Abstract
Kaposi's sarcoma is an angioproliferative tumor caused by Kaposi's sarcoma–associated herpesvirus (KSHV) infection of vascular endothelial cells. Fibulins, proteins that associate with extracellular matrix (ECM) proteins, may have both tumor-suppressive and oncogenic activities. We found that the expression of fibulin-2 protein and mRNA were decreased 50-fold and 26-fold, respectively, in 10-day KSHV-infected dermal microvascular endothelial cells (DMVEC). Using quantitative RT-PCR, we found a fivefold and 25-fold decrease of fibulin-2 extracellular matrix binding partners, fibronectin and tropoelastin, respectively. Time-course transcriptional analyses over 10 days showed that in addition to that of fibulin-2, expression of fibulins 3 and 5 was decreased in KSHV-infected DMVEC, fibulins 1C/1D were increased, and fibulins 4, 6, and 7 were unchanged. KSHV latency-associated nuclear antigen (LANA) transcription levels rose consistently over the same period. Addition of recombinant fibulin-3 or -5 for 48 hours to 10-day KSHV-infected cells caused a suppression of KSHV-induced vascular endothelial growth factor (VEGF) protein and mRNA levels. Recombinant fibulin-3 also significantly reduced VEGF receptor 3 expression. In pleural effusion lymphoma cell lines that express variable levels of KSHV lytic replication, we observed no detectable fibulin-2 or -5 expression. Finally, fibulin-2 expression was decreased in tissue microarrays from KSHV-infected, LANA-positive patient cells as compared to that in patient nontumor controls. Understanding the interactions between KSHV and the fibulins may lead to the development of novel therapies for treatment of Kaposi's sarcoma.
Before highly active antiretroviral treatment in 1996, at the height of the HIV/AIDS epidemic, the incidence of Kaposi's sarcoma (KS) increased 20,000-fold among gay males in the United States1,2 and is now the predominant HIV/AIDS-related malignancy in Southern Africa and, hence, the world.3 Kaposi's sarcoma–associated herpesvirus (KSHV) or Human Herpesvirus type 8 (HHV8) is the etiological agent of KS.4,5 KS is characterized as an angioproliferative tumor caused by KSHV infection of vascular endothelial cells and produces rare B-cell lymphoproliferative diseases in the form of pleural effusion lymphomas (PEL) and some forms of Multicentric Castleman's Disease. KS lesions are usually papular in nature, but can be plaque-like as well, and in a more advanced stage, they develop into nodular tumors with extensive organ involvement.6
Fibulins comprise a seven-member family of extracellular matrix glycoproteins that share a carboxyl-terminal globular domain plus a series of calcium-binding epidermal growth factor (EGF)-like motifs, and are known to be highly conserved throughout evolution.7,8 The human fibulin-1 protein contains 20 exons and, by the process of alternative splicing, yields four separate transcript variants designated 1A to 1D.7–9 Fibulin-2, the primary focus of this study, is the second largest member of the fibulin family because of a unique 400-amino acid N-terminal domain.10 Fibulins 3, 4, and 5 are relatively smaller in size (50 to 60 kDa) and share a higher degree of structural similarity to one another.11 They are characterized by a modified EGF-like motif at the N-terminus upstream of five tandem EGF repeat elements and a fibulin-type C terminal domain. Fibulin-6 is the largest of the fibulin family members and is distinguished by its large N-terminal immunoglobulin C-2 domain.12 Fibulin-7 is the most recently discovered member of this family and is the human homolog of the mouse fibulin-7 gene also known as TM14.13 The smallest of the fibulins, human fibulin-7 isoform-1, is only 439 amino acids and has a unique sushi domain with only three EGF-like motifs.13
Fibulins are reported to have both tumor-suppressive and oncogenic activities as indicated by the following examples: i) fibulin-1 is overexpressed in ovarian epithelial tumors and breast cancer,14,15 whereas loss of fibulin-2 expression is linked to breast cancer progression16; ii) fibulin-3 antagonizes tumor angiogenesis in vivo, and its expression is decreased in various tumors17; iii) fibulin-4 expression is increased in colon cancer18; and iv) fibulin-5 expression is increased in the HT1080 fibrosarcoma cell line and might inhibit angiogenesis.17,19 However, fibulin-5 expression is also reported to be decreased in ovarian, colon, kidney, and breast cancer.19 Fibulin-6 and -7 have yet to be linked to cancer.
Thus fibulins are ECM proteins involved in cell adhesion, proliferation, migration, invasion, and angiogenesis that have been linked to progression of several cancer types, but have not been studied in KS. We examined fibulin-2 in studies leading up to this report because we found it to be significantly down-regulated by microarray analysis in KSHV-infected DMVEC cells. Because of our results, we also assayed other fibulins for their activities after KSHV infection of DMVEC cells, as well as examined KS archival tissue for expression of fibulin-2. Notably, we found fibulins 2, 3, and 5 to be down-regulated over time in KSHV-infected DMVEC, whereas fibulins 1C and 1D were up-regulated, with no change in fibulins 4, 6, and 7. This represents the first study that examines fibulin-2 expression in KSHV-infected DMVEC and KS. Our results suggest that changes in expression of these ECM proteins may play a role in tumor progression.
Materials and Methods
Cells and Viruses
The BCBL-1 cell line originally isolated from a body cavity–based lymphoma was cultured in RPMI 1640 medium (Gibco, Grand Island, NY; supplemented with 10% FBS, 10 mmol/L HEPES, l-glutamine) until the cell density reach 3 × 106 cells/mL. Lytic cycle virus replication was then induced with TPA (20 ng/mL) and sodium butyrate (0.3 ng/mL). Twenty-four hours after induction, cells were washed twice in PBS to remove butyrate, and induction was continued with TPA for 5 days. Cell-free virus was isolated and concentrated by differential centrifugation. DMVEC maintained in complete EMB-2 medium (Lonza, Basel, Switzerland) obtained from Lonza Corporation were infected at passage 4 at a multiplicity of infection of 0.01; mock-infected cells were used as controls. BJAB (B-cell control) and PEL cell lines JSC-1, BC3, BC2, BCBL-1, and HBL6 were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum and 100 μg/mL of penicillin/streptomycin.
Giemsa Staining
Normal and KSHV-infected DMVEC cells were cultivated in chamber slides at 80% confluence. Medium was removed, and cells were washed three times with PBS (pH 7.4) then fixed for 10 minutes in absolute methanol at −20°C for 15 minutes. Giemsa stock stain was diluted in Giemsa buffer and cells were stained according to the manufacturer's recommendations (Invitrogen, Carlsbad, CA). Stained cells were dried and sealed with a glass coverslip using permanent mounting medium. Slides were viewed on a Nikon TE2000S microscope (Nikon, Tokyo, Japan) under bright-field illumination at a total magnification of ×200.
Western Blot
To determine levels of fibulin-2 protein expressed by DMVEC cells before and after KSHV infection, extracts from infected and mock-infected cells were prepared using radioimmunoprecipitation assay lysis buffer [50 mmol/L Tris (pH 7.5), 150 mmol/L NaCl, 2 mmol/L EDTA (pH 8.0), 1% NP40, 0.5% deoxycholate sodium, 0.1% SDS, and 0.25 mg/mL proteinase inhibitor]. Lysates were placed on ice for 30 minutes and then clarified by centrifugation. Supernatants were removed and total protein measured by BCA assay (Pierce; Thermo Fisher Scientific, Waltham, MA). Fifteen micrograms of protein lysates for mock-infected and infected paired samples were fractionated in 4% to 20% SDS-PAGE gels, transferred to nitrocellulose membranes, blocked with 5% milk powder, 0.1% TBST (0.1% Tween 20, 20 mmol/L Tris, 150 mmol/L NaCl) and incubated separately at 4°C overnight with a primary rabbit antibody fibulin-2, LANA, and β actin, all at a 1:2000 dilution. Membranes were washed five times in washing buffer (0.1% TBST) and incubated for 1 hour, followed by a secondary mouse anti-rabbit peroxidase conjugate (Jackson ImmunoResearch Laboratories, West Grove, PA), at a dilution of 1:10,000. Immunoreactive bands were detected with SuperSignal West-Dura Extended Duration Substrate (Pierce; Thermo Fisher Scientific) following exposure to X-ray film.
Southern Blot Hybridization
Cultured PEL cell lines JSC-1, BC3, BC2, BCBL-1, and HBL6, and BJAB cells were washed three times with PBS, pelleted by centrifugation, and lysed with lysis buffer (1% gelatin, 25 mmol/L EDTA, 1% SDS). PEL lysates were extracted with phenol-chloroform and precipitated with 2.5 mol/L ammonium acetate in ethanol. Ten micrograms of total genomic DNA from PEL cells were electrophoresed in 0.8% agarose gels, transferred to nylon membranes, and then hybridized to a 32P-labeled DNA probe representing an 800-bp NotI fragment of the terminal repeat region of the KSHV genome. Denatured DNA fragments were labeled according to the manufacturer's recommendations using a random primer DNA labeling kit (Invitrogen). Hybridized bands were visualized by autoradiography after exposure to X-ray film.
Densitometry
Densitometry analysis was performed on immunoblots from mock- and KSHV-infected DMVEC cells using a Bio-Rad Chemi-Doc XRS gel docking system and Quantity One 4.6.2 software (Bio-Rad Laboratories, Hercules, CA).
Immunohistochemistry
Dual-labeled immunohistochemistry for fibulin-2 and KSHV LANA was performed on nodular archival AIDS-KS tissue fixed in formalin, embedded in paraffin, sectioned, and then placed on chemate slides before immunohistochemistry. For staining, antigen retrieval was performed with microwave oven pretreatment (500 W) in citrate buffer at 95°C for 15 minutes. Cells were then incubated 30 minutes in PBS (pH 7.2) containing 10% normal goat serum with 1% Tween-20 for blocking of nonspecific binding and permeabilization. Cells were then incubated with a rabbit polyclonal antibody to fibulin-2 (Santa Cruz Biotechnology, Santa Cruz, CA) diluted 1:100 in blocking buffer. After 90 minutes of incubation, cells were subsequently washed, and incubated with a biotinylated donkey anti-rabbit IgG antibody (Dako, Glostrup, Denmark) at a 1:100 dilution and then with a horseradish peroxidase–avidin conjugate (Dako) at 1:500 dilution. Color development was achieved by incubating cells with the substrate 3,3-diaminobenzidine (DAB, Sigma, St. Louis, MO). The second labeling was performed by an additional antigen retrieval step in the microwave as described above and a second round of staining with a monoclonal antibody to the KSHV LANA (Vector Laboratories, Burlingame, CA) at a 1:100 dilution. Cells were subsequently washed, incubated with a biotinylated goat anti-mouse/rabbit IgG antibody (Dako) at a 1:100 dilution, and then an alkaline phosphatase–strepavidin conjugate (Vector Laboratories) at a 1:500 dilution. Color development was generated by incubating cells with the substrate Vector Red using a Vector Red Alkaline Phosphatase Substrate Kit (Vector Laboratories) according to the manufacturer's instructions. Images were taken with a Nikon TE-2000S microscope mounted with a charge-coupled device camera.
Microarray Analysis
The GeneChip Human Genome U133 Plus 2.0 array (Affymetrix, Santa Clara, CA) with complete coverage of the human genome containing over 47,000 transcripts was used to identify gene expression changes in primary DMVEC cells 10 days after KSHV infection. Several genes were found to be down-regulated after KSHV infection. Fibulin-2 was identified as being significantly down-regulated and therefore was the first fibulin selected for further study.
RNA and cDNA Amplification
Total RNA was extracted from KSHV-infected DMVEC cells and mock-infected cells using a Qiagen mini RNA isolation kit (Qiagen, Valencia, CA). The RNA was DNAase treated before elution on the column according to the manufacturer's recommendations. mRNA in 1 μg of each sample was primed using oligo-dT and reverse transcribed with a High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA).
Real-Time Quantitative PCR
Primer sets for quantitative PCR (qPCR) included fibulin-3 forward 5′-TGCCATCAGACATCTTCCAG-3′, reverse 5′-TGCCTGTGGTTGACTCTTAGAA-3′ (292 bp); fibulin-4 forward 5′-GCCCAAACCTGTGTCA-3′, reverse 5′-TGATTTGCCTAATGTAAAAGTCC-3′ (288 bp); fibulin-6 forward 5′-TGCTCGGTCCAAAGATTACC-3′, reverse 5′-TGGAGGCCTGTACTGCTTCT-3′ (227 bp); and fibulin-7 forward 5′-CTCTGCCTTCCAACCTGAAG-3′, reverse 5′-CAGGTTATGCACAAGGATCA-3′ (179 bp) were used. Primer sequences for qPCR for fibulin-2 and fibulin-5 were identical to primers sets used in the RT-PCR reactions. qPCR primer sets for fibulin-2 binding protein partners included aggrecan forward 5′-GCCTCAGAAAAGTGCTGACC-3′, reverse 5′-GTCCCTCTGTCTCCTTGCAG-3′ (197 bp); fibronectin forward 5′-CCCAACTGGCATTGACTTTT-3′, reverse 5′-CTCGAGGTCTTTCACTGAAG-3′ (136 bp); fibrillin forward 5′-GTGACTGCCCACCTGATTTT-3′, reverse 5′-AGCAGGAAGCTTTGGAAACA-3′ (156 bp); laminin α2 forward 5′-AAAGTATCTGTGTCTTCAGGAGGTGC-3′, reverse 5′-GCAGCCAGTGAATGTAATCACACGTC-3′ (474 bp); nidogen forward 5′-ATAGAAGTGGCGAAGCTGGA-3′, reverse 5′-GTGCCGTCCATGTAGGAAGT-3′ (161 bp); perlecan forward 5′-GGAGCCTATTTCCACGATGA-3′, reverse 5′-GAGGCTGATGAAGTCCTTGC-3′ (180 bp); tropoelastin forward 5′-ACCTGGGACAACTGGAATCC-3′, reverse 5′-AAAGCAGCAGCAAAGTTCGG-3′ (288 bp); and versican forward 5′-CGGGATCCGGGGTGAGAACCCTGTATCG-3′, reverse 5′-ACTCTAGAGGCCACGCCTAGCTTCTGCAGC-3′ (375 bp) were used.
Primer sets for amplification of VEGF 121, 165, and 189, and VEGF receptor 1 (VEGFR1), VEGF receptor 2 (VEGFR2), and VEGF receptor 3 (VEGFR3) are as follows: VEGF 121, 165, and 189 forward 5′-GAGATGAGCTTCCTACAGCAC-3′, reverse 5′-TCACCGCCTCGGCTTGTCACAT-3′; VEGFR1 forward 5′-GGAACAAGGCAAGAAACCAA-3′, reverse 5′-CGATGAATGCACTTTCTGGA-3′; VEGFR2 forward 5′-ATCCCTGTGGATCTGAAACG-3′, reverse 5′-CCAAGAACTCCATGCCCTTA-3′; and VEGFR3 forward 5′-TGAAAGCATCTTCGACAAGG-3′, reverse 5′-TTCAGCATGATGTGGCGTAT-3′.
Real-time PCR was performed in 96-well optical plates (Sorenson Bioscience, Salt Lake City, UT) with cDNA using the MyiQ Single Color Real-Time PCR Detection System (Bio-Rad Laboratories) in 25-μL reaction volumes. A master mix was made according to the manufacturer's instructions using SYBR Green Supermix (Bio-Rad Laboratories). Veriquest SYBR Green Supermix was substituted when amplifying high GC-rich cDNA templates according to the manufacturer's instructions (Affymetrix). Forward and reverse primers were used at a concentration of 250 nmol/L per well, made in RNAase and DNAase free H2O. Primer sets for fibulin-2 binding proteins included the cDNAs from mock-infected DMVEC cells and KSHV-infected DMVEC were diluted 1:3 using RNAase DNAase free H2O; 3 μL of this dilution was added to each well. Control wells substituted water for cDNA. The cycling sequence included 95°C for 3 minutes, 95°C for 15 seconds, 60°C for 1 minute, 95°C for 1 minute, 55°C for 1 minute, and 55°C for 30 seconds for 81 cycles total. A GAPDH primer set was included for normalization. Data analysis was done with the Bio-Rad iQ5 Optical System Software Version 2.
Human Angiogenesis ELISA
The effects of fibulins on KSHV-induced angiogensis were measured using a human angiogenesis ELISA assay kit profiling six angiogenic cytokines (Signosis, Sunnyvale, CA). Normal and KSHV-infected DMVEC cells were plated at a density of 4 × 105 cells in six-well dishes and grown to 90% confluence (10 days). Normal DMVEC, KSHV-infected DMVEC, and KSHV-infected DMVEC exposed to 10 μg/mL of purified recombinant fibulin-3 protein (Origene, Rockville, MD) or 10 μg/mL of purified recombinant fibulin-5 protein (Origene) were cultured in EBM-2 media (Lonza, Walkersville, MD) supplemented with 2% fetal calf serum only for an additional 48 hours. Fibulin recombinant protein concentrations needed for biological activity were estimated based on prior studies in mice angiogenic sprouting assays.20 Cell supernatants were then assayed for angiogenic cytokines and compared to mock-infected control cells and KSHV-infected DMVEC cells, according to the manufacturer's protocol. Cell monolayers were harvested, washed three times in phosphate-buffered saline (pH 7.4), and total RNA extracted for real-time PCR analysis.
Tissue Microarrays
Tissue microarrays were obtained from AIDS Cancer Specimen Resource Consortium (ACSR, San Francisco, CA). All specimens were procured and used after obtaining approval by the institutional review board of Meharry Medical College. Paraffin-embedded KS tissue of 0.6-mm core sizes representing patch/plaque and nodular forms of KS from multiple patients were placed on slides. KS tumor tissue from the mouth, skin, tongue, soft palate, and neck masses were included on the array. These specimens included areas of surrounding tissue (controls) that were not infected as judged by the lack of a spindle shape morphology and LANA-positive staining. Only those tissues staining positive for LANA were considered KS tumor cores. All tissues were from HIV-positive patients. When we observed a >50% reduction in fibulin-2 expression in LANA-positive spindle cell regions in 50 optical fields compared to LANA-negative regions, then in 50 optical fields that showed a >50% reduction in fibulin-2, then that specific tumor core was scored as being reduced in expression for fibulin-2 in virus-positive cells.
Results
KSHV Infection of DMVEC
Uninfected DMVEC cells displayed a cobblestone appearance as seen by phase microscopy and Giemsa staining, which is consistent with the morphology of normal, early passaged DMVEC cells (Figures 1, A and 1B). These normal DMVEC were infected in vitro with BCBL1 virus, and 10 days after infection, distinct morphological changes were observed that were consistent with the spindle cell phenotype for KSHV-infected DMVEC (Figure 1C) and KS tumors in vivo.6 This swirling cell pattern was more pronounced when the infected cells were stained with Giemsa stain (Figure 1D). We observed a similar spindle cell morphology in an AIDS-KS tumor specimen stained positive (brown) for KSHV LANA (Figure 1E), which also showed a characteristic nuclear punctate staining pattern as shown by immunohistochemical staining of the tissue (Figure 1F).
Figure 1.
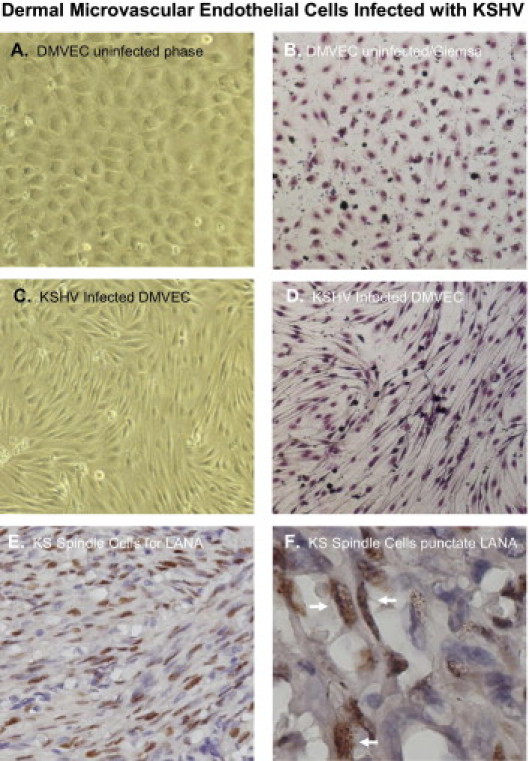
Human dermal microvascular endothelial cell (DMVEC) mock-infected controls and DMVEC cells infected with KSHV for 10 days. For infections, cells were treated with BCBL-1 virus at a multiplicity of infection (moi) of 0.01 and cultivated in EBM-2 complete medium. Phase images were taken with a NIKON TE 2000S microscope mounted with a CCD camera. Shown are phase (A) and Giemsa (B) stained images of mock-infected cells showing a cobblestone-like morphology C: KSHV-infected cells with the characteristic spindle shape morphology. D: This type of swirling/spindling appears more pronounced when a Giemsa stain is performed and appears very distinct from uninfected cells. E: An IHC stain of a nodular AIDS-KS tumor showing KSHV LANA positive spindle cells with a hematoxylin counterstain. F: Shows KS tissue with spindle cells stained positive for KSHV LANA with a nuclear punctate staining pattern (white arrows). All images were, with the exception of Figure 1F, taken at a total magnification of ×200. The image in Figure 1F was taken at a total magnification of ×600 to discern the nuclear punctate staining pattern of KSHV LANA.
KSHV Decreases Fibulin-2 Protein and RNA Expression in DMVEC Cells
Using microarray analysis, we observed that fibulin-2 was significantly down-regulated in KSHV-infected DMVEC cells in comparison to mock-infected control cells (data not shown). To validate our microarray data, we determined the protein expression levels of fibulin-2 in KSHV-infected DMVEC by Western blot analysis and found a significant reduction in fibulin-2 protein (Figure 2A). By densitometry analysis of the Western blot, it was determined that there was a more than 50-fold reduction in fibulin-2 protein expression (Figure 2B).
Figure 2.
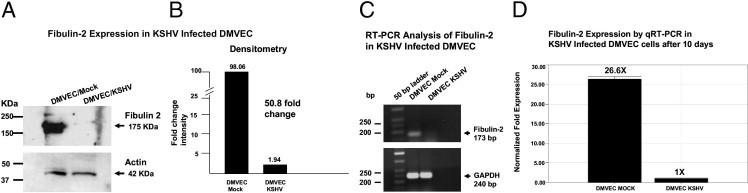
Analyses for fibulin-2 expression at 10 days after mock or KSHV infection of DMVEC. A: Lysates (15 μg) from mock- and KSHV-infected DMVEC were separated by electrophoresis in 4% to 20% PAGE gels transferred to nitrocellulose membranes and screened for expression of fibulin-2 using a rabbit polyclonal antibody (Santa Cruz Biotechnology) at a 1:2,000 dilution. Western blot bands show evidence of a ∼175 kDa protein in lysates from mock-infected DMVEC compared to the mark reduction of this band in KSHV-infected DMVEC. B: Densitometry analysis of the Western blots in Figure 2A reveals a greater than 50-fold reduction in fibulin-2 protein expression in KSHV-infected DMVEC cells at 10 days compared to mock-infected control cells. C: Semiquantitative RT-PCR analysis of fibulin-2 mRNA expression in KSHV-infected DMVEC. Ten nanograms of cDNA from mock- and KSHV-infected DMVEC were amplified by PCR using fibulin-2 gene–specific primers. PCR products were electrophoresed in 1.5% agarose, and PCR DNA fragments were sized using a 50-bp ladder. The expected fragment size for fibulin-2 is 173 bp. GAPDH was amplified as a loading control with a fragment size of 240 bp. D: Real-time RT-PCR analysis of fibulin-2 expression in KSHV-infected DMVEC cells 10 days after infection compared to mock-infected control cells. Relative fold expression was normalized to GAPDH.
To determine whether KSHV infection induced transcriptional down-regulation of fibulin-2 as well, we prepared cDNA from mock- and KSHV-infected DMVEC cells and performed semiquantitative RT-PCR (sqRT-PCR). We show a significant down-regulation of fibulin-2 in KSHV-infected DMVEC by sqRT-PCR 10 days after infection (Figure 2C). After quantitation by real-time RT-PCR (qRT-PCR), we observed a 26-fold reduction in fibulin-2 message in KSHV-infected DMVEC cells 10 days after infection compared to mock-infected controls (Figure 2D). Thus, transcriptional down-regulation of fibulin-2 correlated with the decrease seen in fibulin-2 protein expression observed by immunoblot analysis.
Dysregulation of Fibulin-2 Binding Partners Fibronectin and Tropoelastin in KSHV-Infected DMVEC Cells
After observing a 50-fold reduction in fibulin-2 expression in 10-day KSHV-infected DMVEC cells, we wanted to determine the effects of fibulin-2 dysregulation on its extracellular matrix binding partners aggrecan, fibronectin, fibrillin, laminin α2, nidogen, perlecan, tropoelastin, and versican.21–26 Using quantitative PCR (qRT-PCR), we observed a fivefold and 25-fold down-regulation of fibronectin and tropoelastin, respectively, with no significant changes in the other fibulin-2 ECM binding partners examined (Figure 3).
Figure 3.
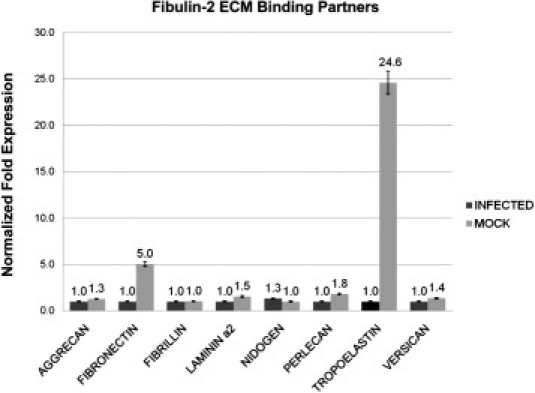
Real-time PCR analysis of fibulin-2 binding partners in KSHV-infected DMVEC cells. cDNA was prepared from total RNA isolated from 10-day KSHV-infected DMVEC cells along with mock-infected control cells. Black bars indicated fold transcription for KSHV-infected DMVEC cells and gray bars represent fold change in transcription of mock-infected cells. This experiment was replicated three times, and amplifications were performed in triplicate and normalized to GAPDH.
Transcription of Other Fibulins in KSHV-Infected DMVEC by qRT-PCR
Because other fibulins have been implicated in cancer and may serve an important role in the establishment and maintenance of the extracellular matrix in these cells, we examined the transcription profile of the seven additional fibulins in 10-day KSHV-infected DMVEC cells. The results are shown in Figure 4. Within this group, we found no significant changes in mRNA expression for fibulins 4, 6, and 7 in infected versus mock-infected cells (Figure 4). We also found no demonstrable levels of either fibulin-1A or -1B in either control or infected DMVEC cells (data not shown). In contrast, fibulin-1C and -1D mRNA expression was up-regulated 10-fold and 47-fold, respectively, 10 days after KSHV infection. Fibulins 3 and 5, like fibulin-2, were down-regulated in KSHV-infected DMVEC. The fibulin-2, -3, and -5 data correlates with previous reports of their down-regulation in several human tumors.27 We observed no significant change in mRNA expression of fibulin-6 and fibulin-7. Because there are no known associations of these two fibulins with cancer, they were excluded from further study (Figure 4). Thus, mRNA levels for fibulins 2, 3, and 5 were all reduced at 10 days after KSHV infection in DMVEC cells, whereas fibulins 1C (10×) and particularly 1D (47×) were significantly up-regulated (Figure 4).
Figure 4.
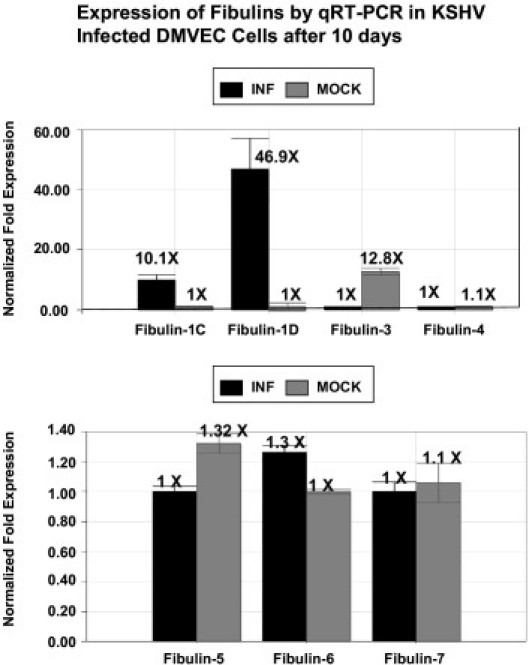
RT-PCR analysis of fibulins. Transcription analysis of fibulins in KSHV-infected DMVEC by RT-PCR. Shown are transcriptional analysis of fibulin family members fibulin-1C, fibulin-1D, fibulin-3, and fibulin-4 (A) as well as fibulin-5, fibulin-6, and fibulin-7 (B). Ten nanograms of cDNA from mock- and KSHV-infected DMVEC (10 days) were amplified by qRT-PCR using a group of fibulin gene–specific primers.
Down-Regulation of Fibulin-2, Fibulin-3, and Fibulin-5 in KSHV-Infected DMVEC Correlates with Increases in Viral LANA Transcription
We examined the temporal expression of fibulin-1C, -1D, -2, -3, and fibulin-5 in KSHV-infected DMVEC by time course transcriptional analysis using quantitative real-time RT-PCR. For fibulin-1C, at 2 days after infection, we observed a marginal down-regulation of mRNA expression in KSHV-infected cells. However, at 5, 7, and 10 days after infection, we observed a consistent increase in fibulin-1C and -1D mRNA expression in KSHV-infected DMVEC (Figure 5, A and B). There is a marginal, but insignificant, increase at 2 days for fibulin-1D. For fibulin-2 at 2 days after infection, we observed a marginal, but insignificant, up-regulation of mRNA expression followed by a consistent decrease in mRNA synthesis at 5, 7, and 10 days after KSHV infection compared to controls (Figure 5C). We observed a similar pattern of mRNA expression for fibulin-3 as we found with fibulin-2 (Figure 5D). In contrast, with fibulin-5, we observed a temporary, but significant, down-regulation in mRNA synthesis at 5 and 7 days after KSHV infection compared to mock-infected control cells. In contrast, the difference in expression between infected and mock cells at day 10 was insignificant in this series of studies (Figure 5E). Taken together, fibulin-3 showed the greatest level of down-regulation followed by fibulins 5 and 2. The greatest level of transcriptional up-regulation was observed for fibulin-1D (46.9-fold) followed by fibulin-1C (10.1-fold) (Figure 4A). Over the same time frame, we observed a consistent increase in LANA transcription in KSHV-infected cells compared to mock-infected control cells (Figure 5F). This suggests that virus-induced changes in these fibulins correlated with the time course of latent viral gene expression.
Figure 5.
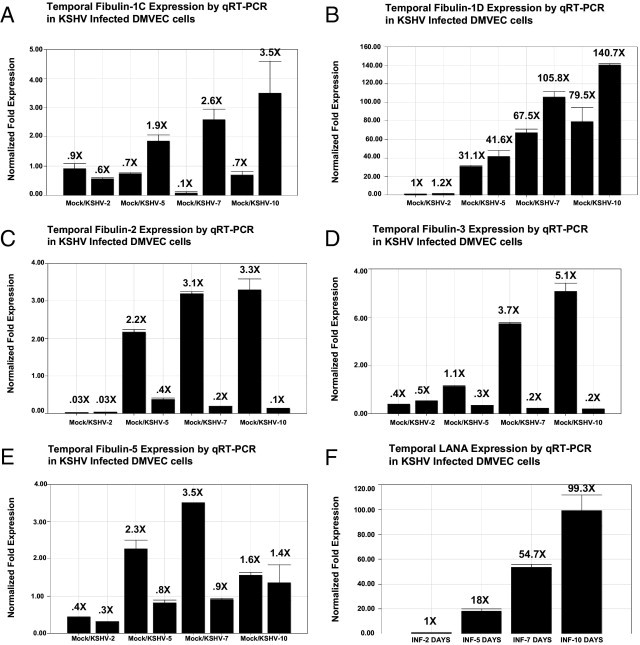
Temporal expression (2 to 10 days) of fibulins IC, ID, 2, 3, and 5 and LANA in KSHV-infected DMVEC determined by RT-PCR. A: Expression of fibulin-1C in KSHV- and mock-infected DMVEC cells after 2, 5, 7, and 10 days after infection. B: Temporal expression of fibulin-1D, mock, or KSHV infection. C: Temporal expression of fibulin 2, mock versus KSHV infection. D: Temporal expression of fibulin-3, mock versus KSHV infection. E: Temporal expression of fibulin-3, mock versus KSHV infection. F: Shows temporal expression of KSHV LANA in KSHV-infected DMVEC after 2, 5, 7, and 10 days after infection. Relative fold expression was normalized to GAPDH.
Fibulin Recombinant Protein Suppression of KSHV-Induced VEGF
Confluent KSHV-infected DMVEC cells were exposed for 48 hours to recombinant fibulin-3 or fibulin-5 and compared to KSHV-infected DMVEC alone or uninfected cells, to determine the ability of these fibulins to alter the levels of angiogenic cytokines that may have been elicited by KSHV (recombinant fibulin-2 is not commercially available). Using a human angiogenic cytokine ELISA screening assay, we observed that KSHV alone increased tumor necrosis factor (TNF), VEGF, IL-6, and basic fibroblast growth factor (FGFβ) supernatant concentrations (Figure 6A). Recombinant fibulin-3 added to KSHV-infected cells reduced TNF and VEGF values to those seen in controls, but had no or little effect on FGFb. Recombinant fibulin-5 reduced VEGF values to those seen in controls, but elevated both TNF and FGFβ levels beyond those seen with KSHV infection alone. IL-6 results were particularly interesting. KHSV infection produced a four- to fivefold increase in IL-6 that was further enhanced by recombinant fibulin-3 addition, and was more than doubled by recombinant fibulin-5. Values between all four comparative groups were similar for IGF and transforming growth factor (TGFβ) concentrations.
Figure 6.
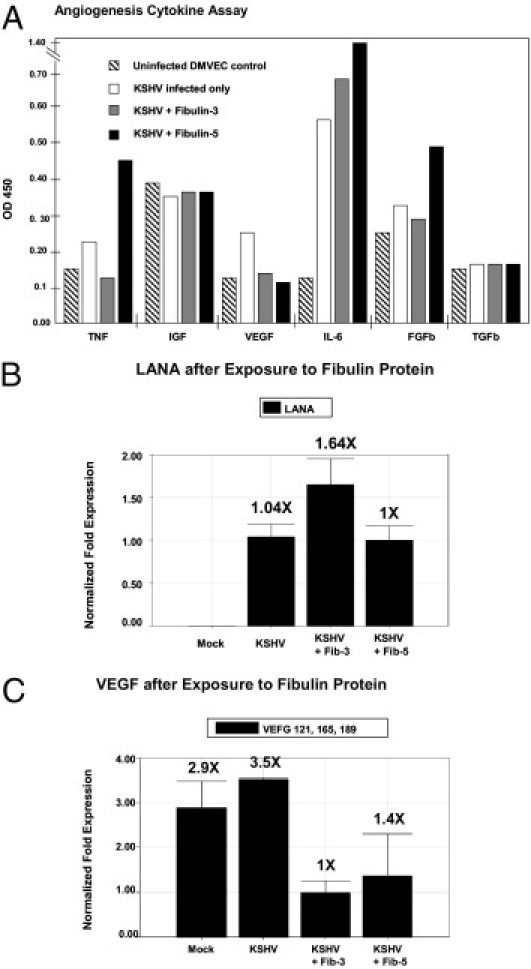
Analysis of angiogenic cytokines after KSHV infection and exposure to fibulins. A: A human angiogenic cytokine ELISA assay was performed with confluent 10-day normal and KSHV-infected cells. Results show the relative amounts of angiogenic cytokines TNFα, IGF, VEGF, IL-6, FGFb, and TGFb in cellular supernatants elicited 48 hours after exposure to fibulin-3 and fibulin-5 recombinant proteins compared to normal DMVEC cultures and KSHV alone. KSHV-infected DMVEC were exposed to 10 μg/mL of purified recombinant fibulin-3 protein or 10 μg/mL of purified recombinant fibulin-5 protein. B: Shows real-time PCR analysis using RNA extracted from cell pellets for KSHV LANA at 48 hours after exposure to fibulin-3 or fibulin-5 recombinant proteins compared to mock-treated and KSHV-treated cells alone. C: Shows real-time PCR analysis of KSHV-infected DMVEC exposed to fibulin-3 or fibulin-5 proteins for VEGF 121, 165, 189, at 48 hours after exposure to these recombinant proteins compared to mock or KSHV alone. Relative fold expression was normalized to GAPDH.
Transcriptional Down-Regulation of VEGF by Fibulin-3 and Fibulin-5 Recombinant Proteins
Transcriptional expression of KSHV LANA was demonstrated in all three KSHV-infected DMVEC cell preparations, and no LANA transcription was observed in mock-infected cells (Figure 6B). Transcriptional expression of VEGF subtypes (VEGF121, VEGF165, VEGF189) were observed to be down-regulated approximately three- and twofold in KSHV-infected DMVEC exposed to fibulin-3 or fibulin-5 recombinant proteins, respectively (Figure 6C).
Fibulin-3 Recombinant Protein Suppression of KSHV Induction of VEGFR3
Zhang et al reported that KSHV infection of endothelial cells leads to the activation of VEGFR3, which alters endothelial cell function and enhances virus infectivity.28 Therefore, after observing a significant reduction of VEGF protein elicited in supernatants of KSHV-infected DMVEC cells, as well as reduced transcriptional expression of VEGF mRNA in infected cells exposed to fibulin-3 and fibulin-5 recombinant proteins, we wanted to determine the effect of these proteins on the expression of VEGF receptors in infected cells. Using qRT-PCR, we observed a reduced transcriptional expression of VEGFR3 in KSHV-infected DMVEC exposed for 48 hours to fibulin-3 recombinant protein with no significant change in the expression of VEGFR1 and VEGFR2 compared to KSHV-infected DMVEC in medium only (Figure 7, A, B, and C). In infected cells exposed to fibulin-5 protein, we observed a significant increase in expression of VEGFR1 with no significant change in expression of VEGFR2 and VEGFR3. We also observed a consistent induction of all three VEGF receptors in KSHV-infected DMVEC cells in comparison to mock-infected controls cells (Figure 7, A, B, and C).
Figure 7.
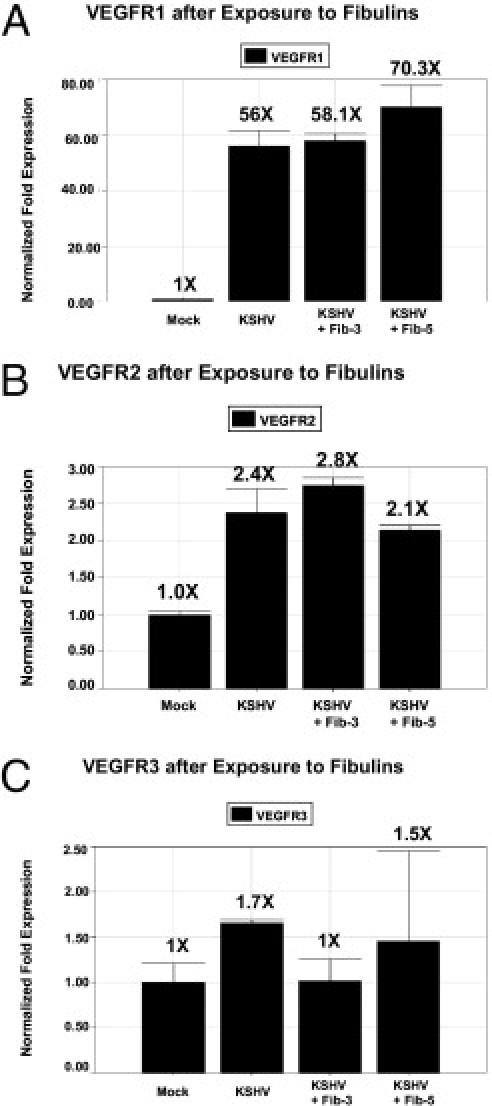
RT-PCR analysis of VEGF receptors. Real-time PCR analysis of KSHV-infected DMVEC exposed to fibulin-3 or fibulin-5 proteins for VEGFR1 (A), VEGFR2 (B), and VEGFR3 (C) at 48 hours after exposure to recombinant fibulin -3 and fibulin-5 proteins compared to mock and KSHV alone infected. Relative fold expression was normalized to GAPDH.
Expression of Fibulin-2 in PEL Cell Line
We then turned to PEL cell lines derived from these B-cell lymphomas. We wanted to determine whether there was an inverse relationship between the basal levels of fibulin-2 in established PEL cell lines and the levels of KSHV replication in those lines.
The levels of KSHV replication in several PEL cell lines (JSC-1, BC3, BC2, BCBL-1, and HBL6) were determined by Southern blot analysis, using 10 μg of total DNA/lane extracted from cell lysates without induction of the virus lytic cycle. We showed variable levels of KSHV genomic DNA replication in PEL cell lines when probed with 32P-labeled, 800-bp terminal repeat fragments from the KSHV genome (Figure 8A). This method measures terminal ends of viral genomes, indicating the degree of lytic viral replication occurring in the PEL cell line in the absence of TPA induction. The relative levels of viral expression found were consistent with those reported in the literature.29 The JSC-1 cell line, established by Cannon et al,29 2000, revealed the highest level of KSHV replication followed by BC3. Similar levels of viral DNA were found in BC2 and HBL6 PELs, but no detectable level of KSHV DNA was observed in the BCBL-1 cell line (Figure 8A). To determine the comparable expression levels of fibulin-2 in several of these PEL lines, we performed Western blot analysis on lysates from cultured primary effusion lymphoma cell lines JSC-1, BC3, BC2, and HBL6 (BJAB cells were used a control). None of these PEL lines had demonstrable fibulin-2 expression in sharp contrast to that observed in DMVEC cells (Figure 8B). We also assayed the PEL cell lines for expression of fibulin-5, and no expression was observed (data not shown). Fibulin-1C and fibulin-1D were not assayed because there are no commercial antibodies available for fibulin-1C and fibulin-1D. Moreover, the transformation of these B cells could negatively regulate fibulin-2 expression because BJAB cells are also negative for fibulin-2 expression.
Figure 8.
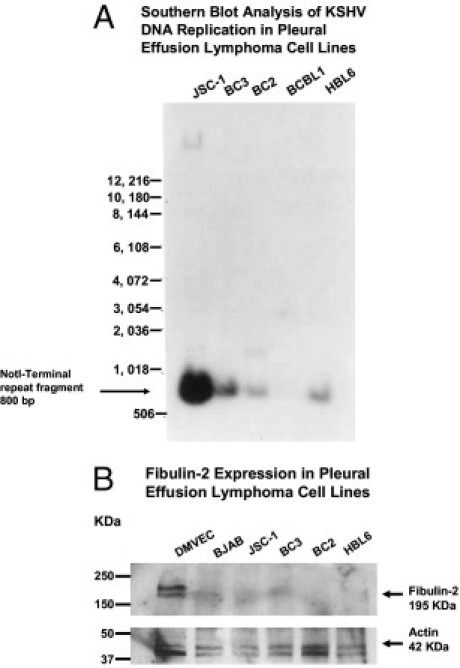
Southern blot screening of KSHV replication and fibulin-2 expression in PEL cell lines. A: DNA from PEL cell lines were screened by Southern blot hybridization using a KSHV terminal repeat probe that was 32P labeled. Hybridized fragment sizes are given in base pairs for the respective lymphoma cell line. A 1-kb DNA ladder was used to determine fragment sizes. B: A Western blot performed with lysates from PEL cell lines probed with a rabbit polyclonal antibody to fibulin-2, compared to DMVEC and a B-cell control, BJAB. Actin was used as a loading control, and protein size makers are given in kilodaltons.
Variable Levels of Viral Burden in KS Tissue
To our knowledge there are no reports of fibulin-2 expression analyses in KS tissue or any reports of fibulin-2 involvement in the pathogenesis of KS. Therefore we first examined archival KS tissue from a lesion that represented the most advanced stage of KS disease, marked by increased levels of virus replication and increased numbers of spindle cells, resembling the nodular form of KS. As shown in Figure 9A-9D, after IHC staining for KSHV LANA, these lesions express variable levels of KSHV infection that suggest a correlation between viral burden and stage of KS disease. Figure 9A and 9B represents low levels of viral burden with little to no spindle cells and Figures 9C and 9D represent intermediate to heavy viral burden with a high number of spindle cells respectively.
Figure 9.
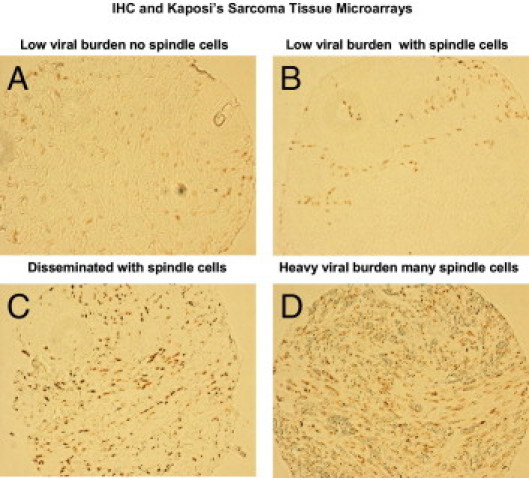
Immunohistochemical analysis of a human KS tissue array, stained for KSHV LANA, and analyzed by IHC. Illustrations represent tumor tissue with different viral burdens and different number of spindle cells. A: Tumor tissue from a patient with low viral burden with little or no spindle cells; B: Tumor tissue with some spindle cells; C: Disseminated KSHV infection with many spindle cells; D: Heavy viral burden with a high number of spindle cells.
Fibulin-2 Expression in Tissue Microarrays
Tissue microarrays were initially stained for KSHV LANA by immunohistochemistry and analyzed by bright-field microscopy. A counterstain was not included to avoid discrepancies with substrate colors. Color development for LANA was performed with DAB, and LANA-positive cells were visualized by an intense brown nuclear punctuate staining pattern. Stained tissue cores were observed at ×200 magnification. Stained tissue cores were divided into KSHV LANA-positive (tumor) and KSHV LANA-negative (nontumor) groups. There were 19 cores designated as being positive for KSHV LANA, and 67 designated as being negative after examination of 50 optical fields for each tissue core examined. Fibulin-2 was stained with DAB, and LANA was stained with the Vector Red substrate for alkaline phosphatase. LANA-positive cells in the tumor were visualized by an intense red nuclear punctuate staining pattern (Figure 10). Fibulin-2–positive cells in the tumors were visualized by brown cytoplasmic staining pattern (Figure 10). Dual-stained tissue cores were observed at ×200 magnification. Although we were supplied with 126 tissue samples, we were only able to analyze 86 that survived intact through tissue processing. We identified 19 (22%) LANA-positive KS tumors and 67 (78%) LANA-negative tissue samples. All 86 samples were found to be fibulin-2 positive, and 19 of 19 LANA-positive cores showed reduced expression of fibulin-2 in LANA-positive spindle cell regions (Figure 1, Table 1). If KSHV LANA-positive tumor core spindle cell regions in 50 optical fields exhibited a > 50% reduction in fibulin-2, they were designated reduced in expression for fibulin-2. The observation of 19 of 19 (100%) KS tumors that were LANA-positive resulting in reduced expression of fibulin-2 is highly significant when compared to LANA-negative tumors. Of the 19 LANA-positive tumors, all 19 had reduced fibulin-2 levels compared to 0 of 67 LANA-negative tumor specimens, which all had high levels of fibulin-2 expression, especially in non-spindle cell regions.
Figure 10.
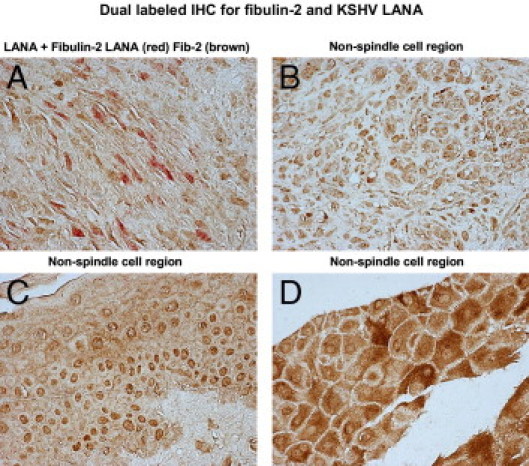
Fibulin-2 expression in human KS tumor tissue. Briefly, paraffin-embedded KS tissue was deparaffinized in xylene and hydrated in graded alcohol, and antigen unmasking was performed in citrate buffer in a microwave. Cells were stained by dual-labeled IHC for KSHV LANA (Vector Red, red) and fibulin-2 (DAB brown). No counterstain was used in this procedure. Bright-field images were photographed on a Nikon TE2000S microscope at total magnification of ×200. A: A spindle cell region; B–D: non-spindle cell regions observed in a representative KS tissue specimen.
Table 1.
Analysis of KS Tissue Microarrays
| Tumor cores spotted on TMA | Intact tumor cores | Total LANA-positive cores | LANA-negative tumor cores | Fibulin-2–positive cores | Tumor cores showing reduced expression of fibulin-2 in LANA+ spindle cell regions |
|---|---|---|---|---|---|
| 126 | 86 | 19 | 67 | 126 | 19/19 |
| 100% | 68% | 22% | 78% | All examined | 100% |
Intact tumor cores are tumor cores examined that were not destroyed by processing during mounting fixation or staining.
Discussion
In this study, we showed the characteristic morphological changes observed in DMVEC cells after KSHV infection in vitro with the similar changes seen in KS tumor tissue (Figure 1). This is the first report indicating down-regulation of fibulin-2 protein and message in KSHV-infected DMVEC cells, which are primary cells that mimic the histopathology of Kaposi's sarcoma in vivo, and in human KS tumor tissue compared to nontumor tissue in the same mix of samples. We also report here the temporal transcriptional down-regulation of fibulin-3 and fibulin-5 in KSHV-infected DMVEC cells. In contrast, we observed the temporal transcriptional up-regulation of fibulin-1C and -1D. We propose that the dysregulation of fibulin family members such as fibulin-2, -3, -5, 1C, and 1D likely contribute to KSHV-induced pathogenesis in KS. Dysregulation of multiple members of the fibulin family of extracellular matrix proteins in KS is a novel observation with implications for viral-induced changes in endothelial cell proliferation, migration, angiogenesis, and invasion that could directly contribute to KS tumorigenesis. We propose that down-regulation or loss of fibulin-2 compromises the structural integrity of the basement membrane due to loss of interactions between fibulin-2 and several ECM proteins.7
In addition, this is the first report of transcriptional down-regulation of fibulin-3 and fibulin-5 and transcriptional up-regulation of fibulin-1C and -1D in KSHV-infected DMVEC. Other fibulin isoforms (fibulin 1A, 1B) were not expressed at detectable levels in our transcription assay. This finding is consistent with results from other laboratories that report fibulin-1C and -1D are the predominantly expressed fibulin-1 spliced variants in various cell types and, further, that other fibulin-1 isoforms are not expressed or poorly expressed in these same cells.30 Different roles have been suggested for fibulin-1C and -1D based on their binding to the basement membrane protein nidogen.31 Fibulin-1C binds to nidogen with a 30-fold higher affinity than does fibulin-1D. More recent studies have shown increased fibulin-1C:1D mRNA ratios in ovarian carcinomas.14 Fibulin1C mRNA expression can be induced by estrogen in estrogen receptor α–positive ovarian tumor cell lines,14,32 and overexpression of fibulin-1C mRNA in ovarian cancer has been associated with tumor progression.14
Fibulin-5 is thought to have an important role in endothelial cell function, vessel repair, and angiogenesis. We observed early down-regulation of fibulin-5 message in KSHV-infected DMVEC, which returned to mock control cell values by day 10. This finding is also intriguing because fibulin-5 mediates endothelial cell adhesion through its integrin-binding RGD domain that interacts with the angiogenic-associated integrin alphavbeta3.33 KSHV is also known to stimulate expression of the proangiogenic cytokine VEGF, a potent inhibitor of fibulin-5 expression.20 This would suggest that fibulin-5 down-regulation in infected DMVEC could be mediated by an increase in expression of VEGF. c-Myc has also been found to repress fibulin-5 expression. Lui et al, 2007, reported that KSHV LANA, which is an abundant latent protein expressed in KSHV-infected PEL cell lines, stabilizes and activates c-Myc.34 This suggests that activation of c-Myc may also enhance suppression of fibulin-5 in KSHV-infected cells.
TGF-β is known to stimulate fibulin-5 expression in endothelial cells,20 and recently, Di Bartolo et al, 2008, showed that KSHV LANA inhibits TGF-β signaling through epigenetic silencing of the TGF-β type II receptor.35 They demonstrated that this pathway is blocked in PEL cells via down-regulation of the TGF-β type II receptor.35 They also concluded that KSHV LANA associates with the TGF-β promoter leading to methylation and deacetylation of proximal histones. Xie et al, 2008, reported that basement membrane–derived fibulins 1 and 5 function as angiogenic inhibitors that suppress tumor growth.36
Temporal expression of fibulin-2, -3, and -5 was largely down-regulated 5, 7, and 10 days after KSHV infection that correlates with time periods in which there was a consistent increase in KSHV LANA transcriptional expression. VEGF protein expression and transcription induced by KSHV was inhibited by addition of recombinant fibulin-3 or fibulin-5 for 48 hours. The fact that IL-6 and TNFα were found to be up-regulated in this assay by fibulin-5 suggests that fibulin-5 may have a role in proliferative responses during KSHV infection.
VEGFR3 activation and expression induced by KSHV glycoprotein B (gB) has been shown to enhance KSHV infectivity and play an important role in KSHV-mediated transformation.28 Down-regulation of VEGFR3 after exposure to fibulin-3 recombinant protein was found to be marginal. This level of suppression was obtained with only 10 μg/mL of purified protein and may require higher levels of fibulin-3 protein that could only be obtained by vector delivery of fibulin-3 to cells or tissue.
We observed no detectable expression of fibulin-2 or fibulin-5 in any of the PEL lines tested regardless of the degree of KSHV lytic replication observed. The absence of demonstrable fibulin-2 or fibulin-5 expression in PELs that express varying levels KSHV replication may suggest low to no demonstrable expression of fibulin-2 or fibulin-5 in PEL cell lines in general, and/or that the presence of KHSV replication reduces endogenous fibulin-2 and fibulin-5 expression to undetectable amounts, independent of KSHV replication levels.
Tissue microarrays revealed that LANA-positive KS tumors showed a reduction in fibulin-2 expression when compared to LANA-negative tissue. Among the 19 LANA-positive tumors observed, all 19 showed reduced expression of fibulin-2 in LANA-positive spindle cell regions. This is a significant finding, and we realize that examination of more tumor specimens is required. We intend to examine tissue fibulin-2 levels directly by real-time PCR in the future after acquiring additional KS tumors along with negative matched control specimens.
The dysregulation of the fibulin-2 extracellular matrix protein binding partners fibronectin and tropoelastin suggest that KSHV and/or down-regulation of fibulin-2 can affect the transcription of these essential ECM binding proteins. Tropoelastin has also been shown to interact with fibulin-5, and this interaction is likely involved in the initial steps of elastic fiber assembly.37
Fibronectin is essential for many cellular functions including migration, proliferation, and differentiation.38 Down-regulation of fibronectin transcription has also been shown to be associated with a highly metastatic form of murine mammary adenocarcinoma cells.39 These authors demonstrated that loss of fibronectin expression in these cells is not due to changes in the fibronectin promoter sequence but is the result of negative transcriptional regulation.39 More recent studies using cDNA arrays to identify genes associated with the more metastatic variant of MM3 adenocarcinoma cells as compared to the parent tumor (M3) found that fibronectin was one of only two genes significantly down-regulated among the genes analyzed in the study.40 They suggested that fibronectin may behave as an important metastasis suppressor protein in mammary cancer.40 In addition, viral proteins have been known to repress fibronectin transcription.41 As an example, the oncogenic adenovirus E1a protein has been shown to suppress fibronectin transcription in 3Y1 rat fibroblasts.41 Finally, a study by Torre et al revealed significantly reduced serum levels of fibronectin in HIV-infected patients with KS.42
Tropoelastin is a soluble precursor of elastin that can be induced by UV irradiation and degraded by metalloproteinases such as macrophage elastase (MMP1243). Loss of tropoelastin can lead to developmental tissue disorders such as aneurysms, atherosclerosis, and loss of skin elasticity. Tropoelastin along with fibronectin are essential ECM proteins critical for wound healing.
Finally, we report here the transcription of fibulin-3, -2, and -5 are down-regulated in DMVEC cells infected with KSHV. The dysregulation of these additional fibulin family members other than fibulin-2 (fibulin-1C, -1D, -3, and -5) after KSHV infection of DMVEC suggests that virus targets fibulins to compromise the integrity of the extracellular matrix thereby altering cell proliferation, migration, invasion, and angiogenesis. This suggests that fibulins can interact directly with viral gene products involved in transformation and may serve as potential targets for a novel treatment strategy for KS disease. The results of this study suggest that the dysregulation of fibulin-2 and its ECM binding partners likely contributes to KS pathogenesis. Understanding the underlying mechanisms involved in fibulin-mediated contributions to KS pathogenesis will require further studies.
Acknowledgments
We thank Drs James E.K. Hildreth and Diana Marver for their advice in the preparation of this manuscript. We also thank Jared Elzey for editing this manuscript.
Footnotes
Supported in part by Vanderbilt-Meharry Center for AIDS Research (CFAR) [National Institutes of Health (NIH) grant P30AI054999]; the Center for AIDS Health Disparities Research (NIH grant U54RR019192); Meharry Center for Clinical Research (CRC); and NIH grant P20RR011792. D.J.A. was funded by pilot grants from the CFAR and the Meharry CRC.
References
- 1.Kedes D.H., Operskalski E., Busch M., Kohn R., Flood J., Ganem D. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat Med. 1996;8:918–924. doi: 10.1038/nm0896-918. [DOI] [PubMed] [Google Scholar]
- 2.Schultz T.F. Kaposis's sarcoma-associated herpesvirus (human herpesvirus 8): epidemiology and pathogenesis. J Antimicrobial Chemother. 2000;45:15–27. doi: 10.1093/jac/45.suppl_4.15. [DOI] [PubMed] [Google Scholar]
- 3.Sitas F., Newton R. Kaposi's sarcoma in South Africa. J Natl Cancer Inst Monogr. 2001;28:1–4. doi: 10.1093/oxfordjournals.jncimonographs.a024250. [DOI] [PubMed] [Google Scholar]
- 4.Chang Y., Cesarman E., Pessin M.S., Lee F., Culpepper J., Knowles D.M., Moore P.S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 5.Hayward G.S., Alcendor D.J., Boger R.A. The role of KSHV in the pathogenesis of Kaposi's sarcoma. In: Khalili K., Jeang K.T., editors. Viral Oncology: Basic Science and Clinical Applications. Wiley-Blackwell; Hoboken, NJ: 2010. pp. 377–407. [Google Scholar]
- 6.Simonart T., Hermans P., Schandene L., Van Vooren J.P. Phenotypic characteristics of Kaposi's sarcoma tumour cells derived from patch-, plaque- and nodular-stage lesions: analysis of cell cultures isolated from AIDS and non-AIDS patients and review of the literature. Br J Dermatol. 2000;143:557–563. doi: 10.1111/j.1365-2133.2000.03709.x. [DOI] [PubMed] [Google Scholar]
- 7.Timpl R., Sasaki T., Kostka G., Chu M.L. Fibulins: a versatile family of extracellular matrix proteins. Nat Rev Mol Cell Biol. 2003;4:479–489. doi: 10.1038/nrm1130. [DOI] [PubMed] [Google Scholar]
- 8.Argraves W.S., Greene L.M., Cooley M.A., Gallagher W.M. Fibulins: physiological and disease perspectives. EMBO Rep. 2003;12:1127–1131. doi: 10.1038/sj.embor.7400033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan T.C., Kostka G., Zhang R.Z., Timpl R., Chu M.L. Complete exon-intron organization of the mouse fibulin-1 gene and its comparison with the human fibulin-1 gene. FEBS Lett. 1999;444:38–42. doi: 10.1016/s0014-5793(99)00024-1. [DOI] [PubMed] [Google Scholar]
- 10.Pan T.C., Sasaki T., Zhang R.Z., Fässler R., Timpl R., Chu M.L. Structure and expression of fibulin-2, a novel extracellular matrix protein with multiple EGF-like repeats and consensus motifs for calcium binding. J Cell Biol. 1993;123:1269–1277. doi: 10.1083/jcb.123.5.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi N., Kostka G., Garbe J.H., Keene D.R., Bächinger H.P., Hanisch F.G., Markova D., Tsuda T., Timpl R., Chu M.L., Sasaki T. A comparative analysis of the fibulin protein family, biochemical characterization, binding interactions and tissue localization. J Biol Chem. 2007;282:11805–11816. doi: 10.1074/jbc.M611029200. [DOI] [PubMed] [Google Scholar]
- 12.Fisher S.A., Rivera A., Fritsche L.G., Keilhauer C.N., Lichtner P., Meitinger T., Rudolph G., Weber B.H. Case-control genetic association study of fibulin-6 (FBLN6 or HMCN1) variants in age-related macular degeneration (AMD) Hum Mutat. 2007;4:406–413. doi: 10.1002/humu.20464. [DOI] [PubMed] [Google Scholar]
- 13.de Vega S., Iwamoto T., Nakamura T., Hozumi K., McKnight D.A., Fisher L.W., Fukumoto S., Yamada Y. TM14 is a new member of the fibulin family (fibulin-7) that interacts with extracellular matrix molecules and is active for cell binding. J Biol Chem. 2007;282:30878–30888. doi: 10.1074/jbc.M705847200. [DOI] [PubMed] [Google Scholar]
- 14.Moll F., Katsaros D., Lazennee G., Hellio N., Giacalone P.L., Chalbos D., Maudelonde T., Rochefort H., Pujol P. Estrogen induction and overexpression of fibulin-1C mRNA in ovarian cancer cells. Oncogene. 2002;7:1097–1107. doi: 10.1038/sj.onc.1205171. [DOI] [PubMed] [Google Scholar]
- 15.Greene L.M., Twal W.O., Duffy M.J., McDermott E.W., Hill A.D., O'Higgins N.J., McCann A.H., Dervan P.A., Argraves W.S., Gallagher W.M. Elevated expression and altered processing of fibulin-1 protein in human breast cancer. Br J Cancer. 2003;6:871–878. doi: 10.1038/sj.bjc.6600802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yi C.H., Smith D.J., West W.W., Hollingsworth M.A. Loss of fibulin-2 expression is associated with breast cancer progression. Am J Pathol. 2007;5:1535–1545. doi: 10.2353/ajpath.2007.060478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albig A.R., Niel J.R., Schiemann W.P. Fibulins 3 and 5 antagonize tumor angiogenesis in vivo. Cancer Res. 2006;5:2621–2629. doi: 10.1158/0008-5472.CAN-04-4096. [DOI] [PubMed] [Google Scholar]
- 18.Gallagher W.M., Greene L.M., Ryan M.P., Sierra V., Berger A., Laurent-Puig P., Conseiller E. Human fibulin-4: analysis of its biosynthetic processing and mRNA expression in normal and tumor tissue. FEBS Lett. 2001;1:59–66. doi: 10.1016/s0014-5793(00)02389-9. [DOI] [PubMed] [Google Scholar]
- 19.Schiemann W.P., Blobe G.C., Kalume D.E., Pandey A., Lodish H.F. Context-specific effects of fibulin-5 (DANCE/EVEC) on cell proliferation, motility, and invasion: Fibulin-5 is induced by transforming growth factor-beta and affects protein kinase cascades. J Biol Chem. 2002;30:27367–27377. doi: 10.1074/jbc.M200148200. [DOI] [PubMed] [Google Scholar]
- 20.Albig A.R., Schiemann W.P. Fibulin-5 antagonizes vascular endothelial growth factor (VEGF) signaling and angiogenic sprouting by endothelial cells. DNA Cell Biol. 2004;23:367–379. doi: 10.1089/104454904323145254. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi N., Kostka G., Garbe J.H., Keene D.R., Bachinger H.P., Hanisch F.G., Markova D., Tsuda T., Timpl R., Chu M.L., Sasaki T. A comparative analysis of the fibulin protein family: Biochemical characterization, binding interactions, and tissue localization. J Biol Chem. 2007;282:11805–11816. doi: 10.1074/jbc.M611029200. [DOI] [PubMed] [Google Scholar]
- 22.Reinhardt D.P., Sasaki T., Dzamba B.J., Keene D.R., Chu M.L., Gohring W., Timpl R., Sakai L.Y. Fibrillin1 and fibulin-2 interact and are colocalized in some tissues. J Biol Chem. 1996;271:19489–19496. doi: 10.1074/jbc.271.32.19489. [DOI] [PubMed] [Google Scholar]
- 23.Olin A.I., Morgelin M., Sasaki T., Timpl R., Heinegard D., Aspberg A. The proteoglycans aggrecan and versican form networks with fibulin-2 through their lectin domain binding. J Biol Chem. 2001;276:1253–1261. doi: 10.1074/jbc.M006783200. [DOI] [PubMed] [Google Scholar]
- 24.Utani A., Nomizu M., Yamada Y. Fibulin-2 binds to the short arms of laminin-5 and laminin-1 via conserved amino acid sequences. J Biol Chem. 1997;272:2814–2820. doi: 10.1074/jbc.272.5.2814. [DOI] [PubMed] [Google Scholar]
- 25.Sasaki T., Gohring W., Pan T.C., Chu M.L., Timpl R. Binding of mouse and human fibulin-2 to extracellular matrix ligands. J Mol Biol. 1995;254:892–899. doi: 10.1006/jmbi.1995.0664. [DOI] [PubMed] [Google Scholar]
- 26.Hopf M., Gohring W., Kohfeldt E., Yamada Y., Timpl R. Recombinant domain IV of perlecan binds to nidogens, laminin-nidogen complex, fibronectin, fibulin-2 and heparin. Eur J Biochem. 1999;259:917–925. doi: 10.1046/j.1432-1327.1999.00127.x. [DOI] [PubMed] [Google Scholar]
- 27.Gallagher W.M., Currid C.A., Whelan L.C. Fibulins and cancer: friend or foe? Trends Mol Med. 2005;11:336–340. doi: 10.1016/j.molmed.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X., Wang J.F., Chandran B., Persaud K., Pytowski B., Fingeroth J., Groupman J.E. Kaposi's sarcoma-asoociated herpesvirus activation of vascular endothelial growth factor receptor 3 alters endothelial function and enhances infection. J Biol Chem. 2005;28:26216–26224. doi: 10.1074/jbc.M411392200. [DOI] [PubMed] [Google Scholar]
- 29.Cannon J.S., Ciufo D., Hawkins A.L., Griffin C.A., Borowitz M.J., Hayward G.S., Ambinder R.F. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi's sarcoma herepesvirus-containing supernatant. J Virol. 2000;21:10187–10193. doi: 10.1128/jvi.74.21.10187-10193.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tran H., Mattei M., Godyna S., Argraves W.S. Human fibulin-1D: molecular cloning, expression and similarity with S1-5 protein, a new member of the fibulin gene family. Matrix Biol. 1997;15:479–493. doi: 10.1016/s0945-053x(97)90021-4. [DOI] [PubMed] [Google Scholar]
- 31.Sasaki T., Gohring W., Pan T.C., Chu M.L., Timpl R. Binding of mouse and human fibulin-2 to extracellular matrix ligands. J Mol Biol. 1995;5:892–899. doi: 10.1006/jmbi.1995.0664. [DOI] [PubMed] [Google Scholar]
- 32.Bardin A., Moll F., Margueron R., Delfour C., Chu M.L., Maudelonde T., Cavailles V., Pujol P. Transcriptional and post-transcriptional regulation of fibulin-1 by estrogen leads to differential induction of messenger ribonucleic acid variants in ovarian and breast cancer. Endocrinology. 2005;2:760–768. doi: 10.1210/en.2004-1239. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura T., Ruiz-Lozano P., Lindner V., Yabe D., Taniwaki M., Furukawa Y., Kobuke K., Tashiro K., Lu Z., Andon N.L., Schaub R., Matsumori A., Sasayama S., Chien K.R., Honjo T. DANCE, a novel secreted RGD protein expressed in developing, atherosclerotic, and balloon-injured arteries. J Biol Chem. 1999;274:22476–22483. doi: 10.1074/jbc.274.32.22476. [DOI] [PubMed] [Google Scholar]
- 34.Liu J., Martin H.J., Liao G., Hayward S.D. The Kaposi's sarcoma-associated herpesvirus LANA protein stabilizes and activates c-Myc. J Virol. 2007;19:10451–10459. doi: 10.1128/JVI.00804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Bartolo D.L., Cannon M., Liu Y.F., Renne R., Chadburn A., Boshoff C., Cesarman E. KSHV LANA inhibits TGF-beta signaling through epigenetic silencing of the TGF-beta type II receptor. Blood. 2008;9:4731–4740. doi: 10.1182/blood-2007-09-110544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie L., Palmsten K., MacDonald B., Kieran M.W., Potenta S., Vong S., Kalluri R. Basement membrane derived fibulin-1 and fibulin-5 function as angiogenesis inhibitors and suppress tumor growth. Exp Biol Med (Maywood) 2008;2:155–162. doi: 10.3181/0706-RM-167. [DOI] [PubMed] [Google Scholar]
- 37.Nonaka R., Onoue S., Wachi H., Sata F., Urban Z., Starcher B.C., Seyama Y. DANCE/fibulin-5 promotes elastic fiber formation in tropoelastin isoform-dependent manner. Clin Biochem. 2009;7–8:713–721. doi: 10.1016/j.clinbiochem.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 38.Brenner K.A., Corbett S.A., Schwarzbauer J.E. Regulation of fibronectin matrix assembly by activated Ras in transformed cells. Oncogene. 2000;19:3156–3163. doi: 10.1038/sj.onc.1203626. [DOI] [PubMed] [Google Scholar]
- 39.Webajh S.E., Urtreger A.J., Puricelli L.I., de Lustig E.S., Joffe E.B.K., Kornblitt A.R. Downregulation of fibronectin transcription in highly metastatic adenocarcinoma cells. FEBS Letters. 1998;440:277–281. doi: 10.1016/s0014-5793(98)01473-2. [DOI] [PubMed] [Google Scholar]
- 40.Urtreger A.J., Werbajh S.E., Verrecchia F., Mauviel A., Puricelli L.I., Kornblihtt A.R., Bal de Kier Joffe E.D. Fibronectin is directly downregulated in murine mammary adenocarcinoma cells with high metatstaic potential. Oncol Rep. 2006;6:1403–1410. [PubMed] [Google Scholar]
- 41.Nakamura T., Nakamura T., Tsunoda S., Nakada S., Oda K., Tsuri H., Wada A. Induction of E1A-responsive negative factors for transcription of the fibronectin gene in adenovirus E1-transformed rat cells. J Virol. 1992;11:6436–6450. doi: 10.1128/jvi.66.11.6436-6450.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torre D., Zeroli C., Martegani R., Pugliese A., Basilico C., Speranza F. Levels of the bcl-2 protein, fibronectin and alpha (5) beta (1) fibronectin receptor in HIV-1-infected patients with Kaposi's sarcoma. Microbes Infect. 2000;15:1831–1843. doi: 10.1016/s1286-4579(00)01342-3. [DOI] [PubMed] [Google Scholar]
- 43.Seo J.Y., Lee S.H., Youn C.S., Choi H.R., Rhie G., Cho K.H., Kim K.H., Park K.C., Eun H.C., Chung J.H. Ultraviolet radiation increases tropoelastin mRNA expression in the epidermis of human skin in vivo. J Invest Dermatol. 2001;6:915–919. doi: 10.1046/j.1523-1747.2001.01358.x. [DOI] [PubMed] [Google Scholar]


