves alternating purine-pyrimidine repeats. Nucleic Acids Res. 1990;18:1–11. doi: 10.1093/nar/18.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeWeese TL, et al. Human papillomavirus E6 and E7 oncoproteins alter cell cycle progression but not radiosensitivity of carcinoma cells treated with low-dose-rate radiation. Int J Radiat Oncol Biol Phys. 1997;37:145–154. doi: 10.1016/s0360-3016(96)00448-8. [DOI] [PubMed] [Google Scholar]
- 48.Gurel B, et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol. 2008;21:1156–1167. doi: 10.1038/modpathol.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 50.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haffner MC, et al. Interaction and functional interference of glucocorticoid receptor and SOCS1. J Biol Chem. 2008;283:22089–22096. doi: 10.1074/jbc.M801041200. [DOI] [PubMed] [Google Scholar]
- 52.Padilla PI, et al. Association of guanine nucleotide-exchange protein BIG1 in HepG2 cell nuclei with nucleolin, U3 snoRNA, and fibrillarin. Proc Natl Acad Sci U S A. 2008;105:3357–3361. doi: 10.1073/pnas.0712387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.