Abstract
Challenges in measurement of epidermal growth factor receptor (EGFR) protein expression have led to conflicting data on its prognostic value and discontinuation of its use for prediction of response. Herein is described a quantitative standardized assay for EGFR and its use in a series of retrospective cohorts of patients with non–small cell lung cancer (NSCLC). The AQUA technology of quantitative immunofluorescence was used in conjunction with Western blot analysis to calculate the absolute concentration of EGFR in two independent NSCLC cohorts (170 from Yale New Haven Hospital and 335 from Sotiria and Patras Hospitals in Greece). EGFR and mutated EGFR were measured using D38B1 antibody and two mutation-specific antibodies. All patients positive or borderline for mutation-specific antibody were genotyped. A threshold for reproducible detection of EGFR was defined as 0.85 ng/μg total protein. EGFR expression demonstrated no prognostic value in either cohort. The mutation rate was 1.79% in the Yale cohort, and 1.52% in the Sotiria/Patras cohort, with no antibody detection–based false-positive cases. No mutations were detected for EGFR concentrations <1.46 ng/μg total protein. In summary, accurate measurement of EGFR still shows no prognostic value in NSCLC. In these two population-based cohorts, the antibody-based EGFR mutation rate was lower than has been frequently reported.
Non–small cell lung cancer (NSCLC) is the leading cause of cancer-related death in the Western world.1 Despite progress in treatment, prognosis of the disease is still poor. Because current treatments expose many patients to adverse effects to help a few, there is a need for diagnostic tests to determine which patients will benefit from each regimen. Administration of tyrosine kinase inhibitors is a relatively new therapy for NSCLC. They initially showed modest efficacy in the general population with NSCLC2; however, the observation of impressive tumor response in a subset of patients with certain demographic characteristics led to discovery of a range of mutations in the tyrosine kinase domain of epidermal growth factor receptor (EGFR) that can predict clinical benefit from tyrosine kinase inhibitors.3,4 The frequency of the mutations varies among different populations. Never smoking status, Asian ethnicity, histologic findings of adenocarcinoma, and female sex are patient characteristics traditionally linked to the mutations.5,6 A deletion in exon 19, DEL746-750, and a point mutation in exon 21, L858R, account for most (85% to 90%) EGFR mutations.6,7
Presence or absence of EGFR mutations has become important baseline information in the treatment of NSCLC because administration of tyrosine kinase inhibitors in the first line of treatment now depends on mutational status.8 The mainstay of determining mutational status in patients with NSCLC is direct DNA sequencing of the tumor. Recently, a set of antibodies that detect EGFR with the DEL746-750 deletion or the L858R point mutation has become available, and was both sensitive and specific in two studies.9,10 However, the checkered history of EGFR IHC may represent a challenge to broad acceptance of these tools. Initially, measurement of EGFR was performed using radioligand binding assays,11 which were difficult to conduct and poorly reproducible. These assays were replaced by IHC as the standard method for assessment of EGFR. However, this assay also demonstrated a marked lack of reproducibility and reliability,12–15 which led to its dramatically decreased use. As a result, neither the prognostic nor the predictive role of EGFR in NSCLC has been definitively determined despite the large number of studies published. For example, in some studies, EGFR predicted a worse prognosis,16–18 whereas in others, it demonstrated no prognostic value.19–21 The wide range of findings reflects the number of different antibodies used (recognizing different epitopes) and the relatively unreliable, nonstandardized, subjective methods used to assess the level of expression of EGFR.
The objective of the present study was to develop and test a method for assessment of the expression of EGFR in a standardized, quantitative, objective manner. Measurement of total EGFR and mutated EGFR was assessed in two independent cohorts of patients with NSCLC to determine prognostic value and mutation frequency in each population.
Materials and Methods
Patient Cohorts
The first cohort was accrued by serial collection of formalin-fixed paraffin-embedded tissue from the Department of Pathology at Yale University (New Haven, CT). Of the lung cancer samples collected, 170 were classified as NSCLC. The second cohort, with 335 patients, was from the Pathology Departments of Sotiria General Hospital (Athens, Greece) and Patras University Hospital (Rion, Greece). In patients in the Yale cohort (median age, 67 years; age range, 42 to 90 years), NSCLC was diagnosed between 1993 and 2003, and median follow-up was 27.4 months (range, 0.1 to 127.79 months). In the Sotiria/Patras cohort (median age, 64 years; age range, 34 to 84 years) NSCLC was diagnosed between 1990 and 2004, and median follow-up was 21 months (range, 0.1 to 223 months). Demographic data for the two cohorts are given in Table 1. The study was approved by the institutional review boards of all centers. Written informed consent was obtained from all patients before inclusion in the study.
Table 1.
Clinicopathologic Characteristics of Yale and Sotiria/Patras Cohorts
| Variable | Yale cohort |
Sotiria/Patras cohort |
||||
|---|---|---|---|---|---|---|
| Total EGFR (ng/μg of total protein) |
Total EGFR (ng/μg of total protein) |
|||||
| No. (%) | Median (range) | P value⁎ | No. (%) | Median (range) | P value⁎ | |
| All patients | 170 | 2.326 (0.21–13.58) | 0.04 | 335 | 1.65 (0.15–89.78) | 0.03 |
| Sex | ||||||
| Male | 85 (50) | 2.718 (0.21–13.58) | 274 (81.8) | 1.93 (0.15–9.54) | ||
| Female | 85 (50) | 2.126 (0.21–8.57) | 41 (12.2) | 0.956 (0.23–9.78) | ||
| NA | 0 | 20 | ||||
| Stage | 0.13 | 0.3 | ||||
| IA | 60 (35.2) | 1.998 (0.29–12.18) | 26 (7.7) | 1.216 (0.15–8.39) | ||
| IB | 27 (15.8) | 2.32 (0.21–9.85) | 76 (22.6) | 1.778 (0.23–9.78) | 87 (25.9) | |
| II | 26 (15.3) | 2.98 (0.26–9.04) | 87 (25.9) | 2.279 (0.15–9.54) | ||
| III | 37 (21.7) | 2.4 (0.21–10.32) | 90 (26.8) | 1.483 (0.21–7.78) | ||
| IV | 16 (9.4) | 2.107 (0.7–13.58) | 35 (10.4) | 1.394 (0.1505.8) | ||
| Data not available | 4 | 21 | ||||
| Differentiation | 0.2 | 0.68 | ||||
| High | 9 (5.3) | 0.86 (0.63–5.37) | 22 (6.5) | 1.2 (0.3–6.38) | ||
| Moderate | 39 (22.9) | 2.90 (0.29–13.58) | 149 (44.4) | 2.14 (0.15–9.78) | ||
| Low | 80 (47) | 2.58 (0.21–12.58) | 133 (39.7) | 1.59 (0.15–9.54) | ||
| Data not available | 63 | 31 | ||||
| Histotype | <0.0001 | <0.0001 | ||||
| Adenocarcinoma | 91 (53.5) | 1.815 (0.91–13.58) | 124 (37) | 0.879 (0.15–9.74) | ||
| Squamous cell carcinoma | 33 (19.4) | 4.238 (0.347–12.785) | 160 (47.7) | 27 (0.23–9.78) | ||
| Bronchioloalveolar carcinoma | 7 (4.1) | 1.498 (0.41–5.37) | 5 (1.5) | 0.65 (0.3–0.69) | ||
| Large cell cancer | 18 (10.5) | 1.847 (0.21–10.42) | 3 (0.9) | 2.32 (2.23–2.68) | ||
| Adenosquamous carcinoma | 13 (7.6) | 3.49 (0.5–9.04) | 0 | |||
| Other/data not available | 5 | 43 | ||||
| Race/ethnicity | 0.03 | NA | ||||
| White | 143 (84.1) | 2.4 (0.21–13.58) | 335 (100) | |||
| African American | 17 (10) | 1.28 (0.29–5.19) | ||||
| Other | 5 (2.9) | |||||
| Unknown | 5 (2.9) | |||||
| Smoking status | 0.5292 | 0.516 | ||||
| Current/former | 158 (92.9) | 2.291 (0.209–12.188) | 252 (75.2) | 1.703 (0.152–9.781) | ||
| Never | 9 (5.3) | 2.807 (0.513–13.58) | 28 (8.3) | 1.391 (0.276–6.849) | ||
| Unknown | 3 | 55 | ||||
NA, not available.
P values calculated using the Mann-Whitney U-test or the Kruskal-Wallis test.
Tissue Microarray Construction
Formalin-fixed paraffin-embedded tissue blocks from the patients were cored twice in representative tumor areas using 0.6-mm cores and arrayed into a recipient block by the Yale Pathology Tissue Services facility. Formalin-fixed paraffin-embedded pellets from the cell lines MCF7, SKBR3, H1299, H1355, H441, H2282, H1666, H193, and HCC2279 were used as positive controls, and HT29, H2126, A549, SW480, HC15, H1819, Calu-1, A431, and H1650 as negative controls. Culture conditions have been published in detail elsewhere.22 An index array containing the cell lines and tissue from 30 patients with a range of EGFR expression in twofold redundancy was assayed with each cohort array.
Western Blot Analysis and Quantification
Cell lines selected to represent the range of EGFR expression in NSCLC were MCF7, SKBR3, H1299, H1355, H441, H2282, H1666, H193, and HCC2279. Whole-cell lysates were prepared, and total protein concentration was measured using the Bradford assay (BioTek Instruments, Winooski, VT). Five micrograms total protein for each lysate was resolved using SDS-PAGE on an 8% Bis-Tris gel (NuPAGE; Invitrogen Corp., Carlsbad, CA) using NuPAGE MOPS [3-(N-morpholino) propane sulfonic acid] SDS running buffer at 45 mA. On each gel, seven dilutions (10, 25, 50, 125, 250, 500, and 1000 ng) of recombinant intracellular domain of EGFR (recEGFR; Cell Signaling Technology, Inc., Beverly, MA) were also resolved for construction of the standard curve. Resolved protein was transferred using NuPAGE transfer buffer at 50V for 2 hours. Western blot analysis was performed according to standard procedures using EGFR rabbit monoclonal D38B1 antibody diluted 1:2000. β -Tubulin (Cell Signaling Technology, Inc.) diluted 1:4000 was used as a loading control. Bands were quantified using ImageJ software (National Institutes of Health, Washington, DC), and were normalized to β-tubulin. The area under the curve was correlated with the nanograms of protein loaded for each band of recEGFR, and linear regression was fit to the linear portion of the curve. This equation was used to transform the normalized area under the curve for each cell line to an EGFR quantity (in nanograms) and to calculate the concentration of EGFR in nanograms per microgram total protein for each cell line by dividing raw EGFR quantity (in nanograms) by total protein loaded (5 μg). Subsequently, a correlation was drawn between the concentrations in nanograms per microgram total protein and the AQUA (Automated Quantitated Analysis technology; Yale University, New Haven, CT) scores of those cell lines. The generated equation (standard curve) was used to transform patient AQUA scores into absolute concentrations of EGFR. The cut-point for the threshold of EGFR detection is addressed in the Results section.
Antibodies and Quantitative Immunofluorescence
Arrays were deparaffinized in xylene (soaking twice for 20 minutes) and rehydrated with alcohol (twice in 100% alcohol for 1 minute, and then in 95%, 85%, and 70% alcohol for 1 minute each). Antigen retrieval was performed using a PT module (Lab Vision Corp., Fremont, CA) with EDTA buffer, pH 8, at 97°C for 20 minutes. Endogenous peroxidase activity was blocked via 30-minute incubation in 2.5% hydrogen peroxide in methanol at room temperature. Nonspecific antigens were blocked via incubation in 0.3% bovine serum albumin in Tris-buffered saline solution and Tween 20 for 30 minutes at room temperature. Slides were then incubated overnight with a cocktail of a primary antibody (details of primary antibodies are given in Table 2) and a mouse monoclonal cytokeratin antibody (Dako Corp., Carpinteria, CA). Ideal titers were determined as an optimal combination of image appearance and a quantitative ratio of signal to background for each of the primary antibodies after trying a range of titers in a series of test arrays. Next, a cocktail of Alexa 546–conjugated goat anti-mouse secondary antibody (Molecular Probes, Inc., Eugene, OR) diluted 1:100 in rabbit EnVision reagent (Dako Corp.) was applied to the slides for 1 hour at room temperature. For signal amplification, cyanine 5–tyramide (PerkinElmer, Inc., Waltham, MA) diluted 1:50 was used at room temperature for 10 minutes. Finally, Prolong Gold (Molecular Probes, Inc.) containing DAPI was used to detect nuclei. An index array was stained aside each cohort array to enable standardization of the assay, along with negative (no primary) and positive controls. Single slides with formalin-fixed paraffin-embedded cell pellets (H1975; Cell Signaling Technology, Inc.) were used as positive controls for the experiments using L858R-specific antibody. All antibodies sources and reagents are given in Table 2.
Table 2.
Antibodies and Reagents
| Variable | Clone | Dilution |
|---|---|---|
| Total EGFR | D38B1 | 1:100 |
| DEL746-750 | 6B6 | 1:500 |
| L858R | 43B2 | 1:100 |
| recEGFR | EGFR kinase | NA |
All antibodies and reagents obtained from Cell Signaling Technology, Inc.
NA, data not available; recEGFR, recombinant EGFR.
Automated Quantitative Analysis
AQUA is a method of calculating protein concentration in subcellular compartments, and has been described in detail elsewhere.23 In brief, monochromatic images of DAPI and of the complexes target–cyanine 5 and cytokeratin–Alexa 546 were captured using a microscope (PM-2000; HistoRx, Inc., New Haven, CT). A tumor “mask” is generated to define the tumor area after binarization of the cytokeratin signal so that every pixel is either on or off on the basis of a clustering algorithm. The AQUA scores in the tumor mask were calculated as the ratio of the sum of the target pixels in the tumor mask divided by the area of the tumor mask. The AQUA scores were normalized for exposure time, bit depth, and lamp hours for optimal standardization and reproducibility. Then, using the standard curve, the AQUA scores were transformed into nanograms per microgram total protein.
Genotyping of EGFR Mutations
Cases that were positive or marginally above the background using quantitative immunofluorescence (QIF) for the 746–750 deletion in exon 19 or the L858R point mutation in exon 21 were genotyped using direct Sanger sequencing for these two specific mutations. Tissue blocks from the corresponding tumors were cored after review of an H&E slide and circling the tumor area to guide the coring. One 0.6-mm core per case was used to extract genomic DNA using the RecoverAll Total Nucleic Acid Isolation kit (Ambion, Inc., Austin, TX). The regions of interest were amplified using PCR, and the amplicons were sequenced. Primers used in PCR and sequencing have been published elsewhere.24,25
Statistical Analysis
Tumor heterogeneity of total EGFR was assessed using Pearson's correlation coefficient (R) between the AQUA scores from redundant cores, and reproducibility of the assay was assessed via correlation between serial cuts of the index array. R2 > 0.5 was considered acceptable core-to-core reproducibility (correlation between redundant tissue cores), and R2 > 0.9 was considered acceptable serial section reproducibility (correlation between serial cuts of the same block). The log-rank test was used to assess statistical significance of Kaplan-Meier curves. The Kruskal-Wallis test and Mann-Whitney U-test were used to compare continuous EGFR concentrations between groups with different clinicopathologic characteristics. All statistical analyses were performed using either JMP or StatView software (both from SAS Institute, Inc., Cary, NC).
Results
Antibody Validation in the Index Array
Reproducibility of total EGFR was assessed via correlations of the AQUA scores between serial cuts and redundant cores of the index array, respectively. These correlations showed good reproducibility between serial cuts (Pearson's r = 0.965; Figure 1A) or different cores (Pearson's r = 0.939; data not shown). Total and mutation-specific antibodies were tested in a panel of cell lines serving as positive and negative controls. H2279 and H1650 were positive using the deletion-specific antibody, whereas H1975 was positive using the L858R-specific antibody. The other cell lines in the index array were negative (Figure 1B). All of the cell lines with the exception of MCF7 were positive for total EGFR. The pattern of both total and mutated EGFR in the positive controls was homogeneous and strongly cytoplasmic (Figure 1C). AQUA scores were calculated for total and mutated EGFR for each of the cell lines of the index array (Figure 1B).
Figure 1.
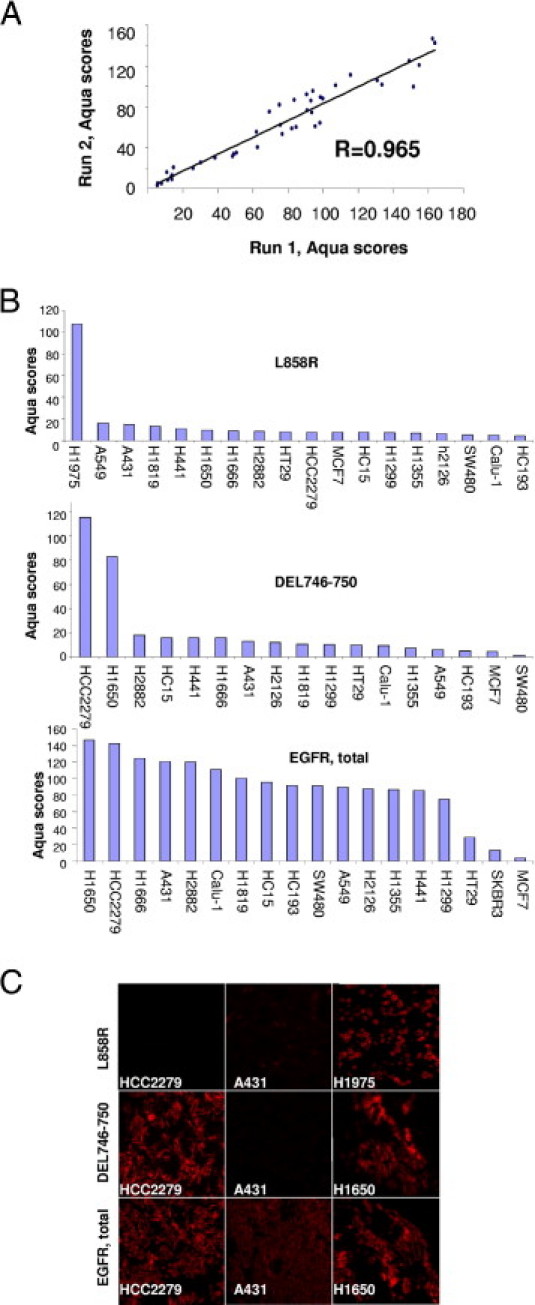
Antibody validation in positive and negative controls. A: Reproducibility of the AQUA assay between experiments performed on different days. AQUA scores of the index array correlate with Pearson's coefficient r = 0.965. B: Quantification (AQUA scores) of immunofluorescence in the cell lines of the index array. Data for H1975 are available only for the L858R-related experiments because H1975 was tested as a separate slide (positive control) in those runs. C: Immunofluorescence in positive and negative controls for total and mutated EGFR. H1975 is positive when stained with the L858R-specific antibody, whereas A431 and HCC2279 are negative (top row). H1650 and HCC2279 are positive when stained with the DEL746-750–specific antibody, whereas A431 is negative (middle row). A431, HCC2279, and H1650 are positive when stained with the total EGFR antibody (bottom row).
Construction of the Standard Curve
A panel of cell lines was selected to reflect the range of EGFR expression in patients with NSCLC. EGFR was detected using Western blot analysis in this panel (Figure 2A), along with a series of different quantities of recEGFR. Band intensity of recEGFR was correlated with the corresponding quantities in nanograms, and the linear portion of the generated curve (including the 10-, 25-, 50-, and 125-ng recEGFR bands) was used to transform the band intensity of each cell line into an absolute EGFR quantity in nanograms. The concentration of EGFR in the cell lines (in nanograms per microgram total protein) was calculated as the ratio of the EGFR quantity to the total protein loaded (5 μg). The experiment was performed twice, and the calculated EGFR concentrations were averaged. Subsequently, a regression of EGFR concentrations with the corresponding AQUA scores (obtained from the index array) was drawn for the cell line panel (Figure 2B). The intercept of the curve was set at zero so that no case would demonstrate a negative EGFR concentration. The best linear fit was (EGFR concentration) = 0.0008 × AQUA score. This standard curve enables transformation of the AQUA scores of the two cohorts into an EGFR concentration (in nanograms per microgram total protein). AQUA scores were normalized for run-to-run variability before transformation to EGFR concentrations by using the correlation of the AQUA scores in the index arrays of each run.
Figure 2.
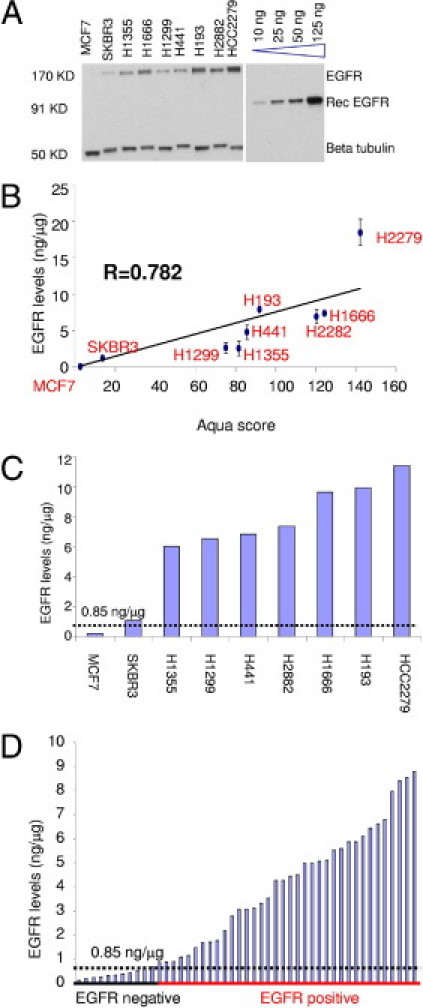
EGFR standardization. A: Immunoblotting of a panel of cell lines (MCF7, SKBR3, H1355, H1666, H1299, H441, H193, H2882, and HCC2279) along with recombinant intracellular domain of EGFR (recEGFR; molecular weight, 91 kDa) at different quantities (10, 25, 50, and 125 ng). For each cell line, 5 μg total protein was loaded. β-Tubulin was used for normalization. This experiment was performed twice. B: EGFR concentration plotted against the AQUA score of each cell line. Intercept was set at zero so that no negative EGFR concentrations would occur at low AQUA scores. Pearson's R coefficient is 0.782. Error bars indicate variance between the two experiments. C: EGFR concentration for each of the cell lines after applying the standard curve generated in B to their AQUA scores. A threshold of 0.85 ng/μg was set between the negative (MCF7) and positive (SKBR3) cell lines. D: EGFR concentration was calculated for each spot of the cases of the index array. The threshold was set to differentiate EGFR-positive from EGFR-negative cases.
The standard curve was applied to the cell lines (Figure 2C), and the cases (Figure 2D) of the index array and EGFR concentrations were calculated. The threshold of EGFR detection was set between the negative (MCF7) and the first positive (SKBR3) cell line in the Western blot analysis and was determined to be 0.85 ng/μg total protein. This level was concordant with our visual impression of the threshold of immunofluorescence images of the negative and positive cases in the index array.
Prognostic Role of EGFR in NSCLC
All EGFR AQUA scores from the Yale and the Sotiria/Patras cohorts were transformed into absolute EGFR concentrations (in nanograms per microgram), and the threshold, defined from the index array, was used to separate positive from negative cases. In the Yale cohort, 35 patients (20.5%) were classified as negative, and 135 (79.5%) as positive (range of expression, 0.864 to 13.58 ng/μg for positive cases). In the Sotiria/Patras cohort, 110 patients (32.8%) were classified as negative, and 225 as positive (range of expression, 0.863 to 9.781 ng/μg for positive cases). No differences in survival between EGFR-positive and EGFR-negative cases were observed between patients in either cohort (P = 0.7265 in the Yale cohort and P = 0.4277 in the Sotiria/Patras cohort; Figure 3,A and C). In the Yale cohort, median survival was 33.9 months for EGFR-positive patients, and 48.43 months for EGFR-negative patients, whereas in the Sotiria/Patras cohort, median survival was 30.5 months for EGFR-positive patients, and 35.5 months for EGFR-negative patients. The same was the case for adenocarcinoma histology (P = 0.8018 for the Yale cohort, and P = 0.7145 for the Sotiria/Patras cohort, Figure 3, B and D). In addition, no difference in survival was observed in the subgroup with squamous cell carcinoma in the Sotiria/Patras cohort (P = 0.497; data not shown). Analysis of squamous cell carcinomas in the Yale cohort was not useful because of the low number of patients and events. Further stratification according to stage and sex did not demonstrate any prognostic potential of EGFR positivity (data not shown).
Figure 3.
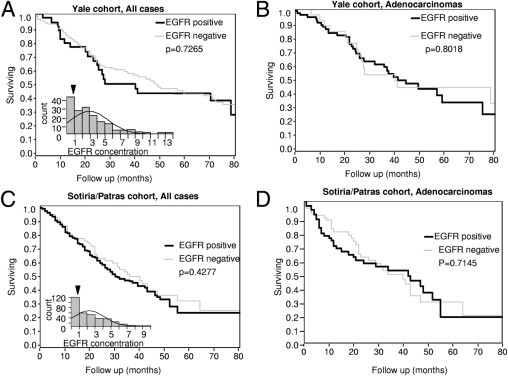
EGFR shows no prognostic value in NSCLC. A: Survival analysis in the entire Yale cohort when patients are grouped according to the presence or absence of EGFR. The inset shows the frequency distribution of EGFR concentration in this cohort, and the arrowhead shows the cut-point. B: Subgroup analysis in patients with histologic findings of adenocarcinoma or bronchioloalveolar disease in the Yale cohort. C: Survival analysis, and frequency distribution (inset), of EGFR for the Sotiria/Patras cohort. D: Subgroup analysis in patients with histologic findings of adenocarcinoma or bronchioloalveolar disease in the Sotiria/Patras cohort.
Mutational Status of EGFR
The mutational status of EGFR, assessed using quantitative immunofluorescence, was available in 167 patients in the Yale cohort and 262 patients in the Sotiria/Patras cohort. Three cases in the Yale cohort were found to be mutated. Specifically, one male patient with large-cell carcinoma tested positive for the deletion between codons 746 and 750 in exon 19, and one male with adenosquamous carcinoma and one female patient with adenocarcinoma tested positive for the L858R point mutation in exon 21. Three cases were found to be mutated in the Sotiria/Patras cohort. One patient with adenocarcinoma harbored the deletion 746–750 in exon 19, and two patients with squamous cell carcinoma tested positive for the L858R point mutation in exon 21. The staining pattern of the mutated cases was homogeneous and strongly cytoplasmic (Figure 4A). Quantification of the signal using AQUA demonstrated a broad dynamic range with a few patients who had scores marginally above background (borderline) in both cohorts (data not shown). Direct sequencing of the regions of interest in exons 19 and 21 was possible for four of six positive cases and eight of eleven borderline cases (Table 3). All four cases that were positive using quantitative immunofluorescence with the mutation-specific antibodies were found to be mutants after sequencing. In addition, one patient from the Yale cohort among those proved to harbor both L858R and P856L (phenylalanine-to-leucine) mutations. P856L is a rare missence mutation that has been reported once previously.26 DNA sequencing revealed one additional mutant (a female patient with adenocarcinoma) among the eight borderline cases tested. For the rest of the positive and borderline cases, there was no available tissue for sequencing. The overall mutation rate was 1.79% in the Yale cohort and 1.52% in the Sotiria/Patras cohort. Matching of EGFR mutational status data with total EGFR quantification demonstrated that in all EGFR mutations detected, total EGFR concentration was at least 1.466 ng/μg total protein (Figure 4, B and C).
Figure 4.
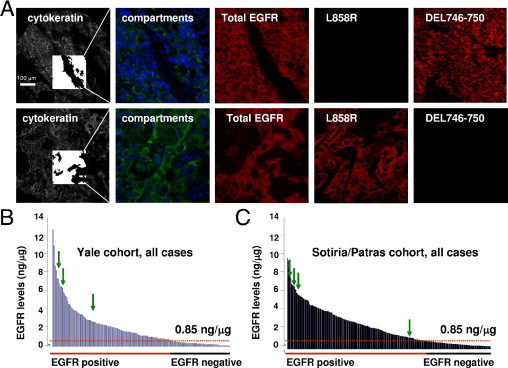
EGFR mutations in the Yale and Sotiria/Patras cohorts A: Immunofluorescence images of a large-cell carcinoma positive for the deletion in exon 19 (top row) and of an adenosquamous cell carcinoma positive for the point mutation in exon 21 (bottom row). In the cytokeratin images (Cy3 channel), the inside box shows the tumor mask that defines the area of the tumor in the AQUA algorithm. In the compartment images, blue represents the nuclear compartment, and green the cytoplasmic compartment. The remainder of the images demonstrate the signal for total EGFR, L858R point mutation, and 746–750 deletion (targets) in the cyanine 5 channel. The compartment and target images are magnified threefold in comparison to the cytokeratin images. B: Distribution of EGFR concentration in the 170 patients in the Yale cohort. Arrows indicate patients with mutated EGFR. C: Distribution of EGFR concentration in the Sotiria/Patras cohort. All mutations were detected in patients with EGFR concentration at least 1.466 ng/μg.
Table 3.
Mutation Detection Using QIF and Sequencing of Positive and Borderline Cases
| Variable | Yale cohort |
Sotiria/Patras cohort |
||
|---|---|---|---|---|
| QIF | DNA sequencing | QIF | DNA sequencing | |
| Positive | ||||
| DEL | 1 | Mutant | 1 | NA |
| L858R | 2 | Mutant | 2 | Mutant (1 tested) |
| Borderline | ||||
| DEL | 2/167 | Wild type | 5/266 | 1 Mutant (2 tested) |
| L858R | 4/167 | Wild type | 0/266 | NA |
| Negative | 158/167 | NA | 254/262 | NA |
| Overall mutant | 3/167 (1.79%) | 4/262 (1.52%) | ||
NA, data not available.
Discussion
In the present study, we developed a quantitative, reproducible, standardized assay to measure EGFR concentration in NSCLC. It was determined that the threshold of EGFR detection for our assay was 0.85 ng/μg total protein, and then this assay was applied to two cohorts of patients with NSCLC. It was demonstrated that the presence or absence of EGFR was not prognostic in either cohort or any subgroup of either cohort. In addition, the two most common mutations in the tyrosine kinase domain of EGFR were detected using mutation-specific antibodies and immunofluorescence in the two cohorts. Only a few patients were found to harbor the mutations. There was no detectable pattern in clinical characteristics of the patients with mutated EGFR. All cases positive in the quantitative immunofluorescence assay were also mutated using direct sequencing. Cases that stained marginally above background using quantitative immunofluorescence were also sequenced, and EGFR was observed to be mutated in one case. Matching of EGFR concentrations with the corresponding mutational status revealed that all mutated cases had EGFR concentrations >1.466 ng/μg.
Several studies have reported that the frequency of mutations in the tyrosine kinase domain of EGFR is 8% to 15%27–31 in white patients with NSCLC, and 15% to 25% in white patients with histologic findings of adenocarcinoma.10,31 However, this effort in two population-based predominantly white cohorts suggests a much lower mutation rate. It is tempting to attribute this discrepancy to lower sensitivity of the mutation-specific antibodies and inability of the antibody-based method to detect mutations other than the two most prominent. Future efforts will be required to determine whether the issue is sensitivity or variability in populations. Although a range of mutation incidence has been reported, large population-based studies such as SEER (Iowa Surveillance, Epidemiology, and End Results registry) have not yet been assessed. A possible explanation of the low frequency of mutation is the low rate of never smokers in our cohorts, which is compatible with previous reports in white populations with NSCLC.32,33 Our work raises the possibility that the EGFR mutation rate in white populations with NSCLC might be less than what has been previously reported. However, inasmuch as we can address only two specific mutations using the antibody approach, further work will be required to compare the sensitivity and specificity of this approach in large populations.
It has been reported that there are NSCLC patients who harbor a mutation in the tyrosine kinase domain of EGFR but in whom no total EGFR is detected using an EGFR specific antibody in quantitative immunofluorescence.10,34 However, in the present study, total EGFR was detected in all patients who were proved to harbor mutated EGFR. A novel antibody for total EGFR was used that has not been used in previous studies. Total EGFR in our cohorts was detected with clone D38B1, which detects the intracellular domain of the receptor and uses heat-induced antigen retrieval rather than protease-mediated methods, which are subject to challenges in standardization. Clones 31G7 and 18C9 were used in previous studies.10,34 Both detect the extracellular domain of EGFR and require protease-mediated antigen retrieval. In our previous work, these and other EGFR antibodies have been historically difficult to validate.12 In addition, an objective standardized assay was used to calculate the EGFR concentration and to set a threshold below which no EGFR mutations were detected. Most patients who were found harbor the mutation exhibited high EGFR expression. Previous reports have demonstrated a positive correlation between EGFR expression, EGFR gene copy number, and the presence of EGFR mutations.35
The prognostic role of EGFR has been studied extensively in NSCLC and other malignant diseases. However, results have been conflicting and diverse, most likely owing to small cohorts, semiqualitative analysis, poorly standardized and nonvalidated methods, and other factors. As a consequence, EGFR has been reported to predict a worse outcome in some studies and to have no prognostic value in other studies. Moreover, the threshold dividing the prognostic groups according to EGFR expression has been variable and not well defined. Thus, the literature on EGFR has been difficult to reproduce, and is not broadly accepted. In the present study, we used the threshold of detection as a cut-point, and found no prognostic value. Prognostic value could be observed in the cut-point was varied. However, the cut-point that showed prognostic value in the first cohort was not validated in the second cohort, and, thus, is not considered meaningful (data not shown). While this work may also not be considered definitive, it demonstrates a standardized method for measurement that is reproducible and can be normalized to both cell lines and recombinant proteins. Given the reproducibility of the method, it is tempting to speculate about potential clinical value. For example, future studies assessing response to EGFR inhibitor therapy, even in the absence of mutations, might be considered.
In the various clinical settings, laboratories provide measurements of clinically important variables that are interpreted as positive or negative and are used to enable decision making. In this context, our assay quantifies the expression of EGFR in tumors in patients with NSCLC and provides an instrument-specific but reproducible sensitivity-based cut-point for defining the group of patients who express the receptor versus those who do not. The quantification is based on the combination of Western blot analysis in a panel of cell lines and AQUA-based assessment of immunofluorescence signal. A standard curve is generated from Western blot analysis to convert AQUA scores into absolute EGFR concentrations. EGFR concentration measured in nanograms per microgram is more generalizable than the raw AQUA score in the clinical setting because the AQUA score depends on the affinity of the receptor to the antibody that was used to detect it. The assay proved to be highly reproducible, both in Western blot analysis and quantitative immunofluorescence, because EGFR AQUA scores and concentrations were close and correlated well when measured on different days and with different experiments. A similar method that combines the AQUA assay with an enzyme ligand immunoassay (enzyme-linked immunosorbent assay) to provide absolute concentrations of HER-2 and β-catenin has been reported.22,36
Use of cell lines to construct the standard curve has certain limitations. Specifically, variables such as cell density, passage number, serum brand and batch, and other variables result in changes in concentration that are not reproducible. These variables decrease the quality of the correlation between EGFR concentrations calculated using Western blot analysis and the corresponding AQUA scores. Finally, the dynamic range of EGFR expression in the cell lines does not necessarily coincide with what is observed in patients. Other limitations of the quantitative immunofluorescence approach are that the current antibodies can detect only the DEL746-750 deletion in exon 19 and the L858R mutation in exon 21. In comparison, genotyping can reveal additional less common mutations in the tyrosine kinase domain of EGFR. The sensitivity of the mutation-specific antibodies, although high, is not 100%, and after the initial reports, more recent studies have calculated the sensitivity to be as low as 80%.37,38 In their cohort, Kitamura et al39 were able to detect only 47% of mutations using the mutation-specific antibodies, probably because of the high frequency of nonclassic deletions in that particular cohort.
Mutations in the tyrosine kinase domain of EGFR have been traditionally detected using direct DNA sequencing40 or other methods.24,41,42 Use of antibodies that are specific to mutated protein present a potentially less expensive and more routine method for mutation detection. However, new technologies and methods may soon neutralize any cost difference between DNA and protein analyses. Protein analysis also has the advantages of revealing spatial and context information that enables assessment of the distribution of the mutations in different areas of the tumors and of demonstrating that mutations are present in sparsely represented epithelial cells. This approach also addresses the problem of low tumor content often encountered in sequencing with potential for false-negative results. With appropriate controls and standardization, this method requires only minimal amounts of tissue, which can be a clinical challenge when only tiny amounts of tissue are obtained using minimally invasive techniques.
In summary, quantitative immunofluorescence can aid in genotyping for determination of EGFR mutational status and enable subsequent decisions about the use of tyrosine kinase inhibitors as first-line therapy in patients with NSCLC. The method can be highly reproducible, inexpensive, and specific, and can be used in any pathology laboratory that performs routine IHC. Our assay reveals no prognostic potential for EGFR in NSCLC, and enables setting a threshold for EGFR that can act as a screening test before genotyping. The low mutation rate in our two cohorts raises the possibility that the frequency of the mutations might be lower than the 8% to 15% often quoted in the general population of white patients with NSCLC.
Footnotes
Supported by a grant from the Oncology Unit, Sotiria General Hospital, Athens, Greece (K.S.), and by grant 2009-0097 from the Connecticut Department of Public Health (D.L.R.).
D.L.R. is a co-founder, consultant, and stockholder of HistoRx. The company was founded by Yale University and is the exclusive Yale licensee of the AQUA technology developed in D.L.R.'s laboratory.
References
- 1.Jemal A., Siegel R., Xu J., Ward E. Cancer Statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Shepherd F.A., Rodrigues Pereira J., Ciuleanu T., Tan E.H., Hirsh V., Thongprasert S., Campos D., Maoleekoonpiroj S., Smylie M., Martins R., van Kooten M., Dediu M., Findlay B., Tu D., Johnston D., Bezjak A., Clark G., Santabarbara P., Seymour L. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 3.Lynch T.J., Bell D.W., Sordella R., Gurubhagavatula S., Okimoto R.A., Brannigan B.W., Harris P.L., Haserlat S.M., Supko J.G., Haluska F.G., Louis D.N., Christiani D.C., Settleman J., Haber D.A. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 4.Pao W., Miller V., Zakowski M., Doherty J., Politi K., Sarkaria I., Singh B., Heelan R., Rusch V., Fulton L., Mardis E., Kupfer D., Wilson R., Kris M., Varmus H. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janne P.A., Engelman J.A., Johnson B.E. Epidermal growth factor receptor mutations in non-small-cell lung cancer: implications for treatment and tumor biology. J Clin Oncol. 2005;23:3227–3234. doi: 10.1200/JCO.2005.09.985. [DOI] [PubMed] [Google Scholar]
- 6.Kosaka T., Yatabe Y., Endoh H., Kuwano H., Takahashi T., Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 7.Riely G.J., Politi K.A., Miller V.A., Pao W. Update on epidermal growth factor receptor mutations in non–small cell lung cancer. Clin Cancer Res. 2006;12:7232–7241. doi: 10.1158/1078-0432.CCR-06-0658. [DOI] [PubMed] [Google Scholar]
- 8.Mok T.S., Wu Y.L., Thongprasert S., Yang C.H., Chu D.T., Saijo N., Sunpaweravong P., Han B., Margono B., Ichinose Y., Nishiwaki Y., Ohe Y., Yang J.J., Chewaskulyong B., Jiang H., Duffield E.L., Watkins C.L., Armour A.A., Fukuoka M. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 9.Yu J., Kane S., Wu J., Benedettini E., Li D., Reeves C., Innocenti G., Wetzel R., Crosby K., Becker A., Ferrante M., Cheung W.C., Hong X., Chirieac L.R., Sholl L.M., Haack H., Smith B.L., Polakiewicz R.D., Tan Y., Gu T.L., Loda M., Zhou X., Comb M.J. Mutation-specific antibodies for the detection of EGFR mutations in non-small-cell lung cancer. Clinical Cancer Res. 2009;15:3023–3028. doi: 10.1158/1078-0432.CCR-08-2739. [DOI] [PubMed] [Google Scholar]
- 10.Brevet M., Arcila M., Ladanyi M. Assessment of EGFR mutation status in lung adenocarcinoma by immunohistochemistry using antibodies specific to the two major forms of mutant EGFR. J Mol Diagn. 2010;12:169–176. doi: 10.2353/jmoldx.2010.090140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veale D., Kerr N., Gibson G.J., Kelly P.J., Harris A.L. The relationship of quantitative epidermal growth factor receptor expression in non–small cell lung cancer to long-term survival. Br J Cancer. 1993;68:162–165. doi: 10.1038/bjc.1993.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anagnostou V.K., Welsh A.W., Giltnane J.M., Siddiqui S., Liceaga C., Gustavson M., Syrigos K.N., Reiter J.L., Rimm D.L. Analytic variability in immunohistochemistry biomarker studies. Cancer Epidemiol Biomarkers Prev. 2010;19:982–991. doi: 10.1158/1055-9965.EPI-10-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allred D.C., Harvey J.M., Berardo M., Clark G.M. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998;11:155–168. [PubMed] [Google Scholar]
- 14.Rhodes A., Jasani B., Balaton A.J., Barnes D.M., Anderson E., Bobrow L.G., Miller K.D. Study of interlaboratory reliability and reproducibility of estrogen and progesterone receptor assays in Europe: documentation of poor reliability and identification of insufficient microwave antigen retrieval time as a major contributory element of unreliable assays. Am J Clin Pathol. 2001;115:44–58. doi: 10.1309/H905-HYC1-6UQQ-981P. [DOI] [PubMed] [Google Scholar]
- 15.Rhodes A., Jasani B., Balaton A.J., Miller K.D. Immunohistochemical demonstration of oestrogen and progesterone receptors: correlation of standards achieved on in house tumours with that achieved on external quality assessment material in over 150 laboratories from 26 countries. J Clin Pathol. 2000;53:292–301. doi: 10.1136/jcp.53.4.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selvaggi G., Novello S., Torri V., Leonardo E., De Giuli P., Borasio P., Mossetti C., Ardissone F., Lausi P., Scagliotti G.V. Epidermal growth factor receptor overexpression correlates with a poor prognosis in completely resected non-small-cell lung cancer. Ann Oncol. 2004;15:28–32. doi: 10.1093/annonc/mdh011. [DOI] [PubMed] [Google Scholar]
- 17.Meert A.P., Martin B., Delmotte P., Berghmans T., Lafitte J.J., Mascaux C., Paesmans M., Steels E., Verdebout J.M., Sculier J.P. The role of EGF-R expression on patient survival in lung cancer: a systematic review with meta-analysis. Eur Respir J. 2002;20:975–981. doi: 10.1183/09031936.02.00296502. [DOI] [PubMed] [Google Scholar]
- 18.Pastorino U., Andreola S., Tagliabue E., Pezzella F., Incarbone M., Sozzi G., Buyse M., Menard S., Pierotti M., Rilke F. Immunocytochemical markers in stage I lung cancer: relevance to prognosis. J Clin Oncol. 1997;15:2858–2865. doi: 10.1200/JCO.1997.15.8.2858. [DOI] [PubMed] [Google Scholar]
- 19.Ceppi P., Volante M., Novello S., Rapa I., Danenberg K.D., Danenberg P.V., Cambieri A., Selvaggi G., Saviozzi S., Calogero R., Papotti M., Scagliotti G.V. ERCC1 and RRM1 gene expressions but not EGFR are predictive of shorter survival in advanced non-small-cell lung cancer treated with cisplatin and gemcitabine. Ann Oncol. 2006;17:1818–1825. doi: 10.1093/annonc/mdl300. [DOI] [PubMed] [Google Scholar]
- 20.Pfeiffer P., Clausen P.P., Andersen K., Rose C. Lack of prognostic significance of epidermal growth factor receptor and the oncoprotein p185HER-2 in patients with systemically untreated non-small-cell lung cancer: an immunohistochemical study on cryosections. Br J Cancer. 1996;74:86–91. doi: 10.1038/bjc.1996.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rusch V., Klimstra D., Venkatraman E., Pisters P.W., Langenfeld J., Dmitrovsky E. Overexpression of the epidermal growth factor receptor and its ligand transforming growth factor alpha is frequent in resectable non–small cell lung cancer but does not predict tumor progression. Clin Cancer Res. 1997;3:515–522. [PubMed] [Google Scholar]
- 22.Dolled-Filhart M., McCabe A., Giltnane J., Cregger M., Camp R.L., Rimm D.L. Quantitative in situ analysis of beta-catenin expression in breast cancer shows decreased expression is associated with poor outcome. Cancer Res. 2006;66:5487–5494. doi: 10.1158/0008-5472.CAN-06-0100. [DOI] [PubMed] [Google Scholar]
- 23.Camp R.L., Chung G.G., Rimm D.L. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 24.Fassina A., Gazziero A., Zardo D., Corradin M., Aldighieri E., Rossi G.P. Detection of EGFR and KRAS mutations on trans-thoracic needle aspiration of lung nodules by high resolution melting analysis. J Clin Pathol. 2009;62:1096–1102. doi: 10.1136/jcp.2009.067587. [DOI] [PubMed] [Google Scholar]
- 25.Schmid K., Oehl N., Wrba F., Pirker R., Pirker C., Filipits M. EGFR/KRAS/BRAF mutations in primary lung adenocarcinomas and corresponding locoregional lymph node metastases. Clin Cancer Res. 2009;15:4554–4560. doi: 10.1158/1078-0432.CCR-09-0089. [DOI] [PubMed] [Google Scholar]
- 26.Oshita F., Matsukuma S., Yoshihara M., Sakuma Y., Ohgane N., Kameda Y., Saito H., Yamada K., Tsuchiya E., Miyagi Y. Novel heteroduplex method using small cytology specimens with a remarkably high success rate for analysing EGFR gene mutations with a significant correlation to gefitinib efficacy in non-small-cell lung cancer. Br J Cancer. 2006;95:1070–1075. doi: 10.1038/sj.bjc.6603396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eberhard D.A., Johnson B.E., Amler L.C., Goddard A.D., Heldens S.L., Herbst R.S., Ince W.L., Janne P.A., Januario T., Johnson D.H., Klein P., Miller V.A., Ostland M.A., Ramies D.A., Sebisanovic D., Stinson J.A., Zhang Y.R., Seshagiri S., Hillan K.J. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 28.Rosell R., Moran T., Queralt C., Porta R., Cardenal F., Camps C., Majem M., Lopez-Vivanco G., Isla D., Provencio M., Insa A., Massuti B., Gonzalez-Larriba J.L., Paz-Ares L., Bover I., Garcia-Campelo R., Moreno M.A., Catot S., Rolfo C., Reguart N., Palmero R., Sanchez J.M., Bastus R., Mayo C., Bertran-Alamillo J., Molina M.A., Sanchez J.J., Taron M. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 29.Hirsch F.R., Varella-Garcia M., Bunn PA J.R., Franklin W.A., Dziadziuszko R., Thatcher N., Chang A., Parikh P., Pereira J.R., Ciuleanu T., von Pawel J., Watkins C., Flannery A., Ellison G., Donald E., Knight L., Parums D., Botwood N., Holloway B. Molecular predictors of outcome with gefitinib in a phase III placebo-controlled study in advanced non-small-cell lung cancer. J Clin Oncol. 2006;24:5034–5042. doi: 10.1200/JCO.2006.06.3958. [DOI] [PubMed] [Google Scholar]
- 30.Tsao A.S., Tang X.M., Sabloff B., Xiao L., Shigematsu H., Roth J., Spitz M., Hong W.K., Gazdar A., Wistuba I. Clinicopathologic characteristics of the EGFR gene mutation in non–small cell lung cancer. J Thorac Oncol. 2006;1:231–239. doi: 10.1016/s1556-0864(15)31573-2. [DOI] [PubMed] [Google Scholar]
- 31.Shigematsu H., Lin L., Takahashi T., Nomura M., Suzuki M., Wistuba I.I., Fong K.M., Lee H., Toyooka S., Shimizu N., Fujisawa T., Feng Z., Roth J.A., Herz J., Minna J.D., Gazdar A.F. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 32.Kawaguchi T., Matsumura A., Fukai S., Tamura A., Saito R., Zell J.A., Maruyama Y., Ziogas A., Kawahara M., Ignatius Ou S.H. Japanese ethnicity compared with Caucasian ethnicity and never-smoking status are independent favorable prognostic factors for overall survival in non–small cell lung cancer: a collaborative epidemiologic study of the National Hospital Organization Study Group for Lung Cancer (NHSGLC) in Japan and a Southern California Regional Cancer Registry databases. J Thorac Oncol. 2010;5:1001–1010. doi: 10.1097/JTO.0b013e3181e2f607. [DOI] [PubMed] [Google Scholar]
- 33.van Puijenbroek R., Bosquee L., Meert A.P., Schallier D., Goeminne J.C., Tits G., Collard P., Nackaerts K., Canon J.L., Duplaquet F., Galdermans D., Germonpre P., Azerad M.A., Vandenhoven G., De Greve J., Vansteenkiste J. Gefitinib monotherapy in advanced nonsmall cell lung cancer: a large Western community implementation study. Eur Respir J. 2007;29:128–133. doi: 10.1183/09031936.00050706. [DOI] [PubMed] [Google Scholar]
- 34.Pinter F., Papay J., Almasi A., Sapi Z., Szabo E., Kanya M., Tamasi A., Jori B., Varkondi E., Moldvay J., Szondy K., Keri G., Dominici M., Conte P., Eckhardt S., Kopper L., Schwab R., Petak I. Epidermal growth factor receptor (EGFR) high gene copy number and activating mutations in lung adenocarcinomas are not consistently accompanied by positivity for EGFR protein by standard immunohistochemistry. J Mol Diagn. 2008;10:160–168. doi: 10.2353/jmoldx.2008.070125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang Z., Zhang J., Zeng X., Gao J., Wu S., Liu T. Relationship between EGFR expression, copy number and mutation in lung adenocarcinomas. BMC Cancer. 2010;10:376. doi: 10.1186/1471-2407-10-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCabe A., Dolled-Filhart M., Camp R.L., Rimm D.L. Automated quantitative analysis (AQUA) of in situ protein expression, antibody concentration, and prognosis. J Natl Cancer Inst. 2005;97:1808–1815. doi: 10.1093/jnci/dji427. [DOI] [PubMed] [Google Scholar]
- 37.Kato Y., Peled N., Wynes M.W., Yoshida K., Pardo M., Mascaux C., Ohira T., Tsuboi M., Matsubayashi J., Nagao T., Ikeda N., Hirsch F.R. Novel epidermal growth factor receptor mutation-specific antibodies for non–small cell lung cancer: immunohistochemistry as a possible screening method for epidermal growth factor receptor mutations. J Thorac Oncol. 2010;5:1551–1558. doi: 10.1097/JTO.0b013e3181e9da60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawahara A., Yamamoto C., Nakashima K., Azuma K., Hattori S., Kashihara M., Aizawa H., Basaki Y., Kuwano M., Kage M., Mitsudomi T., Ono M. Molecular diagnosis of activating EGFR mutations in non–small cell lung cancer using mutation-specific antibodies for immunohistochemical analysis. Clin Cancer Res. 2010;16:3163–3170. doi: 10.1158/1078-0432.CCR-09-3239. [DOI] [PubMed] [Google Scholar]
- 39.Kitamura A., Hosoda W., Sasaki E., Mitsudomi T., Yatabe Y. Immunohistochemical detection of EGFR mutation using mutation-specific antibodies in lung cancer. Clin Cancer Res. 2010;16:3349–3355. doi: 10.1158/1078-0432.CCR-10-0129. [DOI] [PubMed] [Google Scholar]
- 40.John T., Liu G., Tsao M.S. Overview of molecular testing in non-small-cell lung cancer: mutational analysis, gene copy number, protein expression and other biomarkers of EGFR for the prediction of response to tyrosine kinase inhibitors. Oncogene. 2009;28(Suppl 1):S14–S23. doi: 10.1038/onc.2009.197. [DOI] [PubMed] [Google Scholar]
- 41.Willmore-Payne C., Holden J.A., Wittwer C.T., Layfield L.J. The use of EGFR exon 19 and 21 unlabeled DNA probes to screen for activating mutations in non–small cell lung cancer. J Biomol Tech. 2008;19:217–224. [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen V., Agulnik J.S., Ang C., Kasymjanova G., Batist G., Small D., Brandao G., Chong G., Miller W.H., Jr Epidermal growth factor receptor mutations detected by denaturing high-performance liquid chromatography in nonsmall cell lung cancer: impact on response to therapy with epidermal growth factor receptor-tyrosine kinase inhibitors. Cancer. 2010;116:4309–4317. doi: 10.1002/cncr.25214. [DOI] [PubMed] [Google Scholar]


