Abstract
IER3 (formerly IEX-1) encodes a 27kDa glycoprotein that regulates death receptor-induced apoptosis, interacts with NF-κb pathways, and increases expression rapidly in response to cellular stresses such as irradiation. Animal models, gene expression microarray experiments, and functional studies in cell lines have suggested a potential role for IER3 in oncogenesis, but to date no abnormalities of IER3 at the DNA level have been reported in patients with neoplasia. Here we describe breakpoint cloning of a t(6;9)(p21;q34) translocation from a patient with a myelodysplastic syndrome (MDS), facilitated by conversion technology and array-based comparative genomic hybridization, which revealed a rearrangement translocating the IER3 coding region away from critical flanking/regulatory elements and to a transcript-poor chromosomal region, markedly decreasing expression. Using split-signal and locus-specific FISH probes, we analyzed 204 patients with diverse hematologic malignancies accompanied by clonal chromosome 6p21 abnormalities, and found 8 additional patients with MDS with IER3 rearrangements (translocations or amplification). While FISH studies on 157 additional samples from patients with MDS and a normal-karyotype were unrevealing, and sequencing the IER3 coding and proximal promoter regions of 74 MDS patients disclosed no point mutations, RT-PCR results suggested that dysregulated expression of IER3 is common in MDS (61% >4 fold increase or decrease in expression with decreased expression primarily in early MDS and increased expression primarily in later MDS progressing towards leukemia), consistent with findings in previous microarray experiments. These data support involvement of IER3 in the pathobiology of MDS.
Keywords: IER3, myelodysplastic syndromes, array comparative genomic hybridization, chromosomal translocation
INTRODUCTION
IER3 (immediate early response 3), formerly known as IEX-1 or p22/PRG1, was cloned from a squamous carcinoma cell line in 1996 and localized to chromosome 6p21.3.(1) IER3 is expressed in a broad range of human tissues, including hematopoietic cells, and encodes a 156 amino acid (27 kDa glycosylated) protein so named because intracellular levels increase rapidly following ionizing radiation exposure, with maximal expression at ~15 minutes post-exposure.(1, 2) Death receptor agonists (Fas ligand and tumor necrosis factor alpha (TNF-α)) also induce IER3 expression, as does the protein kinase C activator 12-O-tetradecanoylphorbol-13-acetate (TPA), retinoic acid, hydroxytamoxifen, or etoposide, and cellular stresses such as mechanical stretching or viral infection.(3, 4)
The nuclear factor-kappa B (NF-κB) pathway appears to be important in mediating the intracellular effects of IER3; blockade of NF-κB activation abolishes TNF-α-induced IER3 expression and augments TNF-α-induced cell death.(5, 6) IER3 expression is under the control of tumor suppressor p53 and transcription factors Sp1and c-Myc, which bind to the promoter region.(7) Although IER3 has minimal homology to other factors, domains responsible for pro- and anti-apoptotic effects of IER3 under differing cellular stresses have been defined.(8)
Several observations suggest that IER3 may play a role in neoplasia.(3) Changes in IER3 expression alter apoptosis sensitivity (including susceptibility to death receptor ligation) and cell cycle progression or proliferation rates. In mice engineered to overexpress IER3 in immune cells, a lupus-like syndrome resulted, and T cell lymphomas developed in the spleen and lymph nodes.(9, 10) In contrast, IER3−/− mice have hypertension, thickened epidermis with an increased number of cellular layers, and cardiac hypertrophy without immunological abnormalities.(11) Global gene expression microarray experiments in diverse primary cancer cells (12), (13), CD34+ cells from patients with myelodysplastic syndromes(14, 15)) and transformed cell lines (e.g., Colo320 colorectal(16) and MCF-7 breast(17) carcinoma cells) have identified IER3 as an expression outlier that may have prognostic importance.(13, 15)
Despite these observations, no mutations or genomic rearrangements involving IER3 have been reported. Here we describe IER3 rearrangements and amplification for the first time, in 9 patients diagnosed with myelodysplastic syndromes (MDS).
MATERIALS AND METHODS
Propositus
G-banded karyotyping of sequential marrow samples from a 70-year-old man with refractory cytopenia with multilineage dysplasia (RCMD) demonstrated persistent t(6;9)(p21.3;q34), in ~18/20 metaphases (Figure 1A). Testing with fluorescent in-situ hybridization (FISH) probes for genes previously implicated in myeloid neoplasia localized to the 6p21 and 9q34 regions (i.e., DEK, NUP214/CAN, and ABL1 oncogenes) disclosed no rearrangements. Analysis of non-hematopoietic tissue confirmed the translocation was clonally restricted, not constitutional. Because isolated translocations are uncommon in MDS (interstitial deletions and numerical chromosomal abnormalities are more typical), this case seemed likely to be molecularly informative, so we cloned the breakpoint and sought similar rearrangements in other patients. The study protocol was approved by the Institutional Review Board of Mayo Clinic, and patients consented to sample collection and molecular analysis.
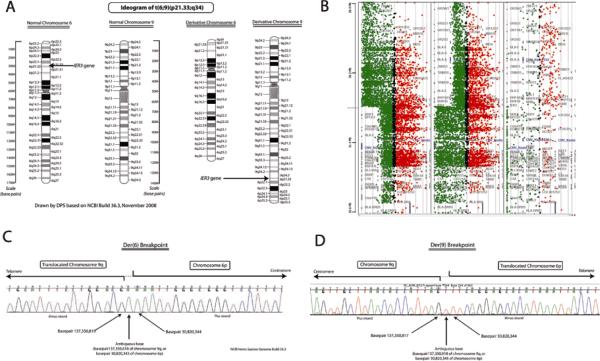
Figure 1.
A, Ideogram of t(6;9)(p23.1;q34), shown to rearrange IER3, in a patient with MDS. Normal chromosomes 6 and 9 are depicted on the left; derivative chromosomes seen in the propositus are depicted on the right. B, Array comparative genomic hybridization (CGH) demonstrating change in DNA copy number for chromosome 6p, centered on IER3, in a patient with MDS. Sample DNA is labeled green, representing intensity of fluorescein signal (proportional to DNA copy number), control DNA is labeled red, representing intensity of Texas Red signal. The IER3 gene is the closest to the apparent breakpoint on chromosome 6p. Array CGH narrowed the breakpoint to within an 800 bp region on chromosome 6p (which included the IER3 transcript initiation site) and 13 kbp on chromosome 9q. C and D, Sequencing chromatogram of breakpoints on derivative chromosome 6p (C) and derivative chromosome 9q (D). Sequencing-defined breakpoints were at base pairs 137,350,819 on chromosome 9 and 30,820,344 on chromosome 6 (NCBI build 36.3), with 1 base pair ambiguity due to a repeated nucleotide. Sequencing was performed in both directions. Southern blotting confirmed that this was a simple, balanced reciprocal translocation.
Somatic hybrid cell lines and chromosome isolation
Using an established conversion technology (18), somatic mouse/human hybrid cell lines were generated for each of the derivative chromosomes (i.e., der(6) and der(9)) and normal chromosomes 6 and 9 from the propositus. Briefly, after electrofusion of propositus leukocytes with E2 murine cells, mouse/human hybrid colonies were grown in selection medium (86% Dulbecco's Modified Eagle's Medium, 10% fetal bovine serum, 1% hypoxanthine-aminopterin-thymidine supplement 1% G418, 1% penicillin-streptomycin, and 1% minimal essential medium with non-essential amino acids; Invitrogen, Carlsbad, CA), selected at day 19 for initial expansion and genotyping, and subcultured. The presence of the derivative chromosomes in clones was confirmed with microsatellite markers (D6S1610, D6S441, D6S1581, and D6S264 and D9S1838 for der(6); D9S288, D9S157, D9S1817, D9S283, D9S1677, D6S309, D6S1574 for der(9)). Clones bearing >40% cells of interest, defined by these markers, were cryopreserved. Derivative and normal comparator chromosomes were then individually isolated by laser capture microdissection.
Array-based comparative genomic hybridization (aCGH)
High-resolution aCGH was performed with a custom 4 × 44K Human Genome CGH Microarray Kit (for derivative chromosomes 6p and 9q) and an off-the-shelf 244A kit (whole genome) (Agilent Technologies, Santa Clara, CA), following the protocol suggested by the manufacturer. Briefly, 800 ng of sample and control DNA, amplified from microdissected chromosomes using degenerate oligonucleotide primers, were labeled with Cy5 or Cy3 (BioPrime Array CGH Genomic Labeling Module, Invitrogen) and purified using Vivaspin 500 columns (Sartorius Stedim Biotech, Aubagne, France). Equal amounts of test and reference DNA were hybridized to the microarray at 65C for 40 hr in a rotation oven at 20 RPM. Slides were washed and then scanned with the G2505B DNA microarray scanner (Agilent). The microarray images were analyzed using Feature Extraction software V8.1 (Agilent), and Log2-transformed ratio data was analyzed with GeneSpring GX V7.3.1 and CGH Analytics V3.4.27 (Agilent).
Southern blotting
In order to determine whether the rearrangement in the propositus was a simple translocation of IER3 or more complex, DNA extracted from der(6) and der(9) somatic hybrid cells, propositus-derived whole marrow cells, and control marrow cells was digested with BamHI and EcoRI restriction endonucleases (New England Biolabs, Cambridge, MA), subject to agar gel electrophoresis for 24 hrs, blotted overnight on a nylon membrane, crosslinked with ultraviolet radiation, hybridized with a radiolabeled IER3 probe and salmon sperm blocking DNA, washed, and visualized by autoradiography. The IER3 probe was generated via polymerase chain reaction (PCR) with primers 5'-TCG GCG CTT GCA AAG ATA CAC T-3' and 5'-GGC GGG CAA ACC AAA TAC CAT A-3', cloned into MaxEfficiency DH5α competent cells (Invitrogen) using a TOPO PCR Cloning kit (Invitrogen), labeled with P32-dCTP using a Megaprime DNA Labeling Kit (Amersham Biosciences, Piscataway, NJ), and purified before hybridization.
Fluorescent in situ hybridization (FISH)
Direct-labeled split-signal and locus-specific FISH probes for the IER3 gene region at 6p21.3 were designed from bacterial artificial chromosomes (BACs), and validated according to established methods. For the 3' portion of the IER3 gene split-signal probe, three BAC clones (CTD-2208E8, RP11–159K11, and CTD-3223M13) were labeled in Spectrum Orange-dUTP (Abbott Molecular, Des Plaines, IL). For the 5' portion of the IER3 gene split-signal probe, three BAC clones (CTD-2017C24, CTD-2643J20, and RP11–100L8) were labeled in Spectrum Green-dUTP (Abbott Molecular). For the locus specific probe, all of the above clones were used, in addition to clone RP11–1039N6. Locus specific clones were labeled in Spectrum Orange-dUTP (Abbott Molecular).
Archived bone marrow and blood cell pellets from patients with a previously defined 6p21.3 chromosome abnormality were stored at −70°C in methanol:glacial acetic acid (2:1) fixative. After a change of fixative, the fixed-cell pellet suspensions were manually dropped onto microscope slides and checked by phase contrast microscopy to verify appropriate cellularity. The samples were then subjected to standard FISH pretreatment, hybridization, post hybridization and fluorescence microscopy methods.
DNA Sequencing
After narrowing of the propositus' chromosome 6p and 9q breakpoints via aCGH, both breakpoint regions were amplified directly by long-range PCR, using a series of primer pairs spaced 0.5–1 kb apart. Amplicons detected by ethidium-bromide-impregnated agarose gel electrophoresis were then sequenced using fluorescent dye chemistry sequencing to explicitly define the rearrangement (ABI Prism 3730 DNA Analyzer; Applied Biosystems, Foster City, CA), with chromatograms compared to NC_000009.10, NC_000006.10 and NM_003987 (IER3) using Sequencher 4.5 software (Gene Codes, Ann Arbor, MI).
To assess for point mutations in the propositus and other patients with MDS with and without 6p rearrangements, genomic DNA purified with QIAamp DNA Blood Mini Kit (QIAgen, Venlo, The Netherlands) was amplified by PCR on a PTC-200 Peltier thermocycler (MJ Research, Waltham, MA) and sequenced as for breakpoint cloning. Reagents included GeneAmp® PCR Buffer II (Applied Biosystems), 1.5 mM MgCl2 (Applied Biosystems), 200 μM dNTPs (Roche, Mannheim, Germany), forward and reverse oligonucleotide primers (see Supplemental Table S1), 100 ng of template DNA, and AmpliTaq Gold® DNA polymerase (total, 1 U; Applied Biosystems).
Murine hematopoietic colonies
Animal protocols were approved by an Institutional Animal Care and Use Committee. Five IER3−/− mice and 5 IER3+/+ C57/BL6 mice were sacrificed and marrow extracted from femoral heads. Next, 300 μL aliquots of density-centrifugation-separated marrow mononuclear cells were placed in 3 mL Methocult GF M3434 murine methylcellulose medium with cytokines (StemCell Technologies; Vancouver, BC). Duplicate aliquots were plated in 35 mm gridded plates and incubated at 37C in 5% CO2 for 10 days as specified by the media manufacturer, at which time hematopoietic progenitor colonies (BFU-E, CFU-GM, CFU-G, CFU-M, and CFU-GEMM) were enumerated using established morphological criteria.
Quantitative real-time PCR (RT-PCR)
RNA was isolated from propositus blood and marrow mononuclear cells, and from equivalent cells in MDS patients and controls, using an RNEasy Mini Kit (QIAgen). We confirmed RNA quality with an Agilent 2100 Bioanalyzer (Agilent), and generated cDNA using a SuperScript III RTS First-Strand cDNA Synthesis Kit (Invitrogen). RT-PCR was performed using TaqMan Universal Master Mix, an ABI 7900HT Fast RT-PCR system, the Hs00174674_m1 IER3 FAM multiplex primer-probe set (interrogates exons 1–2), and Hs_99999905_m1 glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Hs_00742828_s1 (TUBB) control primer-probes (all Applied Biosystems). Assays were performed in duplicate, and expression ratios calculated using the 2−ΔΔCt method.
RESULTS
Rearrangement of IER3 in the propositus
In the propositus, somatic hybrid array CGH results narrowed the translocation breakpoint to within an 800 bp region on chromosome 6p that included the transcription initiation site of IER3, and a 13 kbp region on chromosome 9q (Figure 1B). PCR and sequencing of these regions demonstrated that the t(6;9)(p21;q34) rearrangement resulted in separation of the IER3 gene at 6p21.33 from its upstream regulatory elements, and translocation to a transcript-poor region of 9q, along with just 40 bp of 5' flanking region adjacent to the IER3 transcript initiation site (Figure 1C) - containing only a TATA box, which is inadequate to drive expression. Previous systematic examination of the IER3 promoter had indicated that at least 279 bp of flanking region are critical for full expression.(7) Both Southern blotting and DNA sequencing results indicated a simple reciprocal translocation. By RT-PCR, expression of IER3 in PBMCs and whole marrow of the propositus was <5% of the mean for healthy controls.
FISH results for other patients
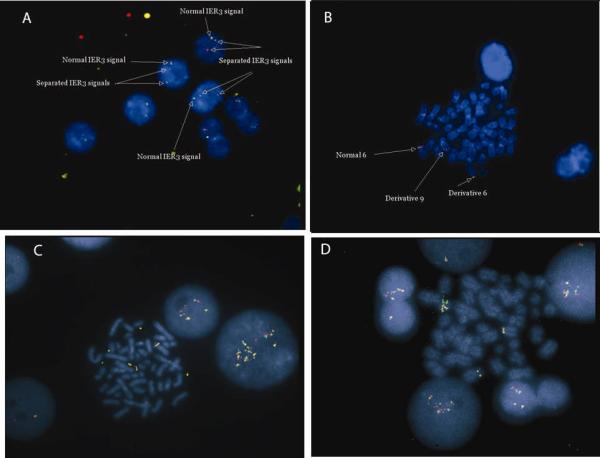
Split-signal and locus specific IER3 FISH probes were incubated with archival cell pellets from 204 patients with diverse hematological malignancies where G-banded karyotyping had detected deletions, translocations, or other rearrangements of chromosome 6p21. An additional 8 patients with abnormalities of IER3 were identified (Figure 2). Including the propositus, there were 4 split signals and 6 amplifications (one patient had both abnormalities), All 9 patients with IER3 breakapart or amplification had forms of MDS, with the 6p21 rearrangements part of a more complex karyotype in 8/9 cases (Table 1). There were not enough patients with IER3 rearrangements or amplification to perform detailed prognostic modeling.
Figure 2. Fluorescent in situ hybridization (FISH) using breakapart probe set for IER3 in MDS samples.
A, Metaphase preparation from patient with t(6;9)(p23.1;q34) demonstrating separation of red signal and green signal. Only a single fusion signal is present from the normal chromosome 6. B, Interphase cells from the same patient. C and D, two other MDS patients in whom IER3 signal was amplified, indicating increased copy number.
Table 1.
Characteristics and G-banded karyotypes of patients with IER3 rearrangements detected by FISH
| Age and Sex | MDS Subtype (WHO classification) | IPSS Risk Categry | Karyotype (ISCN nomenclature) | Split signal (S) or amplification (A) |
|---|---|---|---|---|
| 70 M* | RCMD | Int-1 | 45,X,−Y, t(6;9)(p21;q34)[18]/45,X,−Y[2] | S |
| 75 M | RCMD-RS | Int-2 | 45,XY,del(5)(q13q33), add(6)(p21.3)x2, add(14)(q22), add(19)(p13.3)[5]/45,X, −Y[3]/46,XY[12] | A |
| 80 M | MDS-U | Int-1 | 46,XY,add(1)(p32),add(5)(q11.2),del(6)(p21.3),+8,−17,−18,−18,add(20)(q11.2),+2mar[20] | S and A |
| 64 M | RAEB-2 | High | 41–46,XY,add(3)(p13),−5,del(6)(p21.3),−7[15],+8[15], add(12)(p11.2)[5],der(12)add(12)(p13)add(12)(q22)[15],−14[3],−15,−15[7],add(16)(q22),−17[3],der(17)t(15;17)(q11.2;p13)[18],−22[3],add(22)(q13)[2],+r[10],+0−3mar[cp20] | A |
| 63 M | RCMD | Int-2 | 41–43, XY, add(5)(q11.2), add(6)(p21.3)[5], add(7)(q22)[7], −13, −16, −18, −19[8], add(19)(p13.3)[5], −22[3], add(22)(q11.2)[5], +1−2mar[cp11] | A |
| 37M | Secondary MDS (history of HD) | Int-2 | 45,X,−Y, der(1)add(1)(q25)t(1;1)(p13;q25), add(3)(p13),add(4)(q31.1), add(5)(q11.2), add(6)(p21.3), der(18)t(1;18)(p22;q11.2)[20] | A |
| 70 F | MDS-U | Int-1 | 45,XX,del(5)(q13q33),add(6)(p21.3),−8,add(10)(p1 1.2), −17,add(18)(p11.2),add(22)(q13),+mar[cp30] | A |
| 64 M | MDS evolving to AML | High | 43–46, XY, add(1)(q32), −5, add(6)(p21.3), add(7)(p11.2), add(7)(q11.2), −11, −16, −17, −22, der(22)t(11;22)(q13;q13), +r, +2−4mar[cp18]/46,XY[2] | S |
| 79 F | Secondary MDS (history of CLL and HD) | Int-2 | 52–53, XX, add(6)(p21.3), add(8)(p23), +10, +14, +16, +21, +3mar [cp19]/46,XX[1] | S |
= Propositus.
Abbreviations: WHO = World Health Organization, MDS = myelodysplastic syndromes, ISCN = International System for Cytogenetic Nomenclature, M = male, F = female, FISH = fluorescent in situ hybridization, RCMD = refractory cytopenias with multilineage dysplasia, MDS-U = unclassifiable MDS, RAEB-2 = refractory anemia with excess blasts type 2, RCMD-RS = RCMD with ring sideroblasts, HD = Hodgkin disease, AML = acute myeloid leukemia, CLL = chronic lymphocytic leukemia. IPSS = 1997 International Prognostic Scoring System (note: the IPSS was not validated for patients with secondary MDS or previously treated patients.
In order to assess the frequency of karyotypically occult IER3 genomic rearrangements, 157 normal-karyotype patients representing all forms of World Health Organization-defined MDS (including 90 patients with International Prognostic Scoring System (IPSS(19)) Intermediate-2 or High risk disease, and 67 with IPSS Low or Intermediate-1 risk disease) were studied by FISH. No rearrangements or amplifications were detected in this group.
Sequencing for mutations or polymorphisms
Sequencing of the IER3 coding and promoter regions in 148 alleles from MDS patients without 6p rearrangements revealed no point mutations. An apparently novel polymorphism, a 4 bp deletion in the 3' UTR of IER3 (not a known or predicted area of microRNA binding), was present in 12% of MDS patients and 11% of healthy controls (200 control alleles examined), p=NS. The prevalence of non-synonymous single nucleotide polymorphism rs3094124 (p.A127P) was somewhat higher than expected in patients (variant in 14.6 % of MDS alleles vs. 8.6% of 116 HAP-CEU controls, p=0.01). Other SNPs did not differ in frequency between patients and controls.
RT-PCR
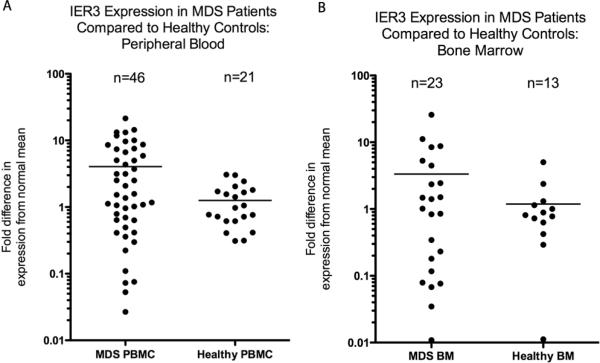
RT-PCR for IER3 in PBMCs (n=46 MDS patients without 6p rearrangements, n=21 healthy controls) and whole marrow (n=23 patients, n=13 controls) (Figure 3) demonstrated both down-regulation and up-regulation of IER3 in subsets of MDS samples compared to the mean for healthy controls, consistent with results from prior microarray experiments using MDS CD34+ cells.(14, 15) Marrow and blood expression of IER3 correlated highly when both types of samples were available. IER3 expression distribution in PBMCs was greater in MDS patients compared to controls, with 16/46 pts (35%; 11 of 16 had higher-risk MDS) demonstrating >4-fold increase in expression compared to mean for healthy controls, and 12/46 (26%; 7 of the 12 with lower-risk MDS) with >4-fold decrease in expression compared to controls.
Figure 3. Real-time polymerase chain reaction (RT-PCR) analysis comparing expression of IER3 at mRNA level in patients with myelodysplastic syndromes (MDS) to controls.
A, Scattergram comparing RT-PCR of peripheral blood mononuclear cells from 46 patients with MDS and 23 healthy controls. Differences are represented in terms of fold change in expression using 2−ΔΔCt method; Y-axis scale is logarithmic (base-10). Median expression of IER3 (horizontal line) was 4.1-fold higher in MDS patients compared to controls, and the distribution was wider in patients compared to controls. B, Scattergram of RT-PCR of ammonium chloride-lysed whole bone marrow from 23 patients with MDS and 13 healthy controls demonstrates a similar expression distribution in patients vs. controls as for the blood results.
Murine hematopoietic colony growth
We noted no significant differences in BFU-E, CFU-E, or CFU-GM growth under unstressed conditions between IER3−/− mice and IER3 wild-type mice.
DISCUSSION
These results demonstrate that IER3 is rearranged or amplified in a subset of patients with MDS, and dysregulated expression of IER3 is common in MDS. To our knowledge, this is the first demonstration of translocations and amplification involving IER3 in a human disease. IER3 is a plausible candidate for involvement in MDS because of its known role in regulating death receptor-induced apoptosis(3, 4), interaction with NF-κb pathways(5, 6) regulation by hematopoietic growth factors including thrombopoietin(20), and importance in immediate response to genotoxic stresses such as ionizing irradiation(1).
While the molecular mechanisms of MDS remain largely obscure, one commonly observed abnormality is dysregulation of apoptosis. Excessive intramedullary apoptosis in early stages of the disease is followed by diminished apoptosis later, as immature blast cells accumulate and the disease progresses towards acute myeloid leukemia (AML). Increased apoptosis of hematopoietic progenitor cells in early MDS may explain the typical “MDS paradox”: a hypercellular bone marrow, coupled with peripheral blood cytopenias. Conversely, the diminished ability of cells to undergo apoptosis late in the course of the disease has been postulated to contribute to the poor treatment response of the forms of leukemia that evolve in the setting of prior MDS.
The dichotomous IER3 RNA expression results reported here are consistent with those reported in 2 global cDNA expression microarray studies with CD34+ cells from MDS patients, and the proposed role of apoptosis in MDS pathobiology - i.e., decreased expression of IER3 early on in lower-risk disease when intramedullary apoptosis is excessive, and increased expression later as MDS progresses towards AML and a frankly neoplastic clone emerges.(14, 15)
It seems likely that IER3 rearrangements or amplification are a secondary, rather than primary, abnormality in MDS. IER−/− mice are not anemic and do not have dysplastic changes in the marrow (although mice >1 year old have not been systematically studied). We observed normal hematopoietic progenitor colony growth from IER−/− mice, at least in the absence of specific cellular stressors. In addition, for 8 of the 9 MDS patients in whom IER3 rearrangements were confirmed, 6p21 abnormalities were present as part of a more complex karyotype.
Array-based CGH is increasingly used to investigate copy-number variation in neoplasia, but unless individual chromosomes are sorted in some way prior to array hybridization, aCGH is not useful for investigation of balanced translocations. Here, we facilitated aCGH mapping of rearranged chromosomal boundaries with somatic hybridization technology, which allowed refinement of translocation breakpoints to within a few kbp - well within the range of long-range PCR - without using a labor-intensive “shotgun” approach requiring dozens of BAC clones. This technique could be applied to other translocations, including those associated with MDS, to discover new neoplasia-associated genes.
Conclusion
IER3 translocations and amplification represent novel, clonally-restricted recurrent genetic abnormalities in a subset of patients with MDS. Additionally, altered IER3 expression is common in MDS, even in patients without 6p rearrangements. Clarification of the role of IER3 in MDS is likely to yield new insights into MDS pathobiology.
Supplementary Material
Acknowledgment
DPS thanks Scott H. Kaufmann, MD PhD, for valuable advice.
Support: K12 CA90628 (National Cancer Institute), Robert A. Kyle Hematologic Malignancies Fund, and the Henry J. Predolin Foundation (DPS); 2 P50 CA100707-067285 to PLB.
Footnotes
Disclosures: No relevant.
REFERENCES
- 1.Kondratyev AD, Chung KN, Jung MO. Identification and characterization of a radiation-inducible glycosylated human early-response gene. Cancer research. 1996;56:1498–502. [PubMed] [Google Scholar]
- 2.Feldmann KA, Pittelkow MR, Roche PC, Kumar R, Grande JP. Expression of an immediate early gene, IEX-1, in human tissues. Histochem Cell Biol. 2001;115:489–97. doi: 10.1007/s004180100284. [DOI] [PubMed] [Google Scholar]
- 3.Wu MX. Roles of the stress-induced gene IEX-1 in regulation of cell death and oncogenesis. Apoptosis. 2003;8:11–8. doi: 10.1023/a:1021688600370. [DOI] [PubMed] [Google Scholar]
- 4.Kumar R, Pittelkow MR, Salisbury JL, et al. A novel vitamin D-regulated immediate-early gene, IEX-1, alters cellular growth and apoptosis. Recent Results Cancer Res. 2003;164:123–34. doi: 10.1007/978-3-642-55580-0_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang YH, Wu JY, Zhang Y, Wu MX. Synergistic and opposing regulation of the stress-responsive gene IEX-1 by p53, c-Myc, and multiple NF-kappaB/rel complexes. Oncogene. 2002;21:6819–28. doi: 10.1038/sj.onc.1205854. [DOI] [PubMed] [Google Scholar]
- 6.Wu MX, Ao Z, Prasad KV, Wu R, Schlossman SF. IEX-1L, an apoptosis inhibitor involved in NF-kappaB-mediated cell survival. Science. 1998;281:998–1001. doi: 10.1126/science.281.5379.998. New York, NY. [DOI] [PubMed] [Google Scholar]
- 7.Im HJ, Pittelkow MR, Kumar R. Divergent regulation of the growth-promoting gene IEX-1 by the p53 tumor suppressor and Sp1. The Journal of biological chemistry. 2002;277:14612–21. doi: 10.1074/jbc.M109414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen L, Guo J, Santos-Berrios C, Wu MX. Distinct domains for anti- and pro-apoptotic activities of IEX-1. The Journal of biological chemistry. 2006;281:15304–11. doi: 10.1074/jbc.M600054200. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Schlossman SF, Edwards RA, Ou CN, Gu J, Wu MX. Impaired apoptosis, extended duration of immune responses, and a lupus-like autoimmune disease in IEX-1-transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:878–83. doi: 10.1073/pnas.022326699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Finegold MJ, Porteu F, Kanteti P, Wu MX. Development of T-cell lymphomas in Emu-IEX-1 mice. Oncogene. 2003;22:6845–51. doi: 10.1038/sj.onc.1206707. [DOI] [PubMed] [Google Scholar]
- 11.Sommer SL, Berndt TJ, Frank E, et al. Elevated blood pressure and cardiac hypertrophy after ablation of the gly96/IEX-1 gene. J Appl Physiol. 2006;100:707–16. doi: 10.1152/japplphysiol.00306.2005. [DOI] [PubMed] [Google Scholar]
- 12.Dilley WG, Kalyanaraman S, Verma S, Cobb JP, Laramie JM, Lairmore TC. Global gene expression in neuroendocrine tumors from patients with the MEN1 syndrome. Mol Cancer. 2005;4:9. doi: 10.1186/1476-4598-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasada T, Azuma K, Hirai T, et al. Prognostic significance of the immediate early response gene X-1 (IEX-1) expression in pancreatic cancer. Ann Surg Oncol. 2008;15:609–17. doi: 10.1245/s10434-007-9669-0. [DOI] [PubMed] [Google Scholar]
- 14.Hofmann WK, de Vos S, Komor M, Hoelzer D, Wachsman W, Koeffler HP. Characterization of gene expression of CD34+ cells from normal and myelodysplastic bone marrow. Blood. 2002;100:3553–60. doi: 10.1182/blood.V100.10.3553. [DOI] [PubMed] [Google Scholar]
- 15.Prall WC, Czibere A, Grall F, et al. Differential gene expression of bone marrow-derived CD34+ cells is associated with survival of patients suffering from myelodysplastic syndrome. Int J Hematol. 2009 doi: 10.1007/s12185-008-0242-9. [DOI] [PubMed] [Google Scholar]
- 16.Sebens Muerkoster S, Rausch AV, Isberner A, et al. The apoptosis-inducing effect of gastrin on colorectal cancer cells relates to an increased IEX-1 expression mediating NF-kappa B inhibition. Oncogene. 2008;27:1122–34. doi: 10.1038/sj.onc.1210728. [DOI] [PubMed] [Google Scholar]
- 17.Semlali A, Oliva J, Badia E, Pons M, Duchesne MJ. Immediate early gene X-1 (IEX-1), a hydroxytamoxifen regulated gene with increased stimulation in MCF-7 derived resistant breast cancer cells. The Journal of steroid biochemistry and molecular biology. 2004;88:247–59. doi: 10.1016/j.jsbmb.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Highsmith WE, Jr., Meyer KJ, Marley VM, Jenkins RB, et al. Current protocols in human genetics / editorial board, Jonathan L Haines. 2007. Conversion Technology for the separation of maternal and paternal copies of any autosomal chromosome in somatic cell hybrids. Chapter 3: Unit 3 6. [DOI] [PubMed] [Google Scholar]
- 19.Greenberg P, Cox C, LeBeau MM, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89:2079–88. [PubMed] [Google Scholar]
- 20.Hamelin V, Letourneux C, Romeo PH, Porteu F, Gaudry M. Thrombopoietin regulates IEX-1 gene expression through ERK-induced AML1 phosphorylation. Blood. 2006;107:3106–13. doi: 10.1182/blood-2005-07-2953. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.