Abstract
Transforming growth factor (TGF)-β is one of the main fibrogenic cytokines that drives the pathophysiology of progressive renal scarring. MicroRNAs (miRNAs) are endogenous non-coding RNAs that post-transcriptionally regulate gene expression. We examined the role of TGF-β–induced expression of miR-21, miRNAs in cell culture models and miRNA expression in relevant models of renal disease. In vitro, TGF-β changed expression of miR-21, miR-214, and miR-145 in rat mesangial cells (CRL-2753) and miR-214, miR-21, miR-30c, miR-200b, and miR-200c during induction of epithelial-mesenchymal transition in rat tubular epithelial cells (NRK52E). miR-214 expression was robustly modulated in both cell types, whereas in tubular epithelial cells miR-21 was increased and miR-200b and miR-200c were decreased by 58% and 48%, respectively, in response to TGF-β. TGF-β receptor-1 was found to be a target of miR-200b/c and was down-regulated after overexpression of miR-200c. To assess the differential expression of these miRNAs in vivo, we used the anti-Thy1.1 mesangial glomerulonephritis model and the unilateral ureteral obstruction model in which TGF-β plays a role and also a genetic model of hypertension, the stroke-prone spontaneously hypertensive rat with and without salt loading. The expressions of miR-214 and miR-21 were significantly increased in all in vivo models, showing a possible miRNA signature of renal damage despite differing causes.
Transforming growth factor (TGF)-β is a critically important mediator of pathophysiological events in renal disease.1,2 Many types of glomerular diseases, including IgA nephropathy and lupus nephritis are characterized by mesangial cell proliferation,3 mesangial matrix expansion, and alterations in extracellular turnover and composition. In the chronic anti-Thy1.1 model of glomerulonephritis, proliferation of mesangial cells is followed by the production of extracellular matrix, leading to chronic glomerulosclerosis and eventually interstitial fibrosis. Tubulo-interstitial fibrosis is the common end point of most progressive kidney diseases and results in loss of renal function. The model of unilateral ureteral obstruction (UUO) has been widely used as an animal model of tubulo-interstitial disease that is characterized by a mononuclear cell infiltration followed by fibroblast proliferation, increased extracellular matrix deposition, and tubule atrophy. These common processes of fibrosis and sclerosis are known to be driven primarily by TGF-β along with a host of other cytokines and growth factors. It is probable therefore that these pathologic processes involve interrelated and complex molecular pathways in which microRNAs (miRNAs) may play an important regulatory role.
In humans, decreased renal function is often a complication of essential hypertension, and hypertension is one of the most common causes of end-stage renal disease in the United States.4 A rat genetic model of essential hypertension the stroke-prone, spontaneously hypertensive rat (SHRSP) also develops renal damage with age.5,6 The damage observed is typically vascular smooth muscle hyperplasia and tubule atrophy and/or dilation.5 Furthermore, in humans salt sensitivity is common in persons with essential hypertension, and, when SHRSPs are challenged with salt, this increases their systolic blood pressure [not observed in Wistar-Kyoto (WKY) rats] and induces further renal damage compared with WKY rats.7
miRNAs are endogenous non-coding RNAs that are ∼22 nucleotides in length and can have structural, enzymatic, and regulatory functions.8 miRNAs act within the RNA-induced silencing complex9 and can down-regulate gene expression by binding to the 3′-untranslated region (UTR) of the mRNA which results in either productive translational repression or target degradation. miRNAs are fundamental in development and are expressed in a tissue-specific manner.10–12 However, they have now been found to play a role in the pathophysiology of many diverse diseases, including cancer,13,14 vascular proliferative disease,15 and cardiac hypertrophy.16 It is clear that many genes contain miRNA binding sequences within their 3′-UTR, and a single miRNA can “hit” multiple genes and influence many pathways.8 With respect to the kidneys, a number of studies of miRNA expression have been conducted.17–22 They have been shown to be fundamentally important in the kidney by several studies.19–21 For example, targeted knockout of DICER, a protein important in miRNA biogenesis, selectively in podocytes leads to severe proteinuria.2,19–22 These animals had marked abnormalities in the glomerulus, including foot process effacement, mesangial expansion, and glomerulosclerosis, which ultimately lead to animal death. From those studies it appears that several miRNAs are important for normal kidney homeostasis; miR-30a was found to be lost in podocytes of DICER knockout mice compared with controls20 and in mutant glomeruli where mature miRNAs were knocked out targets of the miR-30 family were enriched in the up-regulated genes.19
Several miRNAs have also been shown to have specific localization within the kidney. With the use of locked nucleic acid (LNA)—immunostaining in the normal kidney it has been shown that miR-23b, miR-24, and miR-26a show a pan glomerular localization20,21; miR-145 is found in mesangial cells20 and vascular smooth muscle cells20; miR-10a and miR-30c are reported to be tubular specific20,21; and miR-126 is detected in the glomerular and peritubular endothelial cells.20 Furthermore, an integrated study of miRNAs and gene targets found several miRNAs differentially regulated, miR-21, miR-31, miR-128, miR-147, and miR-217, in polycystic kidney disease17; however, the role of these differentially expressed miRNAs in polycystic kidney disease has yet to be shown in vivo.
TGF-β has been shown to stimulate a set of miRNAs.18,22,23 miR-192 and miR-377 are up-regulated in mouse18,22 and human18,22 mesangial cells in vitro in response to TGF-β and also in vivo in mouse diabetic models.18,22 Analysis of the targets has shown that miR-192 targets SIP118 and miR-377 targets p21-activated kinase and superoxide dismutase,22 which results in an increase in collagen and fibronectin, respectively. In contrast TGF-β–stimulated epithelial-mesenchymal transition (EMT) results in loss of the miR-200 family,24–26 and an increase in expression targets the E-cadherin repressors ZEB1 and ZEB2.24–26
Because miRNAs have been shown to be essential in kidney homeostasis, we sought to assign a specific role for TGF-β in modulation of miRNAs in cell culture and in vivo with respect to renal pathology. We therefore focused our analysis on a subset of miRNAs with potential relevance in this setting (miR-21, miR-214, miR-192, miR-26b, miR-145, miR-24, miR-30c, and miR-200b/c). We used two in vitro models to assess TGF-β stimulation of rat mesangial cells and induction of EMT in rat kidney tubular epithelial cells. We then investigated the expression of these miRNAs in the chronic anti-Thy1.1 model of glomerulonephritis, the UUO model of interstitial fibrosis, and a rat genetic model of hypertension, the SHRSP.
Materials and Methods
In Vitro Analysis of miRNA
Rat mesangial cells (CRL-2753) and rat tubular epithelial cells (NRK52E) were cultured according to American Type Culture Collection (ATCC; Manassas, VA) instructions. Cells were serum starved for 48 hours (0.2% fetal calf serum) before stimulation with 10 ng/mL TGF-β(R&D Systems, Minneapolis, MN). Total RNA was extracted from the cells at 24, 48, 72, and 96 hours after stimulation with the use of miRNeasy kit (Qiagen, Inc., Valencia, CA) following the manufacturer's instructions, treated with the Turbo DNase (Ambion, Austin TX), to eliminate genomic DNA contamination, and quantified with the use of the NanoDrop ND-1000 Spectrophotometer (Nano-Drop Technologies, Wilmington, DE).
In Vitro Analysis of EMT
NRK52E cells were plated in 6-well plates at 8 × 104 cells per well. Cells were serum starved for 48 hours (0.2% fetal calf serum) before stimulation with 10 ng/mL TGF-β (R&D Systems). Total RNA and protein were extracted at 24, 48, 72, and 96 hours after stimulation. RNA was extracted and treated as before. Immunocytochemistry was performed with α-smooth muscle actin (α-SMA; 1:500; Abcam, Cambridge, MA) and goat anti-mouse Alexa Fluro 546 secondary antibody (Invitrogen, Paisley, UK) with DAPI Prolong Gold mounting media (Invitrogen). Protein was quantified, and 50 μg was fractionated on 7.5% (for E-cadherin) and 10% (for α-SMA) SDS polyacrylamide gels. Proteins were transferred onto nitrocellulose membranes and probed for E-cadherin (1:2000; BD Biosciences, San Jose, CA) and α-SMA (1:500; Abcam) antibodies. Membranes were exposed with the use of enhanced chemiluminescence ECL (GE Healthcare, Chalfont St Giles, Buckingham, UK) and developed using the X-OMAT 100 (Kodak, Rochester, NY).
TGF-β Inhibition Experiments
TGF-β inhibition experiments were performed with commercially available TGF-β receptor-1 (TGFβR1; Alk5) inhibitors A-83-01 and AB525339 (Tocris Bioscience, Bristol, UK) at a concentration of 1 μmol/L. NRK52E cells were plated at 8 × 104 cells per well. Cells were serum starved for 48 hours (0.2% fetal calf serum) before adding either inhibitor or dimethyl sulfoxide (DMSO; vehicle control) for 1 hour before stimulation with 10 ng/mL TGF-β (R&D Systems). RNA was extracted and treated as before.
Manipulation of miR-200c Levels
NRK52E cells (ATCC) were plated in 6-well plates at 1 × 105 cells per well. Cells were transfected with cytomegalovirus–pri-miR-200c plasmid with the use of 2 μg of DNA/3 μL of lipofectamine per well. Lipofectamine:DNA complex was left on the cells for 5 hours, then supplemented with 20% serum containing media. Twenty-four hours later the cells were washed with PBS, and cells were replenished with fresh serum-free media. After 48 hours serum-starving cells were stimulated with TGF-β (10 ng/mL) and left for 72 hours. Total RNA was extracted as described above, and miR-200c levels were quantified.
Manipulation of miR-21 and miR-214 Levels
Lentiviral vectors were produced by triple transient transfection of HEK293T cells with a packaging plasmid (pCMVΔ8.74), a plasmid encoding the envelope of vesicular stomatitis virus (kind gift from Adrian J. Thrasher, London, UK), and the expression plasmid, using polyethylenimine (Sigma-Aldrich, St Louis, MO) as previously described.27 Lentiviral titers were ascertained by TaqMan real-time quantitative PCR (qPCR) as previously described.28 To overexpress the miR-21 and miR-214 the precursor (pre)–miRNA sequences were obtained from miRBase (http://microrna.sanger.ac.uk), codon-optimized, and manufactured by GeneArt (Regensburg, Germany) and subsequently cloned into the HinDIII-EcoRV sites of pcDNA3.1 (Invitrogen). As the XhoI and BamH1 restriction sites were maintained, the pre-miR sequence was cloned directly into the pLNT/SFFV-MCS plasmid (kind gift from Adrian J. Thrasher, London, UK) to obtain the construct pLNT/SFFV-pre-miR.
Reverse Transcription of miRNA
After quantification of DNase-treated RNA, miRNA reverse transcription was performed with stem-loop reverse transcription primers according to the TaqMan MicroRNA Assay protocol (Applied Biosystems, Foster City, CA). Each reaction contained 5 ng of extracted total RNA and 50 nmol/L stem-looped reverse transcription primer, specific for each miRNA. cDNA synthesis was performed on 1 μg of total RNA with the use of reverse transcription reagents (Applied Biosystems) following the manufacturer's protocol.
miRNA and Gene Expression
Real-time qPCR was performed. The amplification step was performed with specific Taqman miRNA probe or for gene expression inventoried gene expression-specific primers (Applied Biosystems) on the Applied Biosystems 7900 HT real-time PCR system following the manufacturer's instructions. U87 was used as an endogenous control for the miRNA expression, and rat glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous control for gene expression. Results are shown as RQ ± RQ max, calculated with SD.
Northern Blotting
Total RNA was extracted from cells or rat tissues with the use of miRNeasy Mini kit (Qiagen, Dorking, Surrey, UK). Three to 10 μg of total RNA was resolved in a 15% tris-boric acid-EDTA and urea denaturing gel (Invitrogen), transferred onto Hyband-NX membrane (GE Healthcare). After 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC) cross-linking, membranes were hybridized with 5′-DIG–labeled LNA Mercury probes (Exiqon, Vedbaek, Denmark) at 40°C (miR-21 probe) or 55°C (miR-214 probe and U6) overnight. After post-hybridization washes, membranes were incubated with anti-DIG-AP antibody (1:5000; Roche, Burgess Hill, UK) in 1% blocking reagents (Roche), and signals were detected with CDP-Star (Sigma-Aldrich) according to the manufacturer's instructions.
Prediction of Potential miRNA Targets
A list of targets for miR-21 and miR-214 was obtained through searching 3′-UTR sequences of rat mRNAs for the seeds of miRNAs. Rat mRNAs were downloaded from the USCS Genome Browser (http://genome.ucsc.edu). For each 3′-UTR, a number of seeds, its flanking nucleotides, and position were recorded.29,30 Each list of potential targets was screened with specific criteria, including high number of seeds, small distance between seeds, intersection between the two miRNAs, and physiological function of the targets.29,30
Animal Models
All protocols and surgical procedures were approved by the local animal care committee. Animal experiments were in accordance with the Animals Scientific Procedures Act UK 1986.
Induction of Mesangial Proliferative Glomerulonephritis
Male WKY rats (Harlan, Wyton, UK) (7 to 12 weeks old) were used throughout. All protocols and surgical procedures were approved by the local animal care committee. Animals were age matched at sacrifice to minimize age-related differences in miRNA expression. For the induction of mesangial proliferative glomerulonephritis rats were intravenously infused with 2 mg/kg ER4. For the three-injection protocol the ER4 was infused three times, 1 week apart. After sacrifice, kidneys were removed, and portions were fixed in 10% formalin or snap frozen and stored at −80°C.
Histologic scoring was adapted from Tomita et al31 and was performed blind with 50 glomeruli and surrounding tubules scored per section. Glomerular damage was scored from 0 to 4, with 0 being 0% to 4% glomerular lesion, mesangial matrix expansion; 1 being 5% to 24%; 2 being 25% to 49%; 3 being 50% to 74%; and 4 being >75%. Interstitial lesions were counted as tubular dilation, tubular trophy, basophilia, interstitial inflammation, and fibrosis and were scored from 0 to 4, with 0 being no lesion, 1 being <5%, 2 being 5% to 24%, 3 being 25% to 49%, and 4 being ≥50%.
Unilateral Ureteral Obstruction
Male Wistar rats weighing 200 g were used. Each rat was initially placed in an induction chamber and exposed to 4% isoflurane and oxygen at a flow rate of 4 L/minute. Once anesthetized, the rat was transferred to a heated mat with continued administration of isoflurane at 2% to 2.5%. A midline incision was made with aseptic technique, and the left kidney was exposed. The left ureter was isolated by blunt dissection, and two 6-0 silk ligatures were tied around it. The incision was re-sutured, and buprenorphine was administered subcutaneously at a dose of 30 μg/kg. As a control, a sham operation was performed, including a laparotomy and handling of the ureter, only it was not ligated. All rats were sacrificed on postoperative day 16. After sacrifice, kidneys were removed, and portions were fixed in 10% formalin or snap frozen and stored at −80°C.
Genetic Rat Model of Hypertension
Inbred colonies of SHRSP and WKY rats have been developed and maintained at the University of Glasgow since 1991, as described previously.32 For salt-loading experiments, at 18 weeks of age, rats were given a salt challenge (1% NaCl in drinking water) for 3 weeks. All animals were sacrificed at 21 weeks. After sacrifice, kidneys were removed, snap frozen, and stored at −80°C.
Statistical Analysis
All in vitro experiments were performed in triplicate on three separate occasions. All in vivo work was conducted in groups of four to six. A standard Student's t-test was used to designate significant differences between control and other groups.
Results
TGF-β Induces Changes in miRNA Profiles in Renal Cells in Vitro
We first investigated the expression of miRNAs modulated by TGF-β in the rat mesangial cell line CRL-2573 and tubular epithelial cell line NRK52E. Both were stimulated with rTGF-β (10 ng/mL) and assessed for individual miRNA levels compared with no stimulation. The response of mesangial cells to TGF-β stimulation was to significantly proliferate to levels similar to that seen after incubation in growth media (see Supplemental Figure S1A at http://ajp.amjpathol.org). We also found significant increase in the gene expression of TGF-β and TGFβ-R1 (see Supplemental Figure S1, B and C, at http://ajp.amjpathol.org), showing that cellular levels of TGF-β were increased after stimulation. Notably, in the mesangial cell line miR-214 was significantly up-regulated 2 days after stimulation (P < 0.01) and remained elevated for the time course with the peak expression of ∼16-fold increase at 4 days after stimulation when evaluated in a detailed time course (Figure 1). miR-21 expression showed a sustained significant increase ∼twofold from day 1 to day 3 before dropping to levels below that expressed in time-matched control cells (Figure 1). No increase in miR-192 was observed (Figure 1), which is in contrast to previous reports.18,22 This suggests that miR-192 may be differentially regulated or expressed in rat mesangial cells as assessed by real-time qPCR. Interestingly, miR-145 was significantly elevated 24 hours after stimulation before returning to unstimulated levels (Figure 1).
Figure 1.
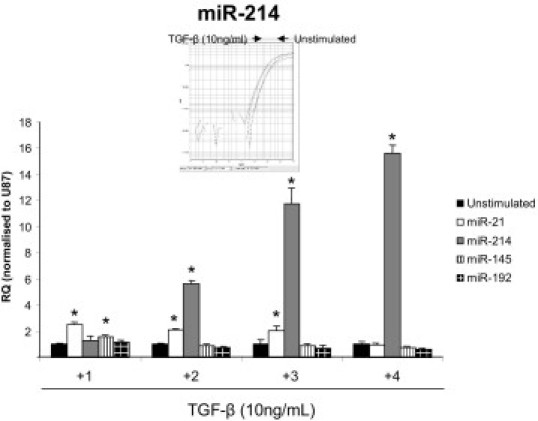
Effect of TGF-β on microRNA expression in cultured rat mesangial cells. Rat mesangial cells (CRL-2573; RMCs) were stimulated with 10 ng/mL rTGF-β (R&D Systems) over a 4-day time course. RNA was harvested from the cells by miReasy kit (Qiagen), and miRNA expression was measured by specific miRNA Taqman probes (Applied Biosystems) and normalized to U87. RQ ± RQ max. *P < 0.05 versus unstimulated time-matched controls. Inset is a representative Taqman amplification curve from one unstimulated versus a TGF-β sample; N = 3.
In tubular epithelial cells, TGF-β stimulation resulted in EMT, characterized by loss of E-cadherin expression, gain of α-SMA expression and increased collagen production (Figure 2, A–D). In response to TGF-β stimulation the NRK52E (NRK) cells also increased the expression of both TGF-β and TGF-βR1 gene expression (see Supplemental Figure S2 at http://ajp.amjpathol.org). TGF-β stimulation of the NRK cells resulted in miR-200b and miR-200c being significantly down-regulated by day 4 of TGF-β stimulation, with miR-200b and miR-200c being reduced 58% and 48%, respectively, compared with time-matched unstimulated controls (Figure 3). miR-192 was not significantly different over the time course (Figure 3), but miR-30c had a significant decrease in expression 24 hours after TGF-β stimulation, which remained for the time course. However, and similar to data obtained in the mesangial cells, miR-214 and miR-21 were significantly up-regulated over the time course in the NRK cells (Figure 3). To further examine miR-21 and miR-214 expression we performed Northern blot analyses and found that over the 5-day time course stimulation of NRK cells with TGF-β resulted in increased expression of the mature miR-21 and miR-214 (see Supplemental Figure S3 at http://ajp.amjpathol.org). Interestingly, when the precursors of the miRs are examined, there is increased expression of pre-miR-21 after stimulation of cells with TGF-β (see Supplemental Figure S3 at http://ajp.amjpathol.org). In contrast no pre-miR-214 could be detected, but there was very high expression of primary (pri)–miR-214 expressed in NRK cells at all time points in both unstimulated and stimulated cells (see Supplemental Figure S3 at http://ajp.amjpathol.org).
Figure 2.
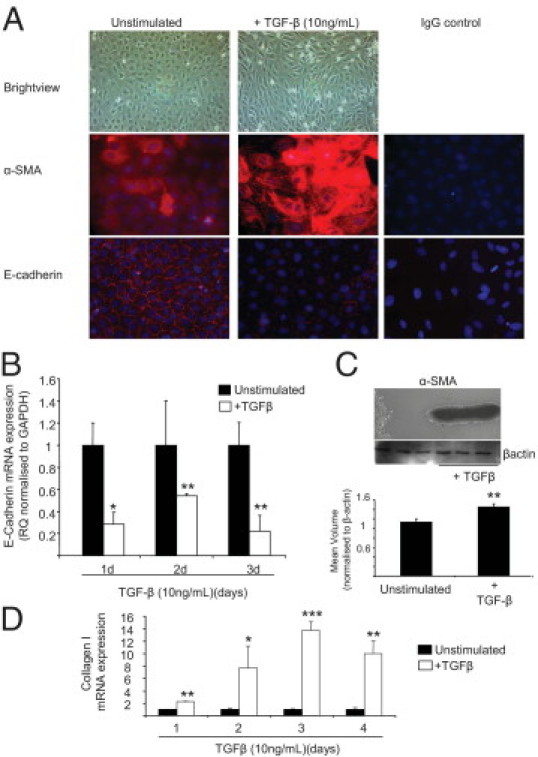
TGF-β–induced EMT in cultured rat tubular epithelial cells. Rat tubular epithelial cells (NRK52E; NRK) were stimulated with 10 ng/mL rTGF-β (R&D Systems) over a 4-day time course. A: Immunocytochemistry for α-SMA and E-cadherin expression in unstimulated and TGF-β–stimulated cells (5 days); original magnification, ×40. B: E-cadherin gene expression. Total RNA was extracted with the miRNeasy kit (Qiagen), and E-cadherin expression was measured by specific rat Taqman probes (Applied Biosystems) and normalized to rat GAPDH. RQ ± RQ max. *P < 0.05 and **P < 0.01 versus unstimulated time-matched controls. C: α-SMA protein expression in cells undergoing EMT. Western blot analysis of cell lysates probed for α-SMA and β-actin as a loading control. Images underwent densitometry. Mean volume ± SD. **P < 0.01. D: Gene expression of collagen 1 in unstimulated and TGF-β–stimulated cells. RQ ± RQ max. *P < 0.05, **P < 0.01, and ***P < 0.001 compared with unstimulated time-matched control cells.
Figure 3.
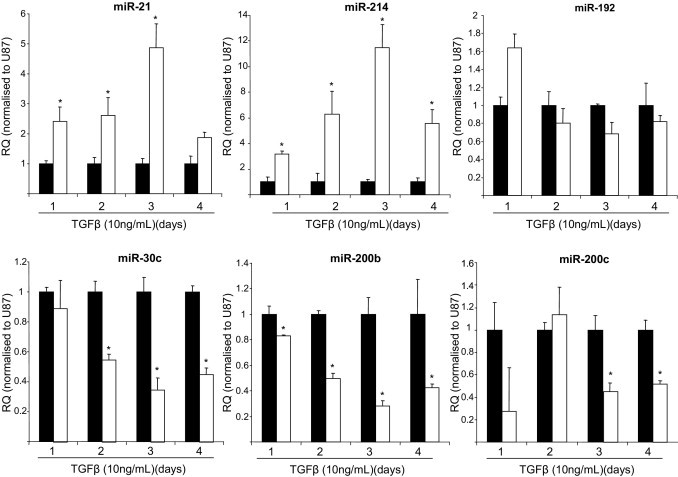
miRNA expression in tubular epithelial cells undergoing EMT. Rat tubular epithelial cells (NRK52E; NRK) were stimulated with 10 ng/mL rTGF-β (R&D Systems) over a 4-day time course. RNA was harvested from the cells by miReasy kit (Qiagen), and miRNA expression was measured by specific miRNA Taqman probes (Applied Biosystems) and normalized to U87. Each graph represents an individual miRNA. RQ ± RQ max. *P < 0.05 compared with unstimulated time-matched control cells.
To examine whether TGF-β directly affected the expression of miR-21 and miR-214, we sought to block TGF-β signaling with the use of TGF-βR1 signaling inhibitors (SB525339 and A-83-01). Both inhibitors block downstream activation of the TGF-β signaling cascade, but they allow binding of TGF-β to TGFRII.33 With the use of the rat tubular epithelial cells, we serum starved the cells for 48 hours then treated the cells with SB525339, A-83-01, or DMSO (vehicle control) for 1 hour before stimulating the cells with TGF-β (10 ng/mL) for 4 days. As expected cells treated with DMSO had a significant increase in miR-21 and miR-214 expressions in response to TGF-β stimulation, which was abolished by both inhibitors (see Supplemental Figure S4A at http://ajp.amjpathol.org). Similarly, the TGF-βRI inhibitors also blocked the loss of the miR-200 family and other TGF-β–modulated miRNAs in these cells (see Supplemental Figure S4B at http://ajp.amjpathol.org). The inhibitor A-83-01 has previously been shown to block EMT induced by TGF-β,34 and we found that both inhibitors prevented the loss of E-cadherin expression and increased in expression of α-SMA and collagen 1a, observed when the cells undergo EMT (see Supplemental Figure S4C at http://ajp.amjpathol.org).
To examine the effects of miR-21 and miR-214 alone, we constructed lentiviruses expressing pre-miR-21 (LNT-miR-21) and pre-miR-214 (LNT-miR-214) to allow overexpression of each miRNA and performed experiments in culture in the absence of TGF-β. Infection of rat tubular epithelial cells with LNT-miR-21 resulted in an increase of mature miR-21 expression by ∼3.5-fold compared with uninfected cells or LNT-egfp–infected cells (Figure 4A). Infection with LNT-miR-214 resulted in a significant increase of ∼400-fold in the levels of mature miR-214 (Figure 4B). The effect of overexpressing either miR-21 or miR-214 was to induce a change in EMT-associated markers E-cadherin, α-SMA, and collagen I, similar to that observed when these cells undergo EMT in response to TGF-β (Figure 4C). In summary, our in vitro data show that TGF-β modulates the expression of several miRNAs in both cell types (miR-214, miR-21, miR-30c, miR-200b, and miR 200c).
Figure 4.
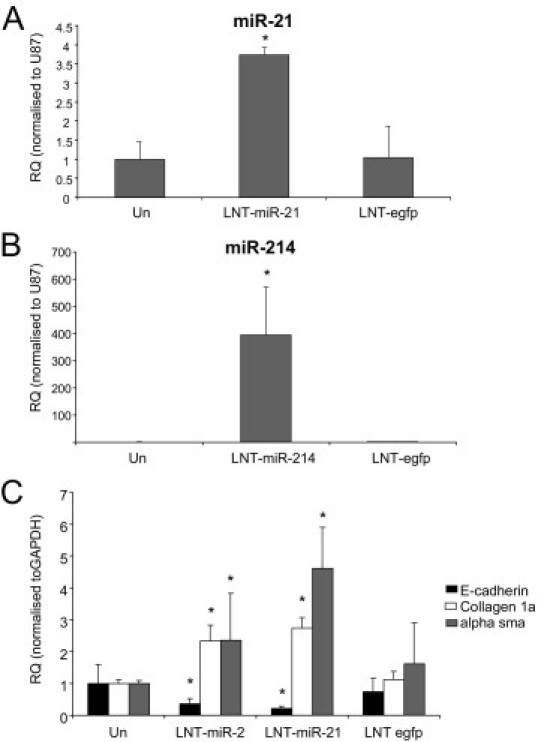
Effect of miR-21 and miR-214 overexpression on rat tubular epithelial cells. NRK52E cells were infected with Lenti-(LNT) miR-21, LNT-miR-214, or LNT-egfp at a multiplicity of infection of 5 for 24 hours. Cells were then serum starved for 2 days and then cultured for 4 days in serum-free media. RNA was harvested from the cells by miReasy kit (Qiagen), and miRNA expression was measured by specific miRNA Taqman probes (Applied Biosystems) and normalized to U87. Gene expression was measured with the use of gene-specific probes (rat) and normalized to GAPDH. A: Expression of miR-21 by LNT-miR-21. RQ ± RQ max. B: Expression of miR-214 by LNT-miR-214. RQ ± RQ max. C: Modulation of EMT-associated genes by LNT-miR-21, LNT-miR-214, and LNT-egfp. RQ ± RQ max. Un, unifected. *P < 0.05 versus Un.
TGFβR1 Is a Target for miR-200c and Is Modulated by Dynamic Changes in miR-200c Levels
We examined the potential targets of miR-200b/c (because these share the same seed sequence). The 3′-UTR sequences of rat mRNAs for the seeds of miRNAs (nucleotides 2 to 7) that we were interested in were downloaded from the USCS Genome Browser and were searched for potential targets.18,24 Interestingly, when a search of targets of miR-200b/c was performed, many other targets along with ZEB1 and ZEB224–26 were found (see Supplemental Table S1 at http://ajp.amjpathol.org). This list included TGFβR1, which has six seeds for miR-200b/c within its 3′-UTR, one of which is a 7mer-m8 (position 746) and another a 7mer-1A (position 3755) (see Supplemental Table S1 at http://ajp.amjpathol.org; Figure 5A). To investigate whether the miR-200b/c members regulate the expression of TGFβR1 we initially used the in vitro model of TGF-β–stimulated EMT in rat tubular epithelial cells. We used plasmid-based transfection to overexpress miR-200c. miR-21 and miR-200b were used as controls because TGF-β induced miR-21 expression and reduced miR-200b expression (Figure 3). In cells transfected with pri-miR-200c, stimulation with TGF-β resulted in the same pattern, an increase in miR-21 (2.7-fold increase) and a 64% decrease in miR-200c expression (Figure 5B). When EMT markers were examined, overexpression of pri-miR-200c resulted in a significant decrease in α-SMA and collagen I after stimulation with TGF-β, which was similar to that observed before.26 Stimulation with TGF-β resulted in a marked reduction in miR-200c (75%) and a significant induction of TGFβ-R1 (P < 0.05; Figure 5B). Overexpression of pri-miR-200c resulted in an approximate sevenfold increase in miR-200c expression and a marked inhibition (∼48%) of TGFβR1 expression (Figure 5). Therefore, the changes in miRNA associated with TGF-β are reflected in changes in TGFβR1.
Figure 5.
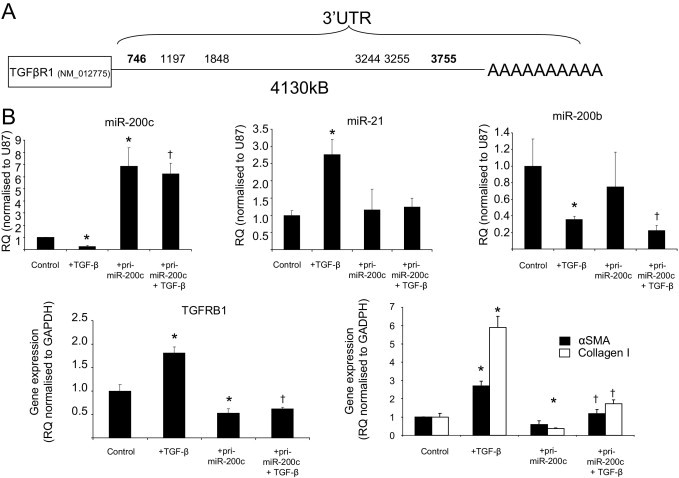
Effect of manipulation of miR-200c on TGFβ-R1 gene expression. A: Schematic diagram of 3′-UTR of rat TGFβR1 and miRNA seed sequence matches (nucleotide 2 to 7) for miR-200b/c. B: NRK52E cells were transfected with a plasmid containing cytomegalovirus promoter-driven pri-miR-200c by lipofectamine 2000 (2 μg of DNA:3 μl of lipofectamine), serum starved for 48 hours, and then stimulated with TGF-β. Total RNA was extracted with the miRNeasy kit (Qiagen), and miRNA expression was measured by specific miRNA Taqman probes (Applied Biosystems) and normalized to U87. TGF-βR1, α-SMA, and collagen I expressions were measured by specific rat gene expression probes (Applied Biosystems) and normalized to GAPDH (rat). RQ ± RQ max. *P < 0.05 versus control cells and †P < 0.05 compared with control cells stimulated with TGF-β.
Correlation of miRNA Modulation in Vitro in Models of Renal Disease in Vivo
To address whether miRNAs are modulated during the development of glomerulonephritis, we used the rat anti-Thy1.1 model of mesangial proliferative glomerulonephritis, which is self-limiting and has a well-characterized progression.35 We induced mesangial proliferative glomerulonephritis in WKY rats, and after sacrifice histologic evaluation was performed to look at glomerular and tubular lesions. A single administration of ER4 resulted in mild glomerular damage (Table 1), which was resolved by 14 days (data not shown). Kidneys from these animals had limited glomerular lesions that featured global or segmental mesangial proliferation (Table 1; Figure 6A). Multiple administration of the ER4 protocol induced more severe histologic changes with ∼50% of glomeruli affected and some evidence of tubular damage in the form of tubular atrophy (Table 1 and Figure 6A). To quantify temporal changes in miRNA we sacrificed a further group of animals 14 days after final administration of ER4, which histologically appeared similar to those sacrificed at 7 days with the glomerular and tubular scores remaining similar (Table 1).
Table 1.
Histologic Evaluation of Kidney Damage in Anti-Thy1.1 Animals
| Group | Glomerular score | Tubular score | Total |
|---|---|---|---|
| Control | 0.25 | 0.25 | 0.5 |
| 1 × ER4, 7 days | 1 | 0.5 | 1.5 |
| 3 × ER4, 7 days | 2 | 2 | 4 |
| 3 × ER4, 14 days | 2.1 | 1.3 | 3.4 |
Histologic scoring of animals used in this study. Scoring was adapted from Tomita et al.29 Scoring was performed blind (N = 4 to 6 animals per group) with 50 glomeruli and surrounding tubules scored per section. Glomerular damage was scored from 0 to 4, with 0 being 0% to 4% glomerular lesion, mesangial matrix expansion; 1 being 5% to 24%; 2 being 25% to 49%; 3 being 50% to 74%; and 4 being >75%. Interstitial lesions were counted as tubular dilation, tubular trophy, basophilia, interstitial inflammation, and fibrosis and scored from 0 to 4, with 0 being no lesion; 1 being <5%, 2 being 5% to 24%, 3 being 25% to 49%, and 4 being ≥50%.
Figure 6.
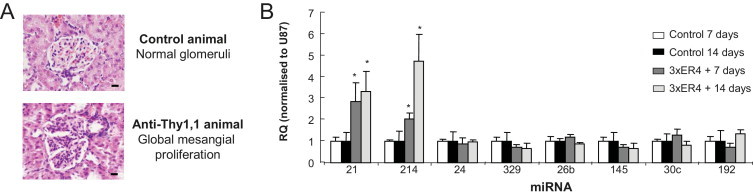
Validation of miRNA expression after induction of mesangial proliferative glomerulonephritis. Validation of miRNAs expressed in animals sacrificed 7 and 14 days after three ER4 injections 1 week apart compared with age-matched controls. A: Representative histology of normal glomeruli from control animals and the global mesangial proliferation observed in glomeruli in kidneys from animals that received three ER4 injections. B: miRNA expression measured by specific miRNA Taqman probes (Applied Biosystems) and normalized to U87. RQ ± RQ max. *P < 0.05 versus age-matched control animals (N = 4 to 6).
We performed TaqMan analysis for miRNA expression in animals that received three injections and sacrificed at 7 days but also in the second group which were sacrificed at 14 days and compared this with age-matched control animals (Figure 6B). miR-21 and miR-214 expression was significantly increased at 7 and 14 days compared with controls (Figure 6B). No other miRs tested in the in vitro setting showed altered expression in this pathological setting (Figure 6B). Because we have demonstrated in vitro that TGF-β was stimulating expression of miR-21 and miR-214, we sought to evaluate the levels of TGF-β expression in this animal model. We found that at 7 days and 14 days after injection of the third ER4 there was a significant increase in TGF-β but no change in TGFβRI expression (see Supplemental Figure S5 at http://ajp.amjpathol.org).
Similarly to the chronic glomerulonephritis model, TGF-β is the main profibrotic mediator in the UUO model.36 To examine miRNA expression in the UUO model, we performed a time course. Histologic evaluation of tissue taken from animals sacrificed at 16 days after surgery showed moderate interstitial fibrosis (Figure 7A) and a high level of α-SMA staining in the interstitium (Figure 7A; see also Supplemental Figure S6 at http://ajp.amjpathol.org), suggestive of a large increase in the number of active myofibroblasts but not the origin. Because EMT is reported to contribute to the fibroblast population in this model,37 we examined the expression of a panel of markers known to be indicative of the EMT process. E-cadherin loss is an integral component of EMT in our in vitro model; yet, the effects of pathological insults in vivo on E-cadherin regulation and association with EMT are less clear. Therefore, we examined E-cadherin gene expression and found that it was up-regulated at this time point along with matrix metalloproteinase 2 and matrix metalloproteinase 14 (see Supplemental Figure S6 at http://ajp.amjpathol.org). We wanted to detail how TGF-β expression in the kidney was changing in this model, so we preformed real-time qPCR and found that within 3 days of surgery the level of TGF-β expression was significantly increased to ∼3.5-fold that of sham-operated animals (Figure 7B). Furthermore, the level of expression increased further when examined at 7 and 14 days after surgery. This confirmed that in the kidneys of UUO animals the levels of TGF-β have increased (Figure 7B). On the basis of our in vitro model results, we assessed the expression levels of a panel of miRNAs in whole kidney extracts from UUO and sham-operated animals at 16 days. In similarity with the anti-Thy1.1 animals miR-21 and miR-214 were significantly up-regulated (sixfold for miR-21 and 2.5-fold for miR-214) (Figure 7C). miR-200b was found to be decreased in the UUO kidneys compared with controls; in addition, several other miRNAs were also found to be significantly down-regulated, miR-192, miR-329, miR-24, miR-26b, and miR-30c (Figure 7C). TGFβ-R1 was also found to be up-regulated at the gene expression level, which correlates with members of the miR-200 family being down-regulated (see Supplemental Figure S6 at http://ajp.amjpathol.org). To examine what happened to miR-21 and miR-214 expression over time in this model we examined their expression and found that miR-21 expression had doubled within 3 days and continued to increase over time (Figure 7D). miR-214 expression was also significantly increased but only significantly from 7 days after surgery when miR-214 expression was ∼2.5-fold higher than that observed in sham-operated animals. The levels of miR-214 were then sustained for the time course (Figure 7D). Because we had observed previously in the in vitro models differences in the precursors of the miR-21 and miR-214 we performed Northern blot analyses on kidney RNA extracted from the animals 7 days after surgery (Figure 7E) that we had also demonstrated had significantly increased TGF-β expression (Figure 7B). For miR-21 the primary form of miR was present in all samples but the precursor form was only present in the UUO samples (Figure 7E). Mature miR-21 could be detected in all samples but was expressed higher in the UUO samples (Figure 7E). In contrast, similar levels of pre- and pri-miR-214 were present in both UUO and sham-operated animals. However, the mature miR-214 levels were increased significantly only in the UUO animals (Figure 7E).
Figure 7.
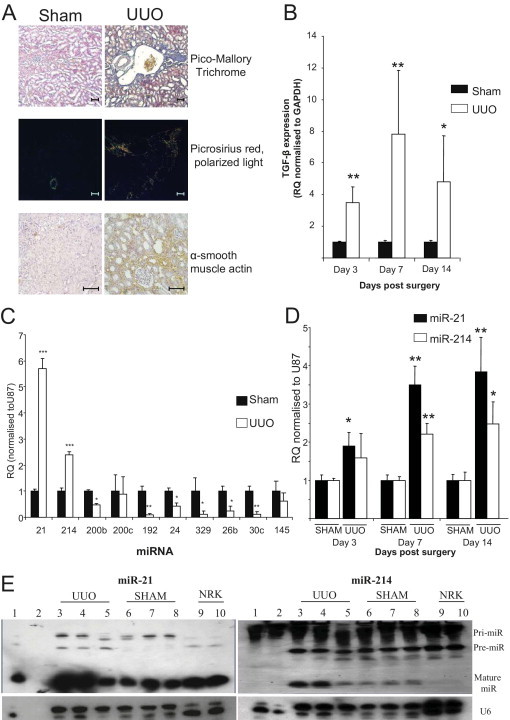
miRNA expression after UUO. A: Histologic analysis of kidneys from sham and UUO animals for tubulo-interstitial fibrosis by Pico-Mallory Trichrome and Picro Sirius red (viewed under cross-polarized light) and α-SMA staining 16 days after surgery. Scale bars: 100 μm. B: TGF-β gene expression in animals undergoing UUO. Total kidney RNA was extracted with the use of the miRNeasy kit (Qiagen), and TGF-β gene expression was measured by specific rat TGF-β gene expression probe (Applied Biosystems) and normalized to GAPDH (rat). RQ ± RQ max. *P < 0.05 and **P < 0.01. C: miRNA expression in kidneys of sham or UUO animals was measured by specific miRNA Taqman probes (Applied Biosystems) and normalized to U87. RQ ± RQ max. *P < 0.05, **P < 0.01, and ***P < 0.001 compared with sham animals (N = 5). D: miR-21 and miR-214 expression in UUO animals over time. Total RNA was extracted with the use of the miRNeasy kit (Qiagen), and miRNA expression was measured by specific miRNA Taqman probes (Applied Biosystems) and normalized to U87. RQ ± RQ max. *P < 0.05 and **P < 0.01 versus age-matched control sham-operated animals (N = 3). E: miR-21 and miR-214 expression in kidney tissue 7 days after surgery. Northern blot analyses were performed on total kidney RNA extracted with the use of the miRNeasy kit (Qiagen) of tissue from UUO and sham-operated animals at 7 days after surgery and probed with 5′-DIG–labeled LNA Mercury probes (Exiqon) rat miRNA for miR-21 and miR-214. U6 was used as a loading control (Exiquon). Lane 1 indicates HeLa cells; lane 2, RNA ladder; lanes 3 to 5, UUO RNA; lanes 6 to 8, SHAM RNA; and lanes 9 and 10, NRK52E (NRK) control RNA.
Because the causes of these two renal diseases are different but TGF-β plays a role, we examined the expression of miR-21 and miR-214 in the SHRSP rat to identify if there is a common miRNA signature in the renal pathological process. We assessed the expression of miR-21 and miR-214 alongside miR-30c, miR-192, and miR-145. The expression of the miRs was assessed in 21-week SHRSP rats with established hypertension (systolic blood pressure > 170 mmHg)5 and compared with that observed in age-matched normotensive reference strain WKY rats (systolic blood pressure < 125 mmHg).5 Both miR-21 (∼2.25-fold increased) and miR-214 (∼threefold increased) were significantly increased when SHRSP animals were compared with the reference strain (WKY) with no change in the other miRs examined (Figure 8). However, when young 5-week-old animals that are not hypertensive were assessed for miR-21 and miR-214, there was no difference (data not shown), indicating that these miRs are not altered before the onset of hypertension or renal damage. Interestingly, the expression of these miRs was further increased in the SHRSP when they were challenged with salt (1%), with miR-21 levels increasing ∼14-fold and miR-214 levels increasing by fourfold. Surprisingly, in the WKY rats that are not salt sensitive and do not have any renal insufficiency, salt loading significantly increased miR-21 expression by six times compared with age-matched normal WKY rats. The salt challenge also resulted in miR-30c and miR-192 being significantly increased in both WKY and SHRSP rats, and miR-145 only being increased in SHRSP. Because we found increases in miR-21 and miR-214 in the SHRSP compared with the WKY rats and under salt loading, we investigated whether an increase in TGF-β is associated with this increase. We therefore measured the level of mRNA for TGF-β and TGFβRI in tissues and observed that TGF-β was significantly increased in the SHRSP rats compared with the WKY rats and further increased under salt loading (see Supplemental Figure S7 at http://ajp.amjpathol.org). Furthermore, TGFβRI was also increased under salt loading, suggesting an increase in TGF-β signaling (see Supplemental Figure S7 at http://ajp.amjpathol.org).
Figure 8.
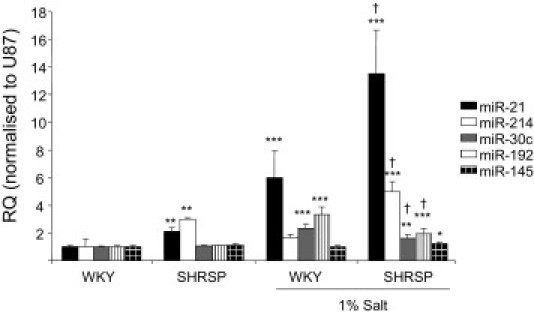
miRNA expression in genetic model of hypertension, the SHRSP. miRNA expression in kidneys of 21-week-old male WKY, SHRSP rats with and without salt loading (1%) was measured by specific miRNA Taqman probes (Applied Biosystems) and normalized to U87. RQ ± RQ max. *P < 0.05, **P < 0.01, and ***P < 0.001 compared with WKY expression levels; †P < 0.05 compared with SHRSP expression levels.
Discussion
miRNAs have recently been shown to be dysregulated in many diseases and are thought to contribute to the pathophysiology of the distinct disease such as cancer and myocardial infarction. Previously, miR-192 and miR-377 have been highlighted as being regulated by TGF-β in mesangial cells and in the diabetic kidney. Here, we establish that the expression of miR-214 and miR-21 are also regulated by TGF-β. We demonstrate in this study that TGF-β–induced EMT in a rat tubular epithelial cell line was blocked by small molecule inhibitors of TGF-β signaling. Induction of miR-21 and miR-214 was also inhibited by these agents. Furthermore, overexpression of either miR-214 or miR-21 in the absence of TGF-β–induced EMT-like changes in culture with associated changes in E-cadherin, α-SMA, and collagen expression. This underpins an important role for these two miRNAs in TGF-β signaling-induced EMT in vitro. To underline the importance of miR-21 and miR-214, both were significantly increased in two models of renal disease in vivo in which TGF-β plays a role in the pathophysiology but also in a rat model of genetic hypertension shown to have predominately vascular renal damage. Interestingly, when these animals were challenged with high salt both WKY and SHRSP animals showed an increase in miR-21, whereas miR-214 was only increased further in the salt-loaded SHRSP animals. Furthermore, because neither miRNA has differential expression in non-hypertensive 5-week-old SHRSP animals, the change in these miRNAs illustrates a role in the pathophysiology of hypertension in this model. The effect of salt loading on WKY miR-21 expression was surprising, but together with the data from the SHRSP on miR-21 and miR-214 it indicates that other pathways beyond TGF-β also probably regulate these miRNAs. Taken together this study shows that the overexpression of both miR-21 and miR-214 represents a common signature for renal damage unrelated to its cause. To further support this a recent study found that miR-214 is increased in monocytes of patients with chronic kidney disease.38
miR-21 overexpression has been shown in mesangial cells stimulated with glucose (to mimic diabetic nephropathy) to protect the cells from proliferation by the down-regulation of PTEN, which was verified as a target of miR-21 in this context.39 miR-21 is frequently found up-regulated in response to stress. In cardiac fibrosis the source of miR-21 has been found to be cardiac fibroblasts.40 miR-214 has been found to be up-regulated in epithelial ovarian carcinomas.41 Furthermore, PTEN has also been confirmed as a target of miR-214, and in human ovarian cancer cells an increase in miR-214 induces cell survival and cisplatin resistance via targeting the PTEN/Akt pathway.41 miR-214 has been shown to be activated by TWIST-1 through an E-box promoter element42 and may be involved in the development of specific neural cell populations; therefore, because TWIST activation is a key event in EMT, it may be through this pathway that miR-214 expression is increased in the tubular epithelial cells. However, TGF-β can also drive cells toward apoptosis, so it is also possible that miR-214 may mediate the switch between these two cell fates.
In mesangial cells, miR-145 expression was also measured, because it has been shown to be a mesangial cell marker,20 and it was rapidly increased in response to TGF-β but returned to normal levels within 24 hours of TGF-β stimulation. miR-145 has been shown to act as a tumor suppressor43 and to prevent proliferation of smooth muscle cells,44 and, because mesangial cells are specialized smooth muscle cells, the increase in miR-145 may be a protective mechanism.
miR-30c expression was lost in the UUO kidney; previously, it has been shown to be essential for normal kidney homoeostasis and targets several genes of importance in structure and function of the kidney. It has a tubular expression within the kidney, and whether the loss of miR-30c was an initiating factor or as a result of the widespread tubular damage would need to be investigated further.
In similarity to other studies, stimulation of the rat tubular epithelial cell line with TGF-β resulted in EMT and a loss of the expression of the miR-200 family members miR-200b and miR-200c. Elegant studies have previously shown that miR-200 family overexpression prevents EMT induced by TGF-β by binding to their targets ZEB1 and ZEB2, which are transcriptional repressors of E-cadherin.24–26 Loss of the miR-200 family results in an increase in ZEB1 and ZEB2 expression and loss of E-cadherin and subsequent induction of EMT.24–26 In the UUO animals, miR-200b was significantly down-regulated; this could be an indicator that EMT is occurring in this model and contributing to the myofibroblast cell number. However, E-cadherin expression was up-regulated in this model at this time point (see Supplemental Figure S6 at http://ajp.amjpathol.org), which has been reported before45 and is in contrast to what was observed in the in vitro model in which E-cadherin was down-regulated. Target analysis of miR-200b and miR-200c found that TGFβR1 was a target for these miRNAs with six seed matches in the rat. This study shows that TGFβR1 is a target of the miR-200 family and suggests the miR-200 family are important regulators of TGF-β signaling, being able to influence multiple parts of the signaling pathway. In the UUO model in which EMT of tubular epithelial cells has been reported to occur37 and contribute to the myofibroblast population, we wanted to examine the expression of miR-200b and miR-200c. In animals 16 days after UUO, the expression of miR-200b was significantly down-regulated, and miR-200c was reduced. This may further suggest that the loss of the miR-200 family is pathophysiologically important in tubule-interstitial fibrosis, especially because miR-200b has been shown to have a tubular location within the kidney.20
This study for the first time shows that miR-21 and miR-214 may represent a miRNA signature of renal damage and both these miRNAs can be regulated by TGF-β in a cell-specific manner.
Acknowledgments
We thank Bruce Hendry for critical appraisal of the manuscript. We also thank Angela McIllhoney, Nicola Britton, and Gregor Aitchison for technical assistance.
Footnotes
Supported by Kidney Research UK, British Heart Foundation Chair, and Programe Grants (RG/09/005/27915 and BHFRG/07/005/23633) and by a pump priming grant from the Integrative Mammalian Biology Initiative at the University of Glasgow.
Supplemental material for this article can be found at http://ajp.amjapthol.org or at doi: 10.1016/j.ajpath.2011.04.021.
Supplementary data
Rat mesangial cell response to TGF-β stimulation. Rat mesangial cells CRL-2573 were serum starved for 48 hours. A: Cells were incubated with TGF-β (10 ng/mL), growth media (10% fetal calf serum) or left in serum-free media for 72 hours. An MTT assay was performed with the CellTiter 96 Non-Radioactive Cell Proliferation Assay (MTT; Promega, Southampton, UK). *P < 0.05. B: Effect of TGF-β (10 ng/mL) stimulation of serum-starved CRL-2573 cells on TGF-β gene expression over a 4-day time course. RNA was harvested from the cells by miReasy kit (Qiagen), and gene expression was measured by rat-specific Taqman probes (Applied Biosystems) and normalized to GAPDH RQ ± RQ max; *P < 0.05 versus unstimulated time-matched controls. C: Effect of TGF-β (10 ng/mL) stimulation of serum-starved CRL-2573 cells on TGF-βR1 gene expression over a 4-day time course. RNA was harvested from the cells by miReasy kit (Qiagen), and gene expression was measured by rat-specific Taqman probes (Applied Biosystems) and normalized to GAPDH RQ ± RQ max; *P < 0.05 versus unstimulated time-matched controls.
Rat tubular epithelial cell response to TGF-β stimulation. Rat tubular epithelial cells (NRK52E) were serum starved for 48 hours. A: Effect of TGF-β (10 ng/mL) stimulation of serum-starved NRK52E cells on TGF-β gene expression over a 4-day time course. RNA was harvested from the cells by miReasy kit (Qiagen), and gene expression was measured by rat specific Taqman probes (Applied Biosystems) and normalized to GAPDH RQ ± RQ max; *P < 0.05 versus unstimulated time-matched controls. B: Effect of TGF-β (10 ng/mL) stimulation of serum-starved CRL-2573 cells on TGF-βR1 gene expression over a 4-day time course. RNA was harvested from the cells by miReasy kit (Qiagen), and gene expression was measured by rat-specific Taqman probes (Applied Biosystems) and normalized to GAPDH RQ +/- RQ max; *P < 0.05 versus unstimulated time-matched controls.
miR-21 and miR-214 expression in NRK52E cells undergoing EMT. Northern blot analyses were performed on total RNA extracted with the miRNeasy kit (Qiagen) of cells stimulated with 10 ng/mL TGF-β undergoing EMT over a 5-day time course and probed with 5′-DIG–labeled LNA Mercury probes (Exiqon) rat miRNA for miR-21 and miR-214. U6 was used as a loading control (Exiquon). Lane 1 indicates HeLa; lane 2, RNA ladder; lane 3, NRK52E (NRK) cells cultured in normal media; lane 4, NRK cells serum starved; lanes 5 and 6, 1 day; lanes 7 and 8, 2 days; lanes 9 and 10, 3 days; lanes 11 and 12, 4 days; lanes 13 and 14, 5 days. The – indicates time-matched controls, serum starved, +, cells stimulated with 10 ng/mL TGF-β. See Supplemental Figure S1.
Effect of TGF-β signaling inhibition on miR-21 and miR-214 expression. Total RNA was extracted with the miRNeasy kit (Qiagen) from cells stimulated with 10 ng/mL TGF-β in the presence or absence of specific ALK5 (TGF-βRI) inhibitors (1 μM) or vehicle control (DMSO). miRNA expression was measured by specific miRNA Taqman probes (Applied Biosystems) and normalized to U87. Gene expression was measured by specific gene expression Taqman probes (Applied Biosystems) and normalized to GAPDH. VC indicates vehicle control RQ ± RQ max; *P < 0.05 versus VC alone and #P < 0.05 versus VC+TGF-β.
TGF-β and TGF-βRI expression in anti-Thy1.1 model. Expression of TGF-β and TGF-βRI in kidneys of animals sacrificed 7 and 14 days after three ER4 injections 1 week apart compared with age-matched controls. Gene expression was measured with specific gene expression Taqman probes (Applied Biosystems) and normalized to GAPDH. *P < 0.05 versus age-matched controls.
Gene expression of EMT markers in UUO animals. RNA was extracted, and cDNA synthesis was performed on 1 μg of RNA with the use of reverse transcription reagents (Applied Biosystems). Briefly, for the quantitative real-time PCR step, amplification was performed with gene expression-specific primers (Applied Biosystems) on the Applied Biosystems 7900 HT real-time PCR system following the manufacturer's instructions. Rat GAPDH was used as an endogenous control RQ ± RQ max; *P < 0.05, **P < 0.01, and ***P < 0.001 versus sham-operated animals.
TGF-β and TGF-βRI expression in a genetic model of hypertension, the SHRSP. Expression of TGF-β and TGF-βRI in kidneys of 21-week-old male WKY, SHRSP rats with and without salt loading (1%) measured by specific gene expression Taqman probes (Applied Biosystems) and normalized to GAPDH RQ ± RQ max; *P < 0.05.
References
- 1.Bottinger E.P., Bitzer M. TGF-{beta} signaling in renal disease. J Am Soc Nephrol. 2002;13:2600–2610. doi: 10.1097/01.asn.0000033611.79556.ae. [DOI] [PubMed] [Google Scholar]
- 2.Liu Y. Renal fibrosis: new insights into the pathogenesis and therapeutics. Kidney Int. 2006;69:213–217. doi: 10.1038/sj.ki.5000054. [DOI] [PubMed] [Google Scholar]
- 3.Schlondorff D. The glomerular mesangial cell: an expanding role for a specialized pericyte. FASEB J. 1987;1:272–281. doi: 10.1096/fasebj.1.4.3308611. [DOI] [PubMed] [Google Scholar]
- 4.Whelton P.K., Klag M.J. Hypertension as a risk factor for renal disease: Review of clinical and epidemiological evidence. Hypertension. 1989;13:I19–I27. doi: 10.1161/01.hyp.13.5_suppl.i19. [DOI] [PubMed] [Google Scholar]
- 5.Koh-Tan H.H., Graham D., Hamilton C.A., Nicoll G., Fields L., McBride M.W., Young B., Dominiczak A.F. Renal and vascular glutathione S-transferase mu is not affected by pharmacological intervention to reduce systolic blood pressure. J Hypertens. 2009;27:1575–1584. doi: 10.1097/HJH.0b013e32832cc5a1. [DOI] [PubMed] [Google Scholar]
- 6.Obata J., Nakamura T., Takano H., Naito A., Kimura H., Yoshida Y., Shimizu F., Guo D.F., Inagami T. Increased gene expression of components of the renin-angiotensin system in glomeruli of genetically hypertensive rats. J Hypertens. 2000;18:1247–1255. doi: 10.1097/00004872-200018090-00011. [DOI] [PubMed] [Google Scholar]
- 7.Graham D., McBride M.W., Gaasenbeek M., Gilday K., Beattie E., Miller W.H., McClure J.D., Polke J.M., Montezano A., Touyz R.M., Dominiczak A.F. Candidate genes that determine response to salt in the stroke-prone spontaneously hypertensive rat: congenic analysis. Hypertension. 2007;50:1134–1141. doi: 10.1161/HYPERTENSIONAHA.107.095349. [DOI] [PubMed] [Google Scholar]
- 8.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 9.Gregory R.I., Chendrimada T.P., Cooch N., Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 10.Chang S., Johnston R.J., Frokjaer-Jensen C., Lockery S., Hobert O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature. 2004;430:785–789. doi: 10.1038/nature02752. [DOI] [PubMed] [Google Scholar]
- 11.Johnston R.J., Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- 12.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 13.Huang Q., Gumireddy K., Schrier M., le Sage C., Nagel R., Nair S., Egan D.A., Li A., Huang G., Klein-Szanto A.J., Gimotty P.A., Katsaros D., Coukos G., Zhang L., Pure E., Agami R. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 14.Ma L., Teruya-Feldstein J., Weinberg R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 15.Ji R., Cheng Y., Yue J., Yang J., Liu X., Chen H., Dean D.B., Zhang C. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of microRNA in vascular neointimal lesion formation. Circ Res. 2007;100:1579–1588. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 16.Care A., Catalucci D., Felicetti F., Bonci D., Addario A., Gallo P., Bang M.-L., Segnalini P., Gu Y., Dalton N.D., Elia L., Latronico M.V.G., Hoydal M., Autore C., Russo M.A., Dorn G.W., Ellingsen O., Ruiz-Lozano P., Peterson K.L., Croce C.M., Peschle C., Condorelli G. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 17.Pandey P., Brors B., Srivastava P.K., Bott A., Boehn S.N., Groene H.J., Gretz N. Microarray-based approach identifies microRNAs and their target functional patterns in polycystic kidney disease. BMC Genomics. 2008;9:624. doi: 10.1186/1471-2164-9-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato M., Zhang J., Wang M., Lanting L., Yuan H., Rossi J.J., Natarajan R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci U S A. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi S., Yu L., Chiu C., Sun Y., Chen J., Khitrov G., Merkenschlager M., Holzman L.B., Zhang W., Mundel P., Bottinger E.P. Podocyte-selective deletion of dicer induces proteinuria and glomerulosclerosis. J Am Soc Nephrol. 2008;19:2159–2169. doi: 10.1681/ASN.2008030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harvey S.J., Jarad G., Cunningham J., Goldberg S., Schermer B., Harfe B.D., McManus M.T., Benzing T., Miner J.H. Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J Am Soc Nephrol. 2008;19:2150–2158. doi: 10.1681/ASN.2008020233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho J., Ng K.H., Rosen S., Dostal A., Gregory R.I., Kreidberg J.A. Podocyte-specific loss of functional microRNAs leads to rapid glomerular and tubular injury. J Am Soc Nephrol. 2008;19:2069–2075. doi: 10.1681/ASN.2008020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q., Wang Y., Minto A.W., Wang J., Shi Q., Li X., Quigg R.J. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J. 2008;22:4126–4135. doi: 10.1096/fj.08-112326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zavadil J., Narasimhan M., Blumenberg M., Schneider R.J. Transforming growth factor-beta and microRNA: mRNA regulatory networks in epithelial plasticity. Cells Tissues Organs. 2007;185:157–161. doi: 10.1159/000101316. [DOI] [PubMed] [Google Scholar]
- 24.Park S.M., Gaur A.B., Lengyel E., Peter M.E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burk U., Schubert J., Wellner U., Schmalhofer O., Vincan E., Spaderna S., Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gregory P.A., Bert A.G., Paterson E.L., Barry S.C., Tsykin A., Farshid G., Vadas M.A., Khew-Goodall Y., Goodall G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 27.Demaison C., Parsley K., Brouns G., Scherr M., Battmer K., Kinnon C., Grez M., Thrasher A.J. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum Gene Ther. 2002;13:803–813. doi: 10.1089/10430340252898984. [DOI] [PubMed] [Google Scholar]
- 28.Butler S.L., Hansen M.S., Bushman F.D. A quantitative assay for HIV DNA integration in vivo. Nat Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 29.Khanin R., Vinciotti V. Computational modeling of post-transcriptional gene regulation by microRNAs. J Comput Biol. 2008;15:305–316. doi: 10.1089/cmb.2007.0184. [DOI] [PubMed] [Google Scholar]
- 30.Selbach M., Schwanhausser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 31.Tomita M., Sogabe H., Nakazato S., Nakatsuji S., Noto T., Hamada K., Kawachi H., Shimizu F., Matsuo M., Mutoh S. Monoclonal antibody 1-22-3-induced glomerulonephritis in uninephrectomized rats as a model of progressive renal failure. Nephrol Dial Transplant. 2005;20:2358–2367. doi: 10.1093/ndt/gfi062. [DOI] [PubMed] [Google Scholar]
- 32.Davidson A.O., Schork N., Jaques B.C., Kelman A.W., Sutcliffe R.G., Reid J.L., Dominiczak A.F. Blood pressure in genetically hypertensive rats: Influence of the Y chromosome. Hypertension. 1995;26:452–459. doi: 10.1161/01.hyp.26.3.452. [DOI] [PubMed] [Google Scholar]
- 33.Rosenbloom J, Castro SV, Jimenez SA: Narrative review: fibrotic diseases: cellular and molecular mechanisms and novel therapies. Ann Intern Med 152:159–166 [DOI] [PubMed]
- 34.Tojo M., Hamashima Y., Hanyu A., Kajimoto T., Saitoh M., Miyazono K., Node M., Imamura T. The ALK-5 inhibitor A-83-01 inhibits Smad signaling and epithelial-to-mesenchymal transition by transforming growth factor-beta. Cancer Sci. 2005;96:791–800. doi: 10.1111/j.1349-7006.2005.00103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bagchus W., Hoedemaeker P., Rozing J., Bakker W. Glomerulonephritis induced by monoclonal anti-Thy 1.1 antibodies: A sequential histological and ultrastructural study in the rat. Lab Invest. 1986;55:680–687. [PubMed] [Google Scholar]
- 36.Kaneto H., Morrissey J., Klahr S. Increased expression of TGF-[beta]1 mRNA in the obstructed kidney of rats with unilateral ureteral ligation. Kidney Int. 1993;44:313–321. doi: 10.1038/ki.1993.246. [DOI] [PubMed] [Google Scholar]
- 37.Iwano M., Plieth D., Danoff T., Xue C., Okada H., Neilson E. Evidence that fibroblasts derive from epithelium during tissue fibrosis. J Clin Invest. 2002;110:341–350. doi: 10.1172/JCI15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li LM, Hou DX, Guo YL, Yang JW, Liu Y, Zhang CY, Zen K: Role of microRNA-214-targeting phosphatase and tensin homolog in advanced glycation end product-induced apoptosis delay in monocytes. J Immunol 186:2552–2560 [DOI] [PubMed]
- 39.Zhang Z., Peng H., Chen J., Chen X., Han F., Xu X., He X., Yan N. MicroRNA-21 protects from mesangial cell proliferation induced by diabetic nephropathy in db/db mice. FEBS Lett. 2009;583:2009–2014. doi: 10.1016/j.febslet.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 40.Thum T., Catalucci D., Bauersachs J. MicroRNAs: novel regulators in cardiac development and disease. Cardiovasc Res. 2008;79:562–570. doi: 10.1093/cvr/cvn137. [DOI] [PubMed] [Google Scholar]
- 41.Yang H., Kong W., He L., Zhao J.-J., O'Donnell J.D., Wang J., Wenham R.M., Coppola D., Kruk P.A., Nicosia S.V., Cheng J.Q. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 42.Lee Y.-B., Bantounas I., Lee D.-Y., Phylactou L., Caldwell M.A., Uney J.B. Twist-1 regulates the miR-199a/214 cluster during development. Nucl Acids Res. 2009;37:123–128. doi: 10.1093/nar/gkn920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.La Rocca G., Badin M., Shi B., Xu S.Q., Deangelis T., Sepp-Lorenzinoi L., Baserga R. Mechanism of growth inhibition by microRNA 145: the role of the IGF-I receptor signaling pathway. J Cellular Physiol. 2009;220:485–491. doi: 10.1002/jcp.21796. [DOI] [PubMed] [Google Scholar]
- 44.Cordes K.R., Sheehy N.T., White M.P., Berry E.C., Morton S.U., Muth A.N., Lee T.-H., Miano J.M., Ivey K.N., Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Docherty N.G., Calvo I.F., Quinlan M.R., Perez-Barriocanal F., McGuire B.B., Fitzpatrick J.M., Watson R.W.G. Increased E-cadherin expression in the ligated kidney following unilateral ureteric obstruction. Kidney Int. 2008;75:205–213. doi: 10.1038/ki.2008.482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rat mesangial cell response to TGF-β stimulation. Rat mesangial cells CRL-2573 were serum starved for 48 hours. A: Cells were incubated with TGF-β (10 ng/mL), growth media (10% fetal calf serum) or left in serum-free media for 72 hours. An MTT assay was performed with the CellTiter 96 Non-Radioactive Cell Proliferation Assay (MTT; Promega, Southampton, UK). *P < 0.05. B: Effect of TGF-β (10 ng/mL) stimulation of serum-starved CRL-2573 cells on TGF-β gene expression over a 4-day time course. RNA was harvested from the cells by miReasy kit (Qiagen), and gene expression was measured by rat-specific Taqman probes (Applied Biosystems) and normalized to GAPDH RQ ± RQ max; *P < 0.05 versus unstimulated time-matched controls. C: Effect of TGF-β (10 ng/mL) stimulation of serum-starved CRL-2573 cells on TGF-βR1 gene expression over a 4-day time course. RNA was harvested from the cells by miReasy kit (Qiagen), and gene expression was measured by rat-specific Taqman probes (Applied Biosystems) and normalized to GAPDH RQ ± RQ max; *P < 0.05 versus unstimulated time-matched controls.
Rat tubular epithelial cell response to TGF-β stimulation. Rat tubular epithelial cells (NRK52E) were serum starved for 48 hours. A: Effect of TGF-β (10 ng/mL) stimulation of serum-starved NRK52E cells on TGF-β gene expression over a 4-day time course. RNA was harvested from the cells by miReasy kit (Qiagen), and gene expression was measured by rat specific Taqman probes (Applied Biosystems) and normalized to GAPDH RQ ± RQ max; *P < 0.05 versus unstimulated time-matched controls. B: Effect of TGF-β (10 ng/mL) stimulation of serum-starved CRL-2573 cells on TGF-βR1 gene expression over a 4-day time course. RNA was harvested from the cells by miReasy kit (Qiagen), and gene expression was measured by rat-specific Taqman probes (Applied Biosystems) and normalized to GAPDH RQ +/- RQ max; *P < 0.05 versus unstimulated time-matched controls.
miR-21 and miR-214 expression in NRK52E cells undergoing EMT. Northern blot analyses were performed on total RNA extracted with the miRNeasy kit (Qiagen) of cells stimulated with 10 ng/mL TGF-β undergoing EMT over a 5-day time course and probed with 5′-DIG–labeled LNA Mercury probes (Exiqon) rat miRNA for miR-21 and miR-214. U6 was used as a loading control (Exiquon). Lane 1 indicates HeLa; lane 2, RNA ladder; lane 3, NRK52E (NRK) cells cultured in normal media; lane 4, NRK cells serum starved; lanes 5 and 6, 1 day; lanes 7 and 8, 2 days; lanes 9 and 10, 3 days; lanes 11 and 12, 4 days; lanes 13 and 14, 5 days. The – indicates time-matched controls, serum starved, +, cells stimulated with 10 ng/mL TGF-β. See Supplemental Figure S1.
Effect of TGF-β signaling inhibition on miR-21 and miR-214 expression. Total RNA was extracted with the miRNeasy kit (Qiagen) from cells stimulated with 10 ng/mL TGF-β in the presence or absence of specific ALK5 (TGF-βRI) inhibitors (1 μM) or vehicle control (DMSO). miRNA expression was measured by specific miRNA Taqman probes (Applied Biosystems) and normalized to U87. Gene expression was measured by specific gene expression Taqman probes (Applied Biosystems) and normalized to GAPDH. VC indicates vehicle control RQ ± RQ max; *P < 0.05 versus VC alone and #P < 0.05 versus VC+TGF-β.
TGF-β and TGF-βRI expression in anti-Thy1.1 model. Expression of TGF-β and TGF-βRI in kidneys of animals sacrificed 7 and 14 days after three ER4 injections 1 week apart compared with age-matched controls. Gene expression was measured with specific gene expression Taqman probes (Applied Biosystems) and normalized to GAPDH. *P < 0.05 versus age-matched controls.
Gene expression of EMT markers in UUO animals. RNA was extracted, and cDNA synthesis was performed on 1 μg of RNA with the use of reverse transcription reagents (Applied Biosystems). Briefly, for the quantitative real-time PCR step, amplification was performed with gene expression-specific primers (Applied Biosystems) on the Applied Biosystems 7900 HT real-time PCR system following the manufacturer's instructions. Rat GAPDH was used as an endogenous control RQ ± RQ max; *P < 0.05, **P < 0.01, and ***P < 0.001 versus sham-operated animals.
TGF-β and TGF-βRI expression in a genetic model of hypertension, the SHRSP. Expression of TGF-β and TGF-βRI in kidneys of 21-week-old male WKY, SHRSP rats with and without salt loading (1%) measured by specific gene expression Taqman probes (Applied Biosystems) and normalized to GAPDH RQ ± RQ max; *P < 0.05.


