Abstract
Objective:
To find out the most common bacterial pathogens responsible for post-operative wound infection and their antibiotic sensitivity profile.
Materials and Methods:
This prospective, observational study was carried out in patients of postoperative wound infection. Samples from wound discharge were collected using a sterile swab and studied for identification of isolates by Gram stains and culture growth followed by in vitro antibiotic susceptibility testing performed by disc diffusion method on Mueller Hinton agar.
Results:
Out of 183 organisms, 126 (68.85%) isolated organisms were gram negative. Staphylococcus aureus, 48 (26.23%), was the predominant organism. S. aureus was sensitive to rifampicin (89.58%), levofloxacin (60.42%), and vancomycin (54.17%). Pseudomonas aeruginosa was sensitive to ciprofloxacin (83.78%), gatifloxacin (51.35%), and meropenem (51.35%). Escherichia coli was sensitive to levofloxacin (72.41%) and ciprofloxacin (62.07%). Klebsiella pneumoniae was sensitive to ciprofloxacin (63.16%), levofloxacin (63.16%), gatifloxacin (63.16%), and linezolid (56.52%). Proteus mirabilis was sensitive to ciprofloxacin (75%) and linezolid (62.50). Proteus vulgaris was sensitive to ampicillin+sulbactam (57.14%) followed by levofloxacin (50%).
Conclusions:
There is an alarming increase of infections caused by antibiotic-resistant bacteria, particularly in the emergence of VRSA/VISA, meropenem, and third generation cephalosporin resistant Pseudomonas aeruginosa. Linezolid showing sensitivity against Gram negative bacteria.
Keywords: Antibiogram, antibiotic sensitivity, bacterial resistance, gram negative bacteria, gram positive bacteria
INTRODUCTION
In 1992, the US Centers for Disease Control (CDC) revised its definition of “wound infection,” creating the definition “surgical site infection” (SSI) to prevent confusion between the infection of a surgical incision and the infection of a traumatic wound. Nosocomial infections occurs worldwide and affect both developed and resource poor countries. Surgical wound infection is a serious hazard to patients, with incidence according to CDC was 15.45%, and according to the UK nosocomial infection surveillance was 11.32%, and according to ASEPSIS was 8.79%.[1] About 77% of the deaths of surgical patients were related to surgical wound infection.[2] SSIs are classified into incisional SSIs, which can be superficial or deep, or organ/space SSIs.
A complex interplay between host, microbial, and surgical factors ultimately determines the prevention or establishment of a wound infection. The most common group of bacteria responsible for SSIs are Staphylococcus aureus. The emergence of resistant strains has increased the morbidity and mortality associated with wound infections. These strains are beginning to develop resistance to vancomycin in North India found in study of Tiwari et al. and they suggested to do such study in other parts of India also because the emergence of VRSA/VISA (Vancomycin resistant Staphylococcus aureus / vancomycin intermediate Styphylococcus aureus) might also be prevalent as antibiotic misuse is equally common there.[3] Vancomycin is currently the most effective antibiotic against MRSA. This new resistance has arisen because another species of bacteria like acinetobacter commonly express vancomycin resistance.[4]
Pseudomonas aeruginosa is an epitome of opportunistic nosocomial pathogen, which causes a wide spectrum of infections and leads to substantial morbidity in immuno compromised patients. Despite therapy the mortality due to nosocomial Pseudomonal pneumonia is approximately 70%.[5] Unfortunately, Pseudomonas aeruginosa developed resistance to most of antibiotics thereby jeopardizing the selection of appropriate treatment.[6] Therefore, in present study, we also find out the antibiotic susceptibility patterns of Pseudomonas aeruginosa.
Risk factors other than microbiology can be due to systemic factors affecting the patient's healing response, local wound characteristics, or operative characteristics. Its risk depends on bleeding, the amount of devitalized tissue created, the need for drains within the wound, obesity and diabetes mellitus.[7]
They are the third most frequent nosocomial infection, associated with increased hospital stay, costs, and use of antimicrobial agents.[8] Antibiotic resistance can be controlled by appropriate antimicrobial prescribing, prudent infection control, new treatment alternatives, and continued surveillance.[9]
Due to significant changes in microbial genetic ecology, as a result of indiscriminate use of anti-microbials, the spread of antimicrobial resistance is now a global problem. Present study was carried out to find out common bacterial pathogens responsible for postoperative wound infection and antibiotic susceptibility of various isolates. It assists the clinicians in appropriate selection of antibiotics especially against hospital acquired infections.
MATERIALS AND METHODS
This prospective, observational, hospital-based cohort study was carried out after prior approval by Institutional Ethics Committee. Total 938 patients of either gender in different age groups admitted to the general surgery wards of Guru Gobindsingh Hospital, Jamnagar, were enrolled in the study between January and September 2006.
In all cases, preoperative, intraoperative, and postoperative details were studied. Information was collected in a case record form for age, sex, date of admission, associated co-morbid condition, reason for admission, type of surgery: emergency or planned, procedure, duration of surgery, preoperative and postoperative stay, preoperative antibiotic prophylaxis and type of wound clean, potentially infected and frankly infected wound.[10] All patients were followed up in wards till discharge from the hospital.
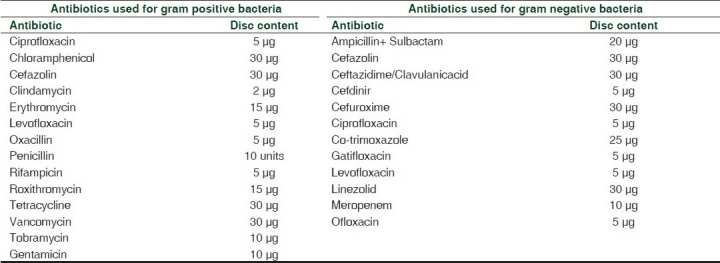
Samples for wound infections were collected from the patients with complaints of discharge, pain, swelling, foul smelling, delayed and non healing wound by using a sterile swab, taking care to avoid contamination of the specimen with commensals from the skin, and were immediately transported to the laboratory. They were studied for identification of isolates by Gram stains and culture growth on nutrient, blood and MacConkey agar. Colonies from nutrient agar were used for biochemical tests and antibiotic sensitivity. On isolation of Gram positive cocci, catalase, and coagulase tests were done. Gram negative bacilli were distinguished using biochemical tests IMViC (indole, methyl red, Voges-Proskauer,citrateutilization),oxidase and triple sugar iron (TSI) agar tests.After confirmation of the organism, culture growth was tested for in vitro antibiotic susceptibility testing performed by disc diffusion method (modified Kirby Bauer method) on Muller Hinton agar.[11] The disc (Eos L0 aboratories, Mumbai) contents are shown in Table 1. Data were expressed as proportions.
Table 1.
Antibiotics used for gram positive and gram negative bacteria with its disc content
RESULTS
Out of 938 surgeries, 110 (11.73%) cases developed postoperative wound infections. Maximum number of patients (62.73%) were from 5th, 6th, and 7th decade of age group. Higher infection rate was noted in males (12.78%) as compared to females (10.60%). Postoperative wound infection rate was 7.28% in clean surgeries, 9.63% in potentially infected surgical wounds and 19.19% in frankly infected wounds. The rate was higher in gangrene (30.0%), diabetic foot (22.45%), surgeries on large bowel (20.0%), abscess (18.18%), and in cellulites (15.52%). Following antibiotics were used for pre-surgical prophylaxis: crystalline penicillin, ampicillin, cloxacillin, cefadroxil, cefotaxime, ceftriaxone, ceftazidime, cotrimoxazole, ciprofloxacin, norfloxacin, gentamicin, amikacin, and metronidazole. All the patients had received two to four antibiotics in varying combinations. Most commonly used combinations were: Beta lactum antibiotics (crystalline penicillin / ampicillin / cefotaxime) ± cloxacillin / aminoglycosides (gentamicin, amikacin) / metronidazole and fluroquinolone (ciprofloxacin) ± cloxacillin / aminoglycosides (gentamicin, amikacin) / metronidazole.
Total 183 organisms were identified by gram staining, 57 were gram positive (31.15%) and 126 were gram negative (68.85%). Predominant organisms isolated were: Staphylococcus aureus 48 (26.23%) Klebsiella pneumoniae 38(20.77%), Pseudomonas aeruginosa 37 (20.22%), and Escherichia coli 29 (15.85%). Staphylococcus aureus was sensitive to rifampicin (89.58%) followed by levofloxacin (60.42%) and vancomycin(54.17%), while it was resistant to oxacillin (29.17%)and penicillin (25%). In one case, Staphylococcus aureus was resistant to all antibiotics used for sensitivity testing. Vancomycin resistant Staphylococcus aureus was sensitive to rifampicin (95%), levofloxacin (35%), tetracycline (25%), erythromycin (25%), ciprofloxacin (20%), and roxithromycin (20%). Staphylococcus epidermis was 100% sensitive to cefazolin, levofloxacin, oxacillin, rifampicin, and vancomycin, while it was less sensitive to gentamicin (77.78%), ciprofloxacin (77.78%), erythromycin (77.78%), penicillin (55.56%), and clindamycin (44.44%). Pseudomonas aeruginosa was sensitive to ciprofloxacin (83.78%) followed by gatifloxacin (51.35%), meropenem (51.35%), ceftazidime (45.95%) and linezolid (43.24%). About 34.37% of isolated strains were resistant to both ceftazidime and meropenem. In these resistant strains, sensitivity of ciprofloxacin, gatifloxacin, cefazolin, and linezolid were 81.18%, 27.27%, 27.27%, and 18.18%, respectively. Escherichia coli was sensitive to levofloxacin (72.41%) and ciprofloxacin (62.07%), while it was less sensitive to ceftazidime (27.59%), cefuroxime (20.69%) and cefdinir (13.79%). Klebsiella pneumoniae was sensitive to gatifloxacin (63.16%), levofloxacin (63.16%), ciprofloxacin (63.16%), and linezolid (56.52%), while it was resistant to ceftazidime (36.84%), cefuroxime (34.21%) and cefdinir (15.79%). Proteus mirabilis was sensitive to ciprofloxacin (75%) and linezolid (62.50%), while it was resistant to cefdinir (25%), cefuroxime (25%), co-trimoxazole (25%), and ampicillin+sulbactam (12.50%). Proteus vulgaris was sensitive to ampicillin+sulbactam (57.14%) followed by levofloxacin (50%), meropenem (42.86%), and ciprofloxacin (42.86%), while it was resistant to cefdinir (7.14%) and cefuroxime (7.14%).
Among the studied antibiotics the gram positive isolates were sensitive to rifampicin (91.23%), levofloxacin (66.67%) and vancomycin (61.40%), whereas they were resistant to penicillin (29.82%), gentamicin (29.82%), and clindamycin (22.81%) [Figure 1]. The gram negative isolates were sensitive to ciprofloxacin (67.46%), levofloxacin (56.35%), gatifloxacin (51.59%), and linezolid (50.79%), and were resistant to cefuroxime (24.6%), co-trimoxazole(19.84%) and cefdinir (16.67%) [Figure 2].
Figure 1.
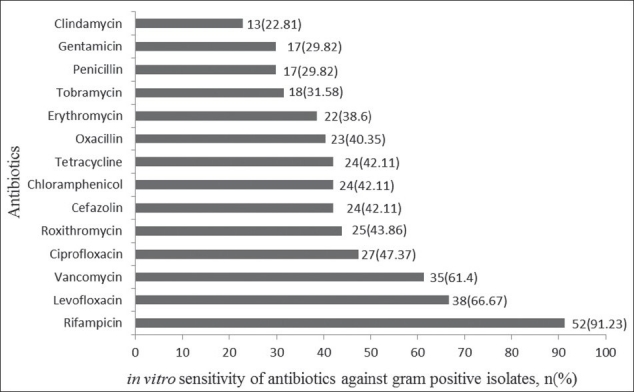
Sensitivity of commonly used antibiotics for gram positive bacteria
Figure 2.
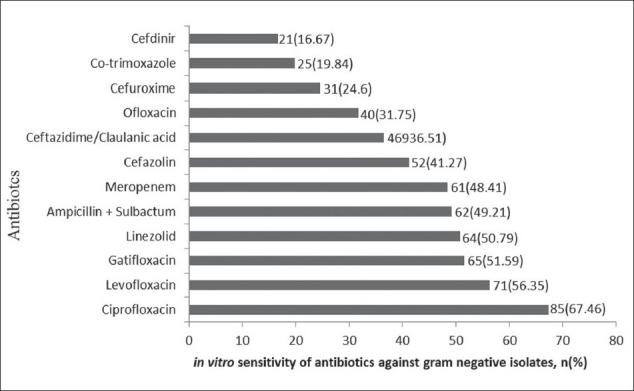
Sensitivity of commonly used antibiotics for gram negative bacteria
DISCUSSION
In present study, rate of postoperative wound infection was 11.73%, that is similar to the other studies conducted in India.[12] We had observed higher infection rate for clean (7.28% vs 2.6% to 2.9%),[10,13] potentially infected (9.63%vs 5.4% to 14%),[10,13] and frankly infected (19.19%vs 3.4% to 9%)[10,13] post-operative wounds as compared to previous studies. Present study supports the conclusion of Malone et al. that diabetes is a significant preoperative risk factor for surgical site infections.[14] On gram stain examination, 31.15% pathogens were gram positive and 68.85% were gram negative, as against 69% for gram positive and 29% for gram negative organisms in other setups.[15] This difference may be due to variation in common nosocomial pathogens inhabitant in different hospital set up. Though percentage of gram negative bacilli from the wounds was more, Staphylococcus aureus was predominant organism isolated followed by Klebsiella pneumoniae, Pseudomonas aeruginosa, and Escherichia coli; similar findings were observed in the studies done previously.[13,15,16,17] The organisms most frequently involved in surgical infections change from time to time, and also vary with hospital settings.
Our study had shown reduced sensitivity of Staphylococcus aureus to most of antibiotics: rifampicin (89.58% vs. 99%),[18] levofloxacin (60.42% vs. 73 %),[19] vancomycin(54.17% vs. 98.7% to 100%),[17–20] oxacillin (29.17% vs. 28% to 61%),[18,20] ciprofloxacin (41.67% vs. 25% to 66.7%),[17,20] erythromycin (33.33% vs. 14%),[18] cefazolin (31.25% vs. 61% to 71.8%),[17,18] and gentamicin (20.83% vs. 77%)[21] as compared to previous studies. Staphylococcus aureus resistance to vancomycin was found to be 45.83%. Incidence of VRSA is higher than other reported studies from India.[22] This should be cautiously interpreted as this might be because of false positive result by bacteria like acinetobacter[4,23] and the resistance to vancomycin was not counterchecked by other laboratory. Other study also shows false positive result by using same disc diffusion method, like IyerRN (2008) studied that Styphylococcus aureus was also capable of transformation into the homogenously resistant strains and adhering to artificial surfaces, 20-fold higher than the usually encountered MRSA isolates. The BSAC (British Society for Antimicrobial Chemotherapy) has standardized method for disc diffusion testing to evaluate vancomycin susceptibility for MRSA isolates.[24] A screening method developed by Hiramatsuet al.[25] appeared to show promise a few years ago, but was found to yield both false positives and false negative results.[26,27] There is a need for the modern day clinical laboratory to develop a definitive method to confirm hetero-resistance to vancomycin among MRSA isolates as this could direct antibiotic therapy. The chief options in this direction are modified PAP method, gradient plates, and the addition of Mu 3 cell wall material to media.[28] Additionally, in 2009, the Clinical and Laboratory Standards Institute (CLSI) altered the guidelines for staphylococci such that disc diffusion was no longer an acceptable means for testing vancomycin susceptibility in these organisms.[29]
This certainly has implications in the management of infections caused by these strains in the nosocomial setting. It is essential for clinical laboratories to screen for and confirm vancomycin resistance in the clinical laboratory. However, there is need for active VRSA surveillance by healthcare institutions in India to enforce infection control measures, rational use of antibiotics and to prevent development of further resistance against this organism.[3] Rifampicin is highly active against staphylococci, but like Mycobacterium tuberculosis resistance develops rapidly if is used as a monotherapy. It should be use with linezolid, quinupristin/dalfopristin, newer fluoroquinolones or other agents to prevent further resistance.
In our study, Staphylococcus epidermis was 100% sensitive to vancomycin, cefazolin, rifampicin, levofloxacin, and ofloxacin, followed by ciprofloxacin (77.78%), tobramycin (77.78%), gentamicin (77.78%), and erythromycin (66.67%). Other studies had shown variable sensitivity pattern with vancomycin (96% to 100%),[17–19] ciprofloxacin (75.8%),[17] erythromycin (43% to 60%),[18–19] and cefazolin (35% to 79.2%).[17,19]
As compared to other studies, in our study Pseudomonas aeruginosa showed reduced sensitivity to commonly used antibiotics except ciprofloxacin (83.78% vs. 76%), meropenem (51.35% vs. 90.5%), ceftazidime/clavulanic acid (45.95% vs. 71.5%), levofloxacin (43.24% vs. 72.5 %), and ofloxacin (32.43% vs. 62.5 %).[30] Ciprofloxacin has been stated to be the most potent drug available for the treatment of P. aeruginosa infections.[31] This report is in conformity with the result of this study in which ciprofloxacin recorded the least resistance (26.22%) to P. aeruginosa isolates from wound infection patients. Similar reduced resistance of P. aeruginosa to ciprofloxacin has been reported in Jamaica(19.6%) (Brown and Izundu,2004), Latin America (28.6%)[32], Ilorin Nigeria (24.7%)[33] and in Kuala Lumpur, Malaysia (11.3%).[34] It is undoubtable that at the present time, ciprofloxacin is the most effective antibiotics against P. aeruginosa involved in wound infection relative to most other commonly used drugs.
Pseudomonas resistant to carbapenem and third generation cephalosporins is real threat. In fact, the irrational and inappropriate use of antibiotics is responsible for the development of resistance of Pseudomonas species to antibiotic monotherapy. Periodic susceptibility testing should be carried out over a period of two to three years to detect the resistance trends. Also, a rational strategy on the limited and prudent use of antipseudomonal agents is urgently required. The incidence of Pseudomonas aeruginosa in postoperative wound infection is becoming more serious in developing countries because of relaxation in general hygienic measures, mass production of low quality antiseptic, and medicinal solutions for treatment, difficulties in proper definition of the responsibility among the hospital staff[35].
Sensitivity pattern of Escherichia coli in our study as compared to others were levofloxacin (72.41% vs. 97 % to 100%),[18,19] ciprofloxacin (62.07% vs. 97% to 100%),[18,20] meropenem (51.72% vs. 100%),[19] ampicillin+sulbactam (51.72% vs.83% to 84%),[18,20] cefazolin (41.38% vs. 92% to 94%),[18,20] ceftazidime/clavulanic acid (27.59% vs. 91% to 99%),[19,20] ofloxacin (27.59% vs. 97%),[19] cefuroxime (20.69% vs. 91%),[20] cefdinir (13.79%), and cotrimoxazole (10.34%). So, reduced antibiotic sensitivity pattern noted for E. coli suggests its importance for hospital acquired infection. Klebsiella pneumoniae was sensitive to linezolid (65.79%) followed by levofloxacin (63.16%), ciprofloxacin (63.16%), and gatifloxacin (63.16%). However, our study had shown reduced sensitivity to ciprofloxacin (63.16% vs. 100%), cefazolin (44.74% vs. 100%), ceftazidime /clavulanic acid (36.84% vs. 100%), cefuroxime (34.21% vs. 100%), cotrimoxazole (26.32% vs. 100%) as compared to previous studies.[20] Proteus mirabilis was sensitive to ciprofloxacin (75%) and linezolid (62.50%), its sensitivity pattern were reduced for ciprofloxacin (75% vs. 95% to 100%),[18,20] cefazolin (37.50% vs. 98% to 100%),[18,20] ceftazidime/clavulanic acid (37.50% vs. 100%),[20] levofloxacin (37.50% vs. 95%),[18] cefuroxime (25% vs. 100%),[17] co-trimoxazole (25% vs.97% to100%),[18,20] ampicillin+sulbactam (12.50% vs. 80% to 95%)[18,20] as compared to previous studies. In our study, Proteus vulgaris was sensitive to ampicillin+sulbactam (57.14%) followed by levofloxacin (50%), meropenem(42.86%), and ciprofloxacin (42.86%). However, compared to previous study sensitivity of organism to most commonly used antibiotics were reduced for levofloxacin (50% vs. 100%), meropenem (42.86% vs. 100%), ceftazidime/clavulanic acid (28.57% vs. 100%), and ofloxacin (28.57% vs.100%)[19] . So, our finding supports the undesirable trends in antibiotic resistance, indicating decreasing efficacy of antibiotic classes such as second and third generation cephalosporins, extended-spectrum beta-lactamase inhibitor, carbapenems, and fluoroquinolones against gram negative bacilli.[36] The increased prevalence of extended-spectrum beta-lactamases may contribute to the finding of multidrug resistance. Linezolid, which is commonly used antibiotic for Gram positive bacteria also showing sensitivity against Gram negative bacteria in our study. This shows need to further study for in vivo effectiveness of Linezolid against Gram negative bacterial infections.[37]
Postoperative sepsis rate in any hospital depends much on the case material, hospital environment, irrational use, and availability of antibiotics. Antibiogram might be varying depending on the study group and hospital set up. So the trend now a day is comparative studies in the same hospital over years.
CONCLUSIONS
There is an alarming increase of infections caused by antibiotic-resistant bacteria. Lack of uniform antibiotic policy and indiscriminate use of antibiotics may have lead to emergence of resistant bacterial strains. Particularly pseudomonas resistance to third generation cephalosporin and meropenem and VRSA/VISA are real threat to control hospital acquired infection. Hence, there should be an immediate response from the concerned authorities to check further emergence and spreading of these notorious VRSA strains and also there is a need to emphasize the rational use of antimicrobials and strictly adhere to the concept of “reserve drugs” to minimize the misuse of available antimicrobials. In our study linezolid shows good sensitivity against gram negative organism so further studies are required to check it. In addition, regular antimicrobial susceptibility surveillance is essential for area-wise monitoring of the resistance patterns. An effective national and state level antibiotic policy and draft guidelines should be introduced to preserve the effectiveness of antibiotics and for better patient management.
Acknowledgment
The authors are thankful to Dr. Manahar V. Mehta, Professor and Head, Dept. of Surgery, K. M. Shah Medical College, Vaghodia and Dr. Malasinha, Shri M. P. Shah Medical College and Guru Gobindsingh Hospital, Jamnagar and EOS laboratory, Mumbai for providing antibiotic sensitivity disc.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Ashby E, Haddad FS, O’Donnell E, Wilson AP. How will surgical site infection be measured to ensure “high quality care for all”? J Bone Joint Surg Br. 2010;92:1294–9. doi: 10.1302/0301-620X.92B9.22401. [DOI] [PubMed] [Google Scholar]
- 2.Mangram AJ, Horan TC, Pearson ML, Silver LC, Jarvis WR. Guideline for prevention of surgical site infection, 1999.Hospital Infection Control Practices Advisory Committee. Infect Control Hosp Epidemiol. 1999;20:250–78. doi: 10.1086/501620. [DOI] [PubMed] [Google Scholar]
- 3.Tiwari HK, Sen MR. Emergence of vancomycin resistant Staphylococcus aureus from a tertiary care hospital from northern part of India. BMC Infect Dis. 2006;6:156. doi: 10.1186/1471-2334-6-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hemant Singhal, Kanchan Kaur. Wound infection. Overview: History. The ancient Egyptians were the first civilization to have trained clinicians to treat physical aliments. eMed General Surg 2009. Surg Infect (Larchmt) 2009;10:323–31. [Google Scholar]
- 5.Chastre J, Trouillet JL. Problem pathogens (Pseudomonas aeruginosa and Acinetobacter) Semin Respir Infect. 2000;15:287–98. doi: 10.1053/srin.2000.20944. [DOI] [PubMed] [Google Scholar]
- 6.Obritsch MD, Fish DN, McLaren R, Jung R. National Surveillance of Antimicrobial Resistance in Pseudomonas aeruginosa isolates obtained from Intensive Care Unit Patients from 1993 to 2002. Antimicrob Agents Chemother. 2004;48:4606–10. doi: 10.1128/AAC.48.12.4606-4610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubin RH. Surgical wound infection: Epidemiology, pathogenesis, diagnosis and management. BMC Infect Dis. 2006;6:171. doi: 10.1186/1471-2334-6-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ducel G, Fabry J, Nicolle L. Prevention of hospital-acquired infections: A practical guide. World Health Organization, Department of Communicable Disease, Surveillance and response. 2nd ed. [Last cited on 2010 Nov 03]. Available from: http://apps.who.int/medicinedocs /documents /s16355e/s16355e.pdf .
- 9.Zhanel GG, DeCorby M, Laing N, Weshnoweski B, Vashisht R, Tailor F, et al. Antimicrobial-resistant pathogens in intensive care units in Canada: results of the Canadian National Intensive Care Unit (CAN-ICU) study, 2005-2006. AntimicrobAgentsChemother. 2008;52:1430–7. doi: 10.1128/AAC.01538-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasselgren PO, Saljo A, Fornander J, Lundstam S, Saretok T, Seeman T. Postoperative wound infections- a prospective study in a newly opened hospital. Ann Chir Gynaecol. 1980;69:269–72. [PubMed] [Google Scholar]
- 11.Wkler Mathew A, Cockerill Franklin R, Bush Karen, Dudley Michael N, Etiopoule George M, Hardy Dhright J, et al. Clinical and laboratory standard institute. Performance standards for an antimicrobial disc susceptibility tests; Approved standard. 10th ed. [Last cited on 2010 Nov 03]. Available from: http:// www.clsi.org/source/orders/free/m02-a10.pdf .
- 12.Suchitra JB, Lakshmidevi N. Surgical site infestions. Assessing risk factors and antimicrobial sensitivity patterns. [Last cited on 2010 Nov 03];Afr J Microbiol Res. 2009 3:185–9. Available from: http://www.academicjournals.org/ajmr . [Google Scholar]
- 13.Abu Hanifah Y. Post-operative surgical wound infection. Med J Malaysia. 1990;45:293–7. [PubMed] [Google Scholar]
- 14.Malone DL, Genuit T, Tracy JK, Gannon C, Napolitano LM. Surgical site infections.Reanalysis of risk Factors. J Surg Res. 2002;103:89–95. doi: 10.1006/jsre.2001.6343. [DOI] [PubMed] [Google Scholar]
- 15.Surucuoglu S, Gazi H, Kurutepe S, Ozkutuk N, Ozbakkaloglu B. Bacteriology of surgical wound infections in a tertiary care hospital in Turkey. East Afr Med J. 2005;82:331–6. [PubMed] [Google Scholar]
- 16.Thanni LO, Osinupebi OA, Deji-Agboola M. Prevalence of bacterial pathogens in infected wounds in a tertiary hospital, 1995-2001: any change in trend? J Natl Med Assoc. 2003;95:1189–95. [PMC free article] [PubMed] [Google Scholar]
- 17.Giacometti A, Cicrioni O, Schimizzi AM, Delprete MS, Barchiesi F, D’Errico MM, et al. Epidemiology and Microbiology of Surgical Wound infections. J Clin Microbiol. 2000;38:918–22. doi: 10.1128/jcm.38.2.918-922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber SG, Miller RR, pereneevich EN, Tolentino J, Meltzer D, Pitrak D, et al. Prevalence of antimicrobial resistant bacteria isolated from older versus younger hospitalized adults: Results of a two centre study. J Antimicrob Chemother. 2009;64:1291–8. doi: 10.1093/jac/dkp349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siegrist HH, Nepa MC, Jacquet A. Susceptibility to Levofloxacin of clinical isolates of bacteria from intensive care and hematological/oncology patients in Switzerland:a multicentre study. J Antimicrob Chemother. 1999;43:51–4. doi: 10.1093/jac/43.suppl_3.51. [DOI] [PubMed] [Google Scholar]
- 20.Kaufman D, Haas CE, Edinger R, Hollick G. Antibiotic Susceptibility in the Surgical Intensive Care Unit Compared With the Hospital-Wide Antibiogram. ArchSurg. 1998;133:1041–5. doi: 10.1001/archsurg.133.10.1041. [DOI] [PubMed] [Google Scholar]
- 21.Schmitz FJ, Fluit AC, Gondolf M, Beyrau R, Lindenlauf E, Verhoef J, et al. The prevalence of aminoglycoside resistance and corresponding resistance genes in clinical isolates of Staphylococci from 19 European hospitals. J Antimicrob Chemother. 1999;43:253–9. [PubMed] [Google Scholar]
- 22.Tiwari HK, Sen MR. Emergence of vancomycin resistant Staphylococcus aureus (VRSA) from a tertiary care hospital from northern part of India. BMC Infect Dis. 2006;6:156. doi: 10.1186/1471-2334-6-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.David EB, Edward RA, Carl AB. Fundamentals Of Molecular Diagnostic - Google Books Result – 2007 – Medical – 267 pages. There have been few cases of infection by VRSA reported in the United… the possibility of falsepositive results in mixed cultures of S. aureus and CoNS…. [Last cited on 2010 Nov 03]. available from: http://www.books.google.co.in/books?isbn=1416037373… .
- 24.Andrews JM. The development of the BSAC standardized method of disc diffusion testing. J Antimicrob Chemother. 2001;48:29–42. doi: 10.1093/jac/48.suppl_1.29. [DOI] [PubMed] [Google Scholar]
- 25.Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, Hori S, Hosoda Y, et al. Dissemiantion in Japanese Hospitals of strains of Staphylkococcus aureus heterogeneously resistant to vancomycin. Lancet. 1997;350:1670–3. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- 26.Howe RA, Wootton M, Bennett PM, Mac Gowan AP, Walsh TR. Interactions between methicillin and vancomycin in methicillin resistant Staphylococcus aureus strains displaying different phenotypes of vancomycin susceptibility. J Clin Microbol. 1999;37:3068–71. doi: 10.1128/jcm.37.9.3068-3071.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wootton M, Walsh TR, Howe RA, White G, Andrews JM, Wise R. Washington DC: American Society for Microbiology; 1996. Methodological factors in the detection of heterogeneously vancomycin resistant Staphylococcus aureus (h VRSA) p. 108. San Diego, CA, USA: Program and Abstracts of the Thirty- eighth Interscience Conference on Antimicrobial Agents and Chemotherapy; Abstract C-139. [Google Scholar]
- 28.Iyer RN, Hittinahalli V. Modified PAP method to detect heteroresistance to vancomycin among methicillin resistant Staphylococcus aureus isolates at a tertiary care hospital. Indian J Med Microbiol. 2008;26:176–9. doi: 10.4103/0255-0857.40537. [DOI] [PubMed] [Google Scholar]
- 29.Burnham CA, Weber CJ, Michael WM., Jr Novel Screening Agar for Detection of Vancomycin- Nonsusceptible Staphylococcus aureus. J Clin Microbiol. 2010;48:949–51. doi: 10.1128/JCM.02295-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Eldere J. Multicentre surveillance of Pseudomonas aeruginosa susceptibility patterns in nosocomial infections. J Antimicrob Chemother. 2003;51:347–52. doi: 10.1093/jac/dkg102. [DOI] [PubMed] [Google Scholar]
- 31.Gales AC, Jones RN, Turnidge J, Rennie R, Ramphal R. Characterization of Pseudomonas aeruginosa: occurrence rate, antimicrobial susceptibility pattern and molecular typing in the global sentry antimicrobial surveillance program 1997-1999. Clinic Infect Dis. 2001;32:S146–55. doi: 10.1086/320186. [DOI] [PubMed] [Google Scholar]
- 32.Jonas RN. Resistance pattern among nosocomial pathogens: trends over the past few years. Chest. 2001;119:397S–404S. doi: 10.1378/chest.119.2_suppl.397s. [DOI] [PubMed] [Google Scholar]
- 33.Fadeyi A, Akanbi AA, 2nd, Nwabuisi C, Onile BA. Antibiotic disc sensitivity pattern of Pseudomonas aeruginosa isolates obtained from clinical specimen in Ilorin, Nigeria. Afr J Med. 2005;34:303–6. [PubMed] [Google Scholar]
- 34.Raja NS, Singh NN. Antimicrobial susceptibility pattern of clinical isolates of Pseudomonas aeruginosa in tertiary care hospital. J Microbiol Immunol Infect. 2007;40:45–9. [PubMed] [Google Scholar]
- 35.Bertrand XM, Thouverez M, Patry C, Balvay P, Talon D. Pseudomonas aeruginosa: Antibiotic susceptibility and genotypic characterization of strains isolated in the intensive care unit. Clin Microbiol Infect. 2002;7:706–8. [PubMed] [Google Scholar]
- 36.Clark NM, Patterson J, Lynch JP., 3rd Antimicrobial resistance among gramnegative organisms in the intensive care unit. Curr OpinCrit Care. 2003;9:413–23. doi: 10.1097/00075198-200310000-00012. [DOI] [PubMed] [Google Scholar]
- 37.Sweeney MT, Zurenko GE. In vitro activities of linezolid combined with other antimicrobial agents against staphylococci, enterococci, pneumococci, and selected gram-negative organisms. Antimicrob Agents Chemother. 2003;47:1902–6. doi: 10.1128/AAC.47.6.1902-1906.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]