Abstract
KISS1 is a metastasis suppressor gene that is lost in several malignancies, including bladder cancer. We tested the epigenetic silencing hypothesis and evaluated the biological influence of KISS1 methylation on its expression and clinical relevance in bladder cancer. KISS1 hypermethylation was frequent in bladder cancer cells analyzed by methylation-specific PCR and bisulfite sequencing and was associated with low gene expression, being restored in vitro by demethylating azacytidine. Hypermethylation was also frequently observed in a large series of bladder tumors (83.1%, n = 804). KISS1 methylation was associated with increasing stage (P = 0.001) and tumor grade (P = 0.010). KISS1 methylation was associated with low KISS1 transcript expression by quantitative RT-PCR (P = 0.037). KISS1 transcript expression was also associated with histopathological tumor stage (P < 0.0005). Low transcript expression alone (P = 0.003) or combined with methylation (P = 0.019) was associated with poor disease-specific survival (n = 205). KISS1 transcript expression remained an independent prognosticator in multivariate analyses (P = 0.017). KISS1 hypermethylation was identified in bladder cancer, providing a potential mechanistic explanation (epigenetic silencing) for the observed loss of KISS1 in uroepithelial malignancies. Associations of KISS1 methylation and its expression with histopathological variables and poor survival suggest the utility of incorporating KISS1 measurement using paraffin-embedded material for tumor stratification and clinical outcome prognosis of patients with uroepithelial neoplasias.
Bladder cancer can be described as a molecular disease, driven by the multistep accumulation of genetic and epigenetic factors.1 The most common epigenetic event is the addition of methyl groups to the carbon-5 position of cytosine nucleotides.2 CpG islands are present in one-half of human genes.3–6 CpG island hypermethylation has been associated with the transcriptional inactivation of cancer-related genes, including in bladder cancer.6–8
KISS1 was identified as suppressing metastases in melanoma and breast cancer cells.9–11 The KISS1 gene maps to chromosome region 1q32,12 and it can be regulated by genes located on chromosome 6.10,11,13 The highest concentrations of the KISS1 protein are found in the placenta, although it is also expressed in the central nervous system, testis, ovary, pancreas, and intestine.9–11 KISS1 encodes a 145-amino acid protein that is processed into kisspeptins of several sizes.14–16 Kisspeptins are physiologically functional at controlling the onset of puberty and at inhibiting cancer metastasis of different tumor types.9–11,13,17–19
Associations between KISS1 expression loss and increased tumor progression and poor prognosis were found in several solid tumors.12,14–16,20–26 In bladder cancer, KISS1 expression was decreased in primary tumors, compared with normal counterparts.12,22 Loss of KISS1 expression was associated with tumor stage, tumor grade, and survival.12,22 In an attempt to uncover the mechanisms by which KISS1 is lost in bladder cancer, we tested the hypothesis of epigenetic silencing after identifying an enriched 5′-CpG island around the promoter region of KISS1, supporting its susceptibility to be epigenetically modified by hypermethylation. Although KISS1 was identified among the genes restoring their transcript expression after azacytidine treatment using oligonucleotide microarrays in gastric cancer cells,26 to our knowledge KISS1 has not been reported to be epigenetically altered in bladder cancer. In this report, the effect of KISS1 methylation on its expression and the clinical relevance were evaluated in bladder cancer. Our results revealed that KISS1 was hypermethylated in bladder cancer and that the methylation of the gene and its transcript expression are potentially useful as tumor stratification biomarkers and clinical outcome prognosticators for bladder cancer patients.
Materials and Methods
Tumor Samples
Primary bladder tumors were collected in accord with the ethical guidelines at the participating hospitals. An initial series included cystectomized invasive bladder tissues, for which both frozen and paraffin-embedded material and paired normal urothelium were available (n = 25). Optimal cutting temperature compound and paraffin-embedded material was macrodissected based on H&E evaluations, to ensure a minimum of 75% of tumor cells.27 These samples served to i) screen KISS1 methylation rates, ii) test the association of KISS1 methylation along bladder cancer progression, and iii) assess the feasibility of KISS1 methylation analyses in matching paraffin-embedded material. Normal urothelium samples obtained from 10 cystoprostatectomized patients with prostate cancer were also evaluated, to test the bladder cancer specificity of KISS1 methylation.
An independent set of 804 paraffin-embedded bladder tumors obtained by transurethral resection for noninvasive lesions and cystectomy for invasive tumors served to i) validate KISS1 methylation rates, ii) test the association of KISS1 transcript expression with its methylation status, and iii) evaluate the association of KISS1 methylation and transcript expression with clinicopathologic variables. This set comprised 481 non-muscle invasive lesions (69 pTa, 402 pT1, and 10 pTis) and 323 muscle-invasive tumors (187 pT2, 93 pT3, and 43 pT4). For the entire set of 804 tumors (ie, both non-muscle invasive and muscle-invasive tumors), the tumor grade distribution was 132 low grade and 672 high grade, according to standard criteria.28 Out of the set of tumors recruited for assessing KISS1 methylation, follow-up data were available for 205 patients: 183 men and 22 women, with a median age of 65 years (range, 25 to 81 years). The tumor stage distribution was 91 pT1, 65 pT2, 33 pT3, and 16 pT4. Tumor grade distribution was 10 low grade and 195 high grade. The histopathological information and annotated follow-up data allowed evaluation of associations of KISS1 methylation with transcript expression and clinical outcome.
Methylation Analyses of KISS1
A specific search for enrichment of CpG in KISS1 (Figure 1A) was performed using the CpG Island Searcher online tool (http://www.cpgislands.com, last accessed October 12, 2010). KISS1 methylation status was analyzed by two PCR analysis strategies using low amounts of bisulfite-modified genomic DNA (500 to 1000 ng), which induces chemical conversion of unmethylated, but not methylated, cytosine to uracil. First, genomic sequencing of both DNA strands of KISS1 was analyzed after bisulfite treatment (BS-SEQ), for at least three clones per bladder cancer cell line, as previously reported.8,29 Confirmation on at least two independent clones was required to assign methylation sites (Figure 1B). A second strategy used PCR with primers (Figure 1A and Table 1) specific for either the methylated or the modified unmethylated DNA (MS-PCR).8,29 PCRs were performed using a final volume of 15 μL for MS-PCR and a final volume of 25 μL for BS-SEQ containing 1× PCR EcoStart buffer (Ecogen, Barcelona and Madrid, Spain), 1.5 mmol/L of MgCl2, 0.2 mmol/L of dNTP, 0.25 μmol/L of each primer and 1.5 U of EcoStart Taq polymerase (Ecogen). Genomic DNA was extracted using standard methods. DNA from normal lymphocytes treated in vitro (IVD) with SssI methyltransferase was used as a positive control for methylated alleles. DNA from normal lymphocytes and B-cell chronic lymphocytic leukemia cells (MEC1) was used as a positive control for unmethylated alleles. PCR products were loaded onto nondenaturing 2% agarose gels, stained with ethidium bromide, and visualized under an UV transilluminator.8,29
Figure 1.
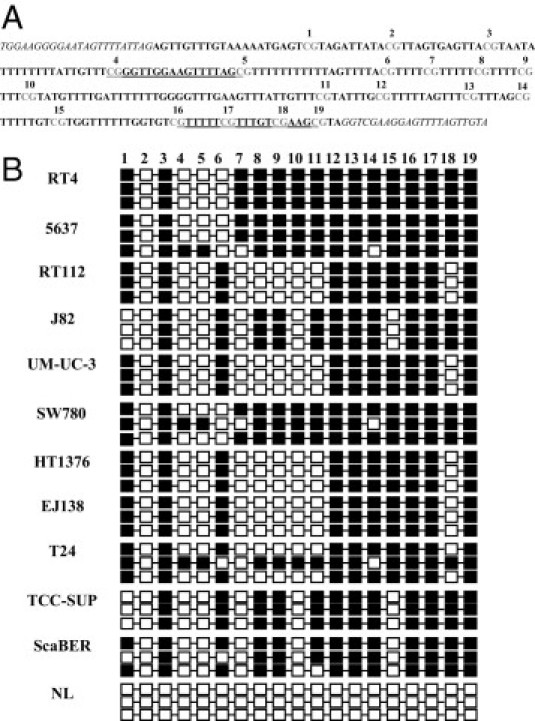
KISS1 was frequently methylated in bladder cancer cells. A: Schematic depiction of the KISS1 CpG islands analyzed as nucleotide sequences of the CpG island region analyzed by BS-SEQ. The sequencing primers are indicated by italics, the primers used for MS-PCR are indicated by underlining, and the CpG islands are numbered and indicated in gray. B: Analysis of CpG island methylation status of KISS1 by bisulfite genomic sequencing in bladder human cancer cell lines. Three individual clones are shown per methylated cell line and per unmethylated normal lymphocytes (NL) control. CpG dinucleotides are represented as dark squares for methylated cytosines and open squares for unmethylated cytosines. For dark squares, the presence of methylation was confirmed in at least two of the clones that were sequenced for the cell lines analyzed.
Table 1.
Primer Sequences and PCR Conditions for KISS1 Analyses
| Analysis | Sense primer | Antisense primer | Product size (bp) | Tanneal (°C) | PCR cycles (no.) |
|---|---|---|---|---|---|
| BS-SEQ | 5′-TGGAAGGGGAATAGTTTTATTAG-3′ | 5′-TACAACTAAAACTCCTTCRACC-3′ | 322 | 52 | 40 |
| MSP | 5′-CGGGTTGGAAGTTTTAGC-3′ | 5′-GCTTCGACAAACGAAAAAC-3′ | 125 | 54 | 37 |
| USP | 5′-TTTTGGGTTGGAAGTTTTAGT-3′ | 5′-ACTTCAACAAACAAAAAACAAC-3′ | 129 | 52 | 37 |
| RT-PCR | 5′-ACTCACTGGTTTCTTGGCAGC-3′ | 5′-ACCTTTTCTAATGGCTCCCCA-3′ | 183 | 60 | 28 |
BS-SEQ, bisulfite sequencing; MSP, methylation-specific PCR; Tanneal, annealing temperature; USP, unmethylated specific PCR.
RNA Analysis of KISS1 in Bladder Cancer Cell Lines
Human bladder cancer cell lines (n = 11) were treated with 1 μmol/L and 5 μmol/L azacytidine (AZA) (Sigma-Aldrich, St. Louis, MO) for 72 hours to achieve demethylation.8,29 RNA was isolated using standard methods. RNA (1 μg) was reverse-transcribed using AMV reverse transcriptase (Promega, Madison, WI) and amplified using specific primers and conditions for KISS1 (Table 1). PCR was performed using a final volume of 15 μL containing 1× PCR EcoStart buffer (Ecogen), 1.5 mmol/L MgCl2, 0.2 mmol/L dNTP, 0.25 μmol/L of each primer, and 1.5 U EcoStart Taq polymerase (Ecogen). For PCR amplification, 0.4 μg cDNA was used. GADPH was used as internal control to ensure cDNA quality and loading accuracy. The amplification product was resolved by 2% agarose gel electrophoresis and visualized by ethidium bromide staining.8,29
Transcript Expression Analysis by Real-Time Quantitative PCR in Bladder Tumors
KISS1 transcript expression was assessed in paraffin-embedded tumors among patients evaluated for KISS1 methylation and with available follow-up data (as described above). RNA was extracted using the TRIzol (Invitrogen, Carlsbad, CA) method. Complementary DNA was synthesized using a ThermoScript RT-PCR system (Invitrogen). Template cDNA was added to TaqMan universal master mix (Applied Biosystems, Foster City, CA) in a 15-μL reaction with specific primers and probes acquired from Applied Biosystems for KISS1 (Hs_00158486_m1), and the chaperonin containing T-complex protein 1 subunit 6A (zeta 1) (CCT6A) (Hs_00798979_s1). Quantification of gene expression was performed using the ABI Prism 7900HT sequence detection system (Applied Biosystems). Relative gene expression quantification was calculated according to the comparative cycle threshold (CT) method using CCT6A as endogenous control. Final results were estimated as 2−(ΔCt KISS1), where the ΔCT KISS1 values for each sample are determined by subtracting the CT value of the target KISS1 gene from the value of the CCT6A control gene. Only triplicates with standard deviations of the CT value of <0.20 were accepted.
Statistical Analysis
Associations among KISS1 methylation and its transcript expression with tumor stage and grade were evaluated using nonparametric Wilcoxon-Mann-Whitney and Kruskal-Wallis tests.30 KISS1 transcript cutoff was selected based on the median values of expression among the groups under analysis. Combinations of the presence or absence of methylation and KISS1 transcript levels higher or lower than the selected cutoff were submitted to clinical outcome assessment. Associations of methylation and KISS1 transcript expression with disease-specific survival were evaluated in those cases with available follow-up data, using the log-rank test.30 Disease-specific survival time was defined as the years elapsed between surgery and death from disease (or the date of the last follow-up visit). Patients who were alive at the last follow-up date or lost were censored. Survival curves were plotted using Kaplan-Meier methodology. To test whether KISS1 methylation and transcript expression were independent risk factors, a multivariate analysis was performed using a Cox model including also variables such as age, sex, tumor stage, and tumor grade. All analyses were performed with the SPSS software package version 17.0 (SPSS, Chicago, IL).
Results
KISS1 Is Epigenetically Silenced in Vitro
Eleven human bladder cancer cell lines were initially screened using bisulfite genomic sequencing and MS-PCR (Figure 1A). The BS-SEQ revealed CpG island methylation for all of the bladder cancer cell lines analyzed (Figure 1B), in agreement with the MS-PCR results (Figure 2A). Methylation analyses were then linked with KISS1 expression estimates by RT-PCR. The hypermethylated cell lines showed low transcript expression (Figure 2A). A further link between hypermethylation and gene silencing was established by treatment of bladder cancer cell lines with a demethylating agent. Exposure of the methylated cells to AZA increased the transcript expression of KISS1 both by qualitative (Figure 2B) and by quantitative RT-PCR analyses (Figure 2C). The degree of demethylation after azacytidine exposure of bladder cancer cells was assessed by BS-SEQ (see Supplemental Figure S1 at http://ajp.amjpathol.org). Overall, these results indicated a high correlation of KISS1 methylation with loss of transcript expression in vitro, observations especially supported by AZA reactivation analyses.
Figure 2.
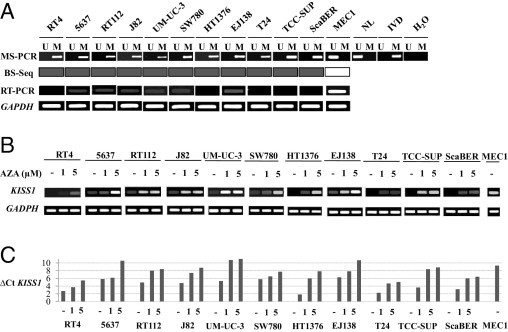
CpG island methylation was associated with KISS1 silencing. A: Methylation-specific PCRs (MS-PCR) for KISS1 in human bladder cancer-cell lines. A PCR band in lane M indicates a methylated KISS1 gene; in lane U, it indicates an unmethylated gene. Normal lymphocytes (NL) and MEC1 were used as controls for unmethylated KISS1; in vitro methylated DNA (IVD) was used as control for methylated KISS1.From bisulfite genomic sequencing (BS-Seq), methylated cell lines are highlighted in dark color. With RT-PCR, GAPDH expression was used as transcript loading control. B: Treatment with the demethylating agent reactivated the transcript expression of KISS1 in methylated bladder cancer cell lines. Qualitative RT-PCR analysis of KISS1 expression, with GAPDH as transcript loading control. After 1 or 5 μmol/L AZA exposure, KISS1 transcript expression increased in all of the methylated bladder cancer cell lines. C: Quantitative RT-PCR analysis of KISS1 expression. ΔCTKISS1 values for each sample were determined by subtracting the CT value of the target KISS1 gene from the value of the CCT6A control gene. KISS1 transcript expression increased in all of the methylated bladder cancer cell lines, after 1 or 5 μmol/L AZA exposure. MEC1, an unmethylated cell line, was used as a positive control for KISS1 expression.
KISS1 Is Frequently Hypermethylated in Primary Bladder Tumors and Is Associated with Tumor Progression
Once the effect of KISS1 CpG island hypermethylation on expression was determined in vitro, we evaluated its clinical impact in human bladder tumors. The methylation status of KISS1 was analyzed by MS-PCR in a set of frozen bladder tumors (n = 25), for which their respective paired normal urothelium was available (Figure 3A). Comparison of methylation of these pairs of 25 bladder tumors and normal urothelium revealed that KISS1 methylation was a frequent cancer-specific epigenetic event (Figure 3B). Importantly, KISS1 was found to be unmethylated in normal urothelium obtained from 10 cystoprostatectomized patients with prostate cancer (Figure 3C), supporting the bladder cancer specificity of KISS1 methylation. High concordance was found between MS-PCR results obtained from genomic DNA from frozen and matching paraffin-embedded tumor material (100%). These observations prompted us to perform further MS-PCR analyses on independent sets of bladder tumors, extracting genomic DNA from paraffin-embedded material. Of note, a high methylation rate for KISS1 was observed (83.1%) using an independent large set of bladder tumors (n = 804). For validation analyses, the methylation status of a subset of the tumors analyzed by MS-PCR was confirmed by BS-SEQ (see Supplemental Figure S2 at http://ajp.amjpathol.org).
Figure 3.
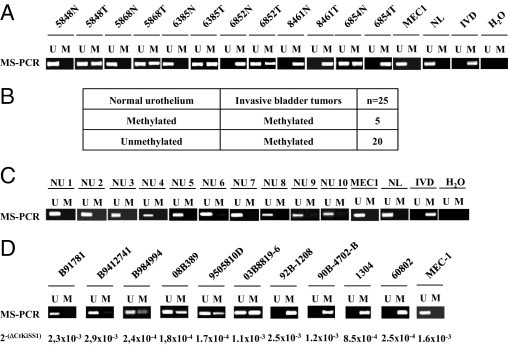
KISS1 hypermethylation is a frequent bladder cancer-specific event. A:KISS1 hypermethylation was shown to be a cancer-specific event, as indicated by representative MS-PCR analyses for KISS1 using primary bladder tumors and normal urothelium. Normal lymphocytes (NL) and MEC1 were used as controls for unmethylated KISS1; in vitro methylated DNA (IVD) was used as control for methylated KISS1. B:KISS1 methylation increased in bladder tumors, compared with their paired normal urothelium, as indicated by methylation status of KISS1 analyzed by MS-PCR in pairs of primary bladder tumors and normal urothelium. C:KISS1 was unmethylated in normal urothelium unrelated to bladder cancer. Representative MS-PCR analyses are shown for KISS1 in normal urothelium (NU) from cystoprostatectomized patients with prostate cancer. MEC1 cells were used as positive control for unmethylated KISS1. D:KISS1 methylation correlated with a low transcript expression. Representative MS-PCR analyses are shown for KISS1 and matching transcript levels of KISS1 in bladder tumors.
The next analyses dealt with evaluating the link between the hypermethylation status of KISS1 and clinicopathological variables. Tumors displaying advanced disease (stage T2 or higher) were more frequently methylated than those presenting non-muscle-invasive lesions (Table 2). Increased methylation rates were observed in high-grade bladder tumors, compared with low-grade lesions (Table 2). Of note, KISS1 methylation was significantly associated with increased stage (Kruskal-Wallis, P = 0.001) and grade (P = 0.010). Overall, these results indicate that KISS1 hypermethylation is a frequent event in bladder cancer in association with pathological indicators of tumor progression.
Table 2.
Summary of the Distribution of CpG Island Methylation Status of KISS1 Obtained by Methylation-Specific PCR with Respect to Tumor Stage and Grade in a Series of 804 Bladder Tumors
| Variable | No. | % |
|---|---|---|
| Stage⁎ | ||
| pTa | 69 | |
| Methylated | 56 | 81.2 |
| Unmethylated | 13 | 18.8 |
| pT1 | 402 | |
| Methylated | 318 | 79.1 |
| Unmethylated | 84 | 20.9 |
| pTis | 10 | |
| Methylated | 8 | 80.0 |
| Unmethylated | 2 | 20.0 |
| pT2 | 187 | |
| Methylated | 160 | 85.6 |
| Unmethylated | 27 | 14.4 |
| pT3 | 93 | |
| Methylated | 86 | 92.5 |
| Unmethylated | 7 | 7.5 |
| pT4 | 43 | |
| Methylated | 40 | 93.1 |
| Unmethylated | 3 | 6.9 |
| Grade† | ||
| Low | 132 | |
| Methylated | 103 | 78.1 |
| Unmethylated | 29 | 21.9 |
| High | 672 | |
| Methylated | 565 | 84.1 |
| Unmethylated | 107 | 15.9 |
KISS1 methylation was associated with increasing tumor stage (P = 0.001).
KISS1 methylation was associated with increasing tumor grade (P = 0.01).
KISS1 Transcript Expression Correlates with Methylation, Tumor Progression, and Clinical Outcome of Patients with Bladder Tumors
The next set of analyses dealt with the assessment of transcript expression of KISS1 by quantitative RT-PCR in bladder tumors for which the KISS1 methylation status was analyzed. Higher KISS1 expression was found in low-grade and early lesions, compared with high-grade and invasive bladder tumors. Low transcript levels of KISS1 were significantly associated with increasing stage (Kruskal-Wallis, P < 0.0005) and grade (Mann-Whitney, P = 0.024). Tumors methylated for KISS1 had lower transcript expression than did unmethylated tumors (P = 0.037; Figure 3D).
In a subset of 205 bladder tumors with available follow-up data, having a transcript expression of KISS1 with a 2−ΔCt value of <0.1 was significantly associated with shorter disease-specific survival (log rank, P = 0.003; Figure 4A). Although KISS1 methylation alone was not significantly associated with disease-specific survival (log rank, P = 0.23; Figure 4B), the combined presence of a KISS1 transcript expression with a 2−ΔCt value of <0.1 and KISS1 methylation was strongly associated with the poorest survival (log rank, P = 0.019; Figure 4C). Lack of statistically significant associations were found when performing log-rank statistics comparing only those patients with low KISS1 expression and methylated for KISS1 versus those with low KISS1 expression and unmethylated for KISS1, as well as only those with high KISS1 expression and methylated for KISS1 versus those with high KISS1 expression and unmethylated for KISS1 (Figure 4C). These analyses indicated the influence of a high expression of KISS1 on a favorable outcome. Multivariate Cox analyses indicated that tumor stage and KISS1 expression were the only independent prognostic factors (P = 0.001), having hazard ratios of death of 2.62 (95% CI = 1.49 to 4.58, P = 0.001) and 0.42 (95% CI = 0.20 to 0.85; P = 0.017), respectively.
Figure 4.
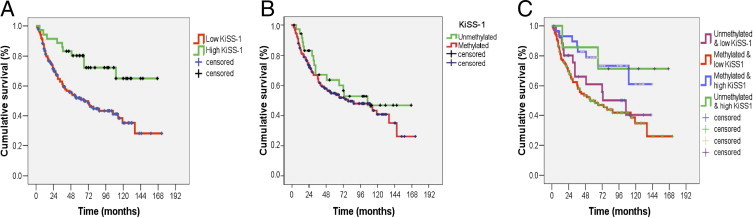
Transcript expression and KISS1 methylation are associated with clinical outcome in bladder cancer. A: Loss of KISS1 expression alone was associated with disease-specific survival. Kaplan-Meier survival curve analysis indicated that low transcript expression of KISS1 with a 2−(ΔCt KISS1) value of < 0.1 measured by RT-PCR was associated with poor survival (log rank, P = 0.003). B:KISS1 methylation alone was not significantly associated with disease-specific survival. Kaplan-Meier survival curve analysis indicated that tumors methylated for KISS1 had poorer survival than unmethylated tumors, but this trend did not reach statistical significance (log rank, P = 0.238). C: Loss of KISS1 transcript expression in combination with KISS1 methylation was associated with disease-specific survival. Kaplan-Meier survival curve analysis indicated that tumors with a transcript expression of KISS1 with a 2−(ΔCt KISS1) value of <0.1 and methylated for KISS1 had the poorest survival (log rank, P = 0.019).
Overall, these analyses revealed that the loss of transcript expression was associated with methylation, tumor stage, and poor survival. The combination of KISS1 methylation and low transcript expression was also related to poor disease-specific survival. Thus, these variables could be considered tumor stratification biomarkers and likely prognosticators of poor outcome for bladder cancer patients.
Discussion
The present study revealed the epigenetic silencing of KISS1 in bladder cancer and showed the clinical relevance of KISS1 methylation in uroepithelial tumors. The consequences of the novel CpG island hypermethylation of KISS1 in bladder cancer can be assessed from the standpoint of mechanistic and translational implications. Mechanistically, it is important to evaluate the cellular consequences of KISS1 methylation. Expression estimates of KISS1 were linked to methylation analyses using several approaches. Methylation status and transcript expression of KISS1 correlated to a high extent among a variety of bladder cancer cell lines. AZA exposure experiments confirmed the biological effect of methylation in KISS1 expression by specifically restoring KISS1 transcript expression in methylated cells. Overall, in vitro analyses revealed that KISS1 was aberrantly silenced by CpG island hypermethylation. Along this line of argument, a statistically significant association was found between methylation and low transcript expression of KISS1 by RT-PCR. In bladder tumors, the increased methylation rate together with the loss of transcript expression of KISS1 was also found to be associated with increasing tumor stage and poor clinical outcome. Thus, increased methylation rates correlated with loss of KISS1 expression and aggressive bladder cancer also in human clinical material. Biologically, KISS1 is reported to play a metastasis suppressor role in neoplastic diseases. Although its role in cancer progression has not been elucidated, the epigenetic silencing of KISS1 might aid understanding as to how KISS1 may contribute to bladder cancer. Further studies are warranted to dissect the influence of KISS1 hypermethylation on its expression in the context of other malignancies.
The translational implications of the discovery of KISS1 methylation have been strongly addressed in the present study. The association of KISS1 methylation with bladder cancer can be justified as follows. First, the cancer specificity was supported by MS-PCR analyses in pairs of bladder tumors and normal urothelium. It is important to be aware of the potential influence of the selected high number of cycles on detecting hypermethylation in normal urothelium counterparts in a tumor characterized by displaying field effect. Second, it was shown, using a large series of bladder cancer patients (n = 804), that the methylation of KISS1 is a frequent event among bladder tumors, regardless of the source of genomic DNA obtained from frozen or paraffin-embedded material. Methylation analyses were performed after bisulfite treatment using low amounts of DNA (500 to 1000 ng). Although bisulfite treatment is feasible using lower amount of nucleic acids, DNA quantity could be a potential limitation for low-stage and low-grade small tumors. Third, KISS1 methylation was significantly associated with increasing tumor stage and tumor grade. In addition to the clinicopathologic stratification of bladder cancer patients, a relevant translational point relates to treatment. In this new scenario, KISS1 represents a potential therapeutic target whose expression could be potentially reactivated by demethylating drugs, and also by means of administration of kisspeptins.18,25
A further step in the translational implications of the clinical evaluation of KISS1 in bladder cancer deals with analyses of its transcript expression by RT-PCR on bladder tumors of known KISS1 methylation status. These analyses served not only to confirm the previously reported clinical relevance of the loss of expression in bladder cancer correlating with increasing tumor stage and grade,12,22 but also to link the low transcript expression with the identified potential methylation mechanism of inactivation. An additional goal was to assess how methylation could further discriminate clinical outcome in combination with transcript expression. Notably, the presence of low transcript levels alone or in combination with methylation was associated with poor survival. KISS1 methylation stratified the discrimination among patients with good or poor outcome obtained by transcript expression alone. The multivariate analysis revealed that KISS1 transcript expression remained an independent prognosticator. Thus, the combination of methylation and transcript analyses has served to discover the epigenetic silencing of a gene with reported low expression. Moreover, these variables were significantly associated with clinicopathologic correlates and clinical outcome.
Conclusions
KISS1 hypermethylation was identified in bladder cancer, providing a potential mechanistic explanation for the observed loss of KISS1 in uroepithelial malignancies by epigenetic silencing. KISS1 expression showed higher correlations with clinical outcome than did methylation, suggesting that DNA methylation correlates with silencing but that it might not be the only mechanism driving silencing. Associations of KISS1 methylation and its expression with histopathological variables and poor survival suggest the utility of incorporating these KISS1 measurements using paraffin-embedded material for tumor stratification and for clinical outcome prognosis of patients affected with uroepithelial neoplasias.
Acknowledgments
We thank all members of the Tumor Markers Group at the CNIO, and especially Noreli Franco, for their technical support and for constructive suggestions in the preparation of this manuscript. We also thank our clinical collaborators at the different institutions involved in this study and the members of the Spanish Oncology Group of Genitourinary Cancer (SOGUG) for their support in making available the tumor specimens and clinical follow-up data for the bladder cancer cases analyzed in this study.
Footnotes
Supported by a grant (SAF2009-13035 to M.S.-C.) from the Spanish Ministry of Science and Innovation.
Supplemental material for this article can be found at http://ajp.amjpathol.org or at doi: 10.1016/j.ajpath.2011.05.009.
Supplementary data
Confirmation of demethylation after azacytidine exposure of bladder cancer cells. A: Analysis of CpG island methylation status of KISS1 by bisulfite genomic sequencing in human bladder cancer cell lines after azacytidine exposure. CpG dinucleotides are represented as dark squares for methylated cytosines and open squares for unmethylated cytosines. For dark squares, the presence of methylation was confirmed in at least two of the clones that were sequenced for the tumors analyzed. B: Chromatograms obtained by bisulfite genomic sequencing in human bladder cancer cell lines. Representative examples are shown of the sequences obtained in the methylated bladder cancer cell line 5637, and in the same cell line unmethylated after azacytidine exposure.
Validation analyses of the methylation status of a subset of the tumors analyzed by MS-PCR and BS-SEQ. A: Methylation-specific PCRs (MS-PCR) for KISS1 in human bladder tumors. A PCR band in lane M indicates a methylated KISS1 gene; in lane U, it indicates an unmethylated gene. Normal lymphocytes (NL) and MEC1 were used as controls for unmethylated KISS1; and in vitro methylated DNA (IVD) was used as control for methylated KISS1. B: Analysis of CpG island methylation status of KISS1 by bisulfite genomic sequencing in human bladder tumors. CpG dinucleotides are represented as dark squares for methylated cytosines and open squares for unmethylated cytosines. For dark squares, the presence of methylation was confirmed in at least two of the clones that were sequenced for the tumors analyzed.
References
- 1.Wolff E.M., Liang G., Jones P.A. Mechanisms of disease: genetic and epigenetic alterations that drive bladder cancer. Nat Clin Pract Urol. 2005;2:502210. doi: 10.1038/ncpuro0318. [DOI] [PubMed] [Google Scholar]
- 2.Jones P.A., Baylin S.B. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 3.Feinberg A.P., Ohlsson R., Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 4.Costello J.F., Frühwald M.C., Smiraglia D.J., Rush L.J., Robertson G.P., Gao X., Wright F.A., Feramisco J.D., Peltomäki P., Lang J.C., Schuller D.E., Yu L., Bloomfield C.D., Caligiuri M.A., Yates A., Nishikawa R., Su Huang H., Petrelli N.J., Zhang X., O'Dorisio M.S., Held W.A., Cavenee W.K., Plass C. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427–5440. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- 6.Herman J.G., Baylin S.B. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 7.Catto J.W., Azzouzi A.R., Rehman I., Feeley K.M., Cross S.S., Amira N., Fromont G., Sibony M., Cussenot O., Meuth M., Hamdy F.C. Promoter hypermethylation is associated with tumor location, stage, and subsequent progression in transitional cell carcinoma. J Clin Oncol. 2005;23:2903–2910. doi: 10.1200/JCO.2005.03.163. [DOI] [PubMed] [Google Scholar]
- 8.Aleman A., Adrien L., Lopez-Serra L., Cordon-Cardo C., Esteller M., Belbin T.J., Sanchez-Carbayo M. Identification of DNA hypermethylation of SOX9 in association with bladder cancer progression using CpG microarrays. Br J Cancer. 2008;98:466–473. doi: 10.1038/sj.bjc.6604143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohtaki T., Shintani Y., Honda S., Matsumoto H., Hori A., Kanehashi K., Terao Y., Kumano S., Takatsu Y., Masuda Y., Ishibashi Y., Watanabe T., Asada M., Yamada T., Suenaga M., Kitada C., Usuki S., Kurokawa T., Onda H., Nishimura O., Fujino M. The metastasis suppressor gene KiSS-1 encodes peptide ligand of a G-protein-coupled receptor. Nature. 2001;411:613–617. doi: 10.1038/35079135. [DOI] [PubMed] [Google Scholar]
- 10.Muir A.I., Chamberlain L., Elshourbagy N.A., Michalovich D., Moore D.J., Calamari A., Szekeres P.G., Sarau H.M., Chambers J.K., Murdock P., Steplewski K., Shabon U., Miller J.E., Middleton S.E., Darker J.G., Larminie C.G., Wilson S., Bergsma D.J., Emson P., Faull R., Philpott K.L., Harrison D.C. AXOR12, a novel human G protein-coupled receptor, activated by the peptide KiSS-1. J Biol Chem. 2001;276:28969–28975. doi: 10.1074/jbc.M102743200. [DOI] [PubMed] [Google Scholar]
- 11.Kotani M., Detheux M., Vandenbogaerde A., Communi D., Vanderwinden J.M., Le Poul E., Brézillon S., Tyldesley R., Suarez-Huerta N., Vandeput F., Blanpain C., Schiffmann S.N., Vassart G., Parmentier M. The metastasis suppressor gene KiSS-1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled receptor GPR54. J Biol Chem. 2001;276:34631–34636. doi: 10.1074/jbc.M104847200. [DOI] [PubMed] [Google Scholar]
- 12.Sanchez-Carbayo M., Capodieci P., Cordon-Cardo C. Tumor suppressor role of KiSS-1 in bladder cancer: loss of KiSS-1 expression in associated with bladder cancer progression and clinical outcome. Am J Pathol. 2003;162:609–617. doi: 10.1016/S0002-9440(10)63854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.West A., Vojta P.J., Welch D.R., Weissman B.E. Chromosome localization and genomic structure of the KiSS-1 metastasis suppressor gene (KISS1) Genomics. 1998;54:145–148. doi: 10.1006/geno.1998.5566. [DOI] [PubMed] [Google Scholar]
- 14.Ikeguchi M., Yamaguchi K., Kaibara N. Clinical significance of the loss of KiSS-1 and orphan G-protein-coupled receptor (hOT7T175) gene expression in esophageal squamous cell carcinoma. Clin Cancer Res. 2004;10:1379–1383. doi: 10.1158/1078-0432.ccr-1519-02. [DOI] [PubMed] [Google Scholar]
- 15.Hesling C., D'Incan M., Mansard S., Franck F., Corbin-Duval A., Chèvenet C., Déchelotte P., Madelmont J.C., Veyre A., Souteyrand P., Bignon Y.J. In vivo and in situ modulation of the expression of genes involved in metastasis and angiogenesis in a patient treated with topical imiquimod for melanoma skin metastasis. Br J Dermatol. 2004;150:761–767. doi: 10.1111/j.0007-0963.2004.05898.x. [DOI] [PubMed] [Google Scholar]
- 16.Dhar D., Naora H., Kubota H., Maruyama R., Yoshimura H., Tonomoto Y., Tachibana M., Ono T., Otani H., Nagasue N. Downregulation of KiSS-1 expression is responsible for tumor invasion and worse prognosis in gastric carcinoma. Int J Cancer. 2004;111:868–872. doi: 10.1002/ijc.20357. [DOI] [PubMed] [Google Scholar]
- 17.Lee J.H., Miele M.E., Hicks D.J., Phillips K.K., Trent J.M., Weissman B.E., Welch D.R. KISS1, a novel human malignant melanoma metastasis-suppressor gene [Erratum appeared in J Natl Cancer Inst 1997, 89:1549] J Natl Cancer Inst. 1996;88:1731–1737. doi: 10.1093/jnci/88.23.1731. [DOI] [PubMed] [Google Scholar]
- 18.Lee J.H., Welch D.R. Identification of highly expressed genes in metastasis-suppressed chromosome 6/human malignant melanoma hybrid cells using subtractive hybridization and differential display. Int J Cancer. 1997;71:1035–1044. doi: 10.1002/(sici)1097-0215(19970611)71:6<1035::aid-ijc20>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 19.Lee J.H., Welch D.R. Suppression of metastasis in human breast carcinoma MDA-MB-435 cells after transfection with the metastasis suppressor gene, KiSS-1. Cancer Res. 1997;57:2384–2387. [PubMed] [Google Scholar]
- 20.Masui T., Doi R., Mori T., Toyoda E., Koizumi M., Kami K., Ito D., Peiper S.C., Broach J.R., Oishi S., Niida A., Fujii N., Imamura M. Metastin and its variant forms suppress migration of pancreatic cancer cells. Biochem Biophys Res Commun. 2004;315:85–92. doi: 10.1016/j.bbrc.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 21.Martin T.A., Watkins G., Jiang W.G. KiSS-1 expression in human breast cancer. Clin Exp Metastasis. 2005;22:503–511. doi: 10.1007/s10585-005-4180-0. [DOI] [PubMed] [Google Scholar]
- 22.Nicolle G., Comperat E., Nicolaïew N., Cancel-Tassin G., Cussenot O. Metastin (KISS-1) and metastin-coupled receptor (GPR54) expression in transitional cell carcinoma of the bladder. Ann Oncol. 2007;18:605–606. doi: 10.1093/annonc/mdl421. [DOI] [PubMed] [Google Scholar]
- 23.Zohrabian V.M., Nandu H., Gulati N., Khitrov G., Zhao C., Mohan A., Demattia J., Braun A., Das K., Murali R., Jhanwar-Uniyal M. Gene expression profiling of metastatic brain cancer. Oncol Rep. 2007;18:321–328. [PubMed] [Google Scholar]
- 24.Hata K., Dhar D.K., Watanabe Y., Nakai H., Hoshiai H. Expression of metastin and a G-protein-coupled receptor (AXOR12) in epithelial ovarian cancer. Eur J Cancer. 2007;43:1452–1459. doi: 10.1016/j.ejca.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Makri A., Pissimissis N., Lembessis P., Polychronakos C., Koutsilieris M. The kisspeptin (KiSS-1)/GPR54 system in cancer biology. Cancer Treat Rev. 2008;34:682–692. doi: 10.1016/j.ctrv.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 26.Yamashita S., Tsujino Y., Moriguchi K., Tatematsu M., Ushijima T. Chemical genomic screening for methylation-silenced genes in gastric cancer cell lines using 5-aza-2′-deoxycytidine treatment and oligonucleotide microarray. Cancer Sci. 2006;97:64–71. doi: 10.1111/j.1349-7006.2006.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanchez-Carbayo M., Socci N.D., Lozano J., Saint F., Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol. 2006;24:778–789. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- 28.Kirkali Z., Chan T., Manoharan M., Algaba F., Busch C., Cheng L., Kiemeney L., Kriegmair M., Montironi R., Murphy W.M., Sesterhenn I.A., Tachibana M., Weider J. Bladder cancer: epidemiology, staging and grading, and diagnosis. Urology. 2005;66(6 Suppl 1):4–34. doi: 10.1016/j.urology.2005.07.062. [DOI] [PubMed] [Google Scholar]
- 29.Paz M.F., Fraga M.F., Avila S., Guo M., Pollan M., Herman J.G., Esteller M. A systematic profile of DNA methylation in human cancer cell lines. Cancer Res. 2003;63:1114–1121. [PubMed] [Google Scholar]
- 30.Dawson-Saunders B., Trapp R.G. ed 2. Appleton & Lange; Norwalk, CT: 1994. Basic & Clinical Biostatistics. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Confirmation of demethylation after azacytidine exposure of bladder cancer cells. A: Analysis of CpG island methylation status of KISS1 by bisulfite genomic sequencing in human bladder cancer cell lines after azacytidine exposure. CpG dinucleotides are represented as dark squares for methylated cytosines and open squares for unmethylated cytosines. For dark squares, the presence of methylation was confirmed in at least two of the clones that were sequenced for the tumors analyzed. B: Chromatograms obtained by bisulfite genomic sequencing in human bladder cancer cell lines. Representative examples are shown of the sequences obtained in the methylated bladder cancer cell line 5637, and in the same cell line unmethylated after azacytidine exposure.
Validation analyses of the methylation status of a subset of the tumors analyzed by MS-PCR and BS-SEQ. A: Methylation-specific PCRs (MS-PCR) for KISS1 in human bladder tumors. A PCR band in lane M indicates a methylated KISS1 gene; in lane U, it indicates an unmethylated gene. Normal lymphocytes (NL) and MEC1 were used as controls for unmethylated KISS1; and in vitro methylated DNA (IVD) was used as control for methylated KISS1. B: Analysis of CpG island methylation status of KISS1 by bisulfite genomic sequencing in human bladder tumors. CpG dinucleotides are represented as dark squares for methylated cytosines and open squares for unmethylated cytosines. For dark squares, the presence of methylation was confirmed in at least two of the clones that were sequenced for the tumors analyzed.


