Abstract
All members of the human herpesvirus protease family are active as weakly associating dimers, but inactive as monomers. A small molecule allosteric inhibitor of Kaposi’s sarcoma-associated herpesvirus protease (KSHV Pr) traps the enzyme in an inactive monomeric state where the C-terminal helices are unfolded and the hydrophobic dimer interface is exposed. NMR titration studies demonstrate that the inhibitor binds to KSHV Pr monomers with low μM affinity. A 2.0 Å resolution X-ray crystal structure of a C-terminal truncated KSHV Pr-inhibitor complex locates the binding pocket at the dimer interface and displays significant conformational perturbations at the active site, 15 Å from the allosteric site. NMR and CD data suggest that the small molecule inhibits human cytomegalovirus protease (HCMV Pr) via a similar mechanism. As all HHV proteases are functionally and structurally homologous, the inhibitor represents a class of compounds that may be developed into broad-spectrum therapeutics which allosterically regulate enzymatic activity by disrupting protein-protein interactions.
Keywords: NMR spectroscopy, X-ray crystallography, protein-protein interactions, small-molecule inhibitor, monomer trap
Introduction
Proteins exhibit conformational selection that can have significant effects on their activity. In some cases, proteins exhibit intrinsic disorder as part of their regulatory mechanism.1,2 Processes of conformational selection, in the form of protein-protein interactions (PPIs), are governed by the internal motions of the individual subunits of the complex. Non-ideal interactions within these macromolecular complexes may result in misregulation of function, and, eventually to disease. Conversely, optimal binding between protein partners may actually enhance a pre-existing condition. To better understand these relationships, a recent upsurge of attention has been paid to linking protein dynamics and regulation of PPIs towards disease and drug discovery.3–6 As a result, PPIs have gained more traction as targets for therapeutics.7,8
Until recently, the most successful mediators of PPIs had been antibody or peptide based.9 Though quite powerful, both modalities have their liabilities. A growing number of examples of preclinical compounds that target PPIs have focused on small molecules that mimic protein structural motifs, such as α-helices.10,11 Although a key feature of PPIs are large, relatively flat hydrophobic surfaces devoid of deep pockets or crevices, recent studies have indicated that these small molecule therapeutics may only need to bind a small subset of the interface residues, termed the “hot spot”.12 Notably, bioinformatic analyses have indicated the presence of aromatic residues at these hot spots, with tryptophan the most commonly occurring.13
PPIs play a significant role in the activity of human herpesvirus (HHV) proteases. Two archetypal members of the HHV protease family include Kaposi’s sarcoma-associated herpesvirus protease (KSHV Pr) and human cytomegalovirus protease (HCMV Pr). As with other structurally and functionally homologous HHV Pr family members, both KSHV Pr and HCMV Pr exist in equilibrium between an inactive monomeric and an active, weakly associating dimeric state. The proteolytically active dimer is critical for the viral lifecycle. The interface of all HHV Pr dimers consists of two α-helices (helix 5, one from each monomer), which buries an approximate 2500 Å2 hydrophobic surface on each partner monomeric unit. Each monomer contains a non-canonical Ser-His-His catalytic triad and an accompanying substrate binding pocket located approximately 15–20 Å from the dimer interface (Fig. 1a–b).14
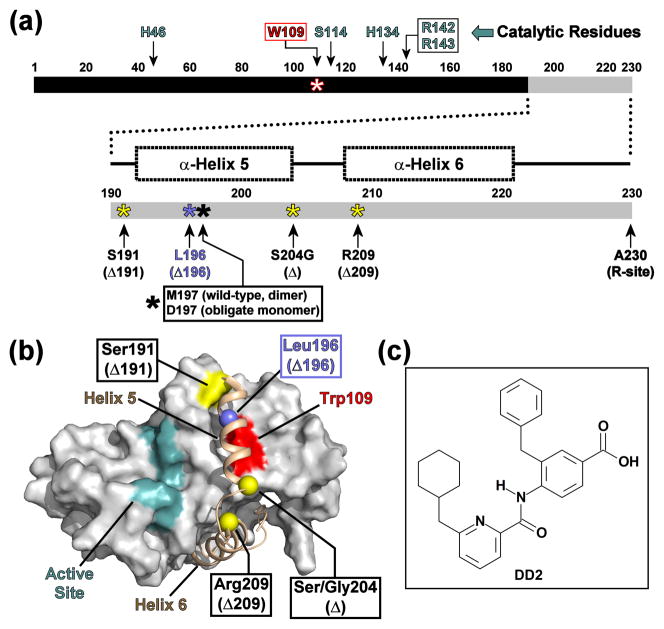
Fig. 1. Domain diagram of KSHV Pr.
(a) Linear domain diagram of KSHV Pr displaying the positions of the “hot spot” Trp109 (red), catalytic residues (cyan) and the conformationally dynamic C-terminus (gray). C-terminal truncations are indicated by yellow or blue asterisks. (b) The dimer interface of a KSHV Pr monomer (2PBK). The partner monomer is omitted for clarity. The active site (cyan), the inhibitor-binding “hot spot” Trp109 (red), and truncation sites (yellow and blue balls) are indicated as in Figure 1a. See also Movie S1. (c) The chemical structure of DD2, an optimized analog of a first generation lead inhibitor of KSHV Pr.21
Previous structural studies performed on KSHV Pr suggested that the trigger for the concentration-dependent dimer formation is a disorder-to-order transition of the C-terminal residues.14,15 Notably, single point mutations of a key residue within helix 5 influence this equilibrium: Met197-to-Asp (M197D) results in an inactive obligate monomer,14 while Met197-to-Leu (M197L) stabilizes the dimer.16 NMR-based chemical shift perturbation mapping and hydrogen-deuterium exchange experiments performed on the KSHV Pr obligate monomer demonstrated that residues 191–230, which constitute the helices 5 and 6 in the dimer, are conformationally dynamic.14 An obligate monomeric version of KSHV Pr in which helix 6 was “stapled” to the core structure via an engineered disulfide bond displayed enhanced enzymatic activity in the oxidized state relative to the reduced state.14 This result suggested that the positioning of helix 6 is also critical for stabilizing the active dimeric conformation of KSHV Pr.
In light of these observations, we focused our efforts towards discovering small molecule ligands that allosterically regulate HHV Pr activity by disrupting dimerization. Since proteins and enzymes are able to sample multiple pre-existing conformational states,6,17 one such inhibitory mechanism is to capture HHV proteases in their inactive monomeric forms. Regulating this conformational switch, which is believed to be conserved across all HHV proteases, represents an unexploited pathway for the development of potential broad-spectrum therapeutics against herpesviruses. In contrast to the successful antivirals used for AIDS treatment that target proteolytic activity of HIV protease, no active-site inhibitors that target HHV proteases have been successfully developed as clinical therapeutics. Successful disruption of the herpesvirus protease dimer offers the potential to resurrect a promising family of drug targets that were deemed undruggable due to the limited efficacy of active-site directed inhibitors.18–20
To validate this approach, we previously demonstrated that addition of a 30-residue helical peptide abolished protease activity by disrupting KSHV Pr dimerization.16 Potential KSHV Pr inhibitors from small molecule helical mimetic libraries were then screened via a high-throughput fluorescence-based activity assay.21 One of the lead candidates, DD2 (Fig. 1c), resulted from chemical optimization of an initial screening hit, and exhibited an IC50 value of 3.1 ± 0.2 μM against KSHV Pr. DD2 is a diaryl-substituted 4-(pyridine-2-amido) benzoic acid. Initial 13C- and 15N-based NMR titration mapping studies indicated that DD2 disrupts dimerization by binding at or near Trp109, which is located in the center of the hydrophobic dimer interface.21 This led to a proposed “monomer trap” model of inhibition: DD2 alters the KSHV Pr monomer-dimer equilibrium by capturing a pre-existing inactive monomer and shifting the population of conformers from the active dimeric state.
In this study, we answer two key questions. (1) How does a small molecule alter the conformation of a dimeric enzyme in order to trap an inactive monomeric conformation? (2) Is this inhibitory mechanism applicable to other members of a family of related enzymes? In particular, we use a C-terminal truncation variant of monomeric KSHV Pr to characterize HHV Pr-DD2 interactions via NMR spectroscopy and X-ray crystallography. By employing this truncated variant, we confirm that KSHV Pr-DD2 binding occurs, even in the absence of the conformationally dynamic C-terminus. We also report the first crystallographic structure of an allosterically inhibited HHV Pr monomer to date. Finally, using the structurally and functionally homologous human cytomegalovirus (HCMV Pr), we demonstrate that, in general, dimer dissociation is a viable allosteric route towards inhibiting HHV Pr activity.
Results And Discussion
Truncation of the C-terminal helices does not affect the core KSHV Pr structure
Residues 191 – 230 constitute helices 5 and 6 in the KSHV Pr dimer, but are conformationally dynamic and only partially structured in the case of the KSHV Pr M197D obligate monomer.14 Truncation of the monomeric KSHV Pr sequence to its native autolysis site, S204, (henceforth, Δ, Fig. 1a–b) displayed no significant chemical shift perturbations with respect to the full-length obligate monomer in the 15N-1H HSQC spectra.15 This suggested that the core structure of the KSHV Pr monomer (residues 1–191) remains intact, even with the absence of helix 6. Determining the minimal construct required for the correct fold of the monomeric protease core was deemed necessary for optimizing both NMR and crystallographic studies. Other constructs were therefore designed to remove portions of the C-terminal helices (Fig. 1a–b, Movie S1): KSHV Pr M197D Δ209 (residues 1–209; Δ209); KSHV Pr Δ196 (residues 1–196; Δ196); and KSHV Pr Δ191 (residues 1–191; Δ191). As with the Δ construct, size exclusion chromatogram elution profiles and 15N-1H HSQC spectra indicate that the resulting truncations are expressed as monomers, with the core protease structure intact and well-folded (Fig. S1). Since DD2 binds to the obligate monomer,21 these newly created truncations can be used to simplify further small-molecule inhibitor binding studies.
Isoleucine δ1-methyl groups act as KSHV Pr dimer interface binding probes
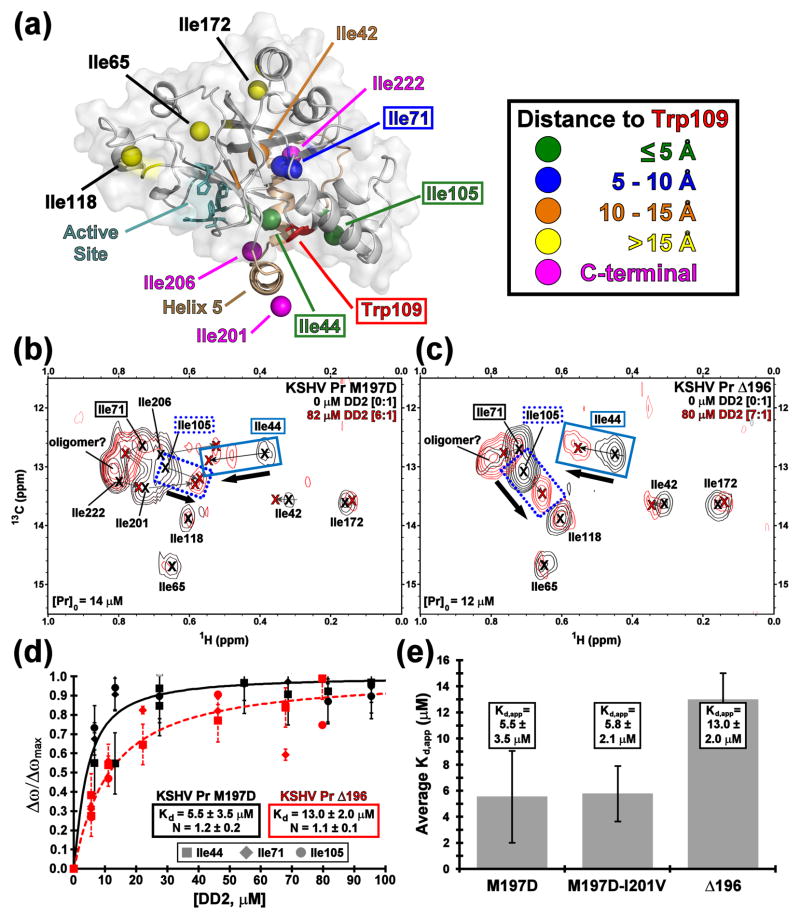
Previous NMR studies of the KSHV Pr M197D monomer reported the hydrophobic dimer interface anchored by Trp109 as the DD2 binding “hot spot”.21 However, significant backbone amide peak broadening was observed in the 15N-1H HSQC spectra during the titration. This result is indicative of conformational or chemical exchange on an intermediate NMR timescale, and precludes quantification of the binding effect. Subsequently, the non branched δ1-methyl groups of isoleucine residues were used as NMR binding probes. Each monomeric unit of the KSHV Pr dimer structure (PDB accession code 2PBK)22 contains ten isoleucine residues, which can be separated into five “zones” in relation to their distance from Trp109 (Fig. 2a). The 13C-1H HSQC spectrum of selective [13C-1H methyl] Ile-Leu-Val (ILV) labeled KSHV Pr M197D displays 10 relatively well-dispersed δ1-methyl isoleucine resonances (Fig. S2). In order to assign the resonance peaks, Ile-to-Val mutations were performed on the M197D and Δ196 constructs. As with the truncations, both 13C-1H (Fig. S3) and 15N-1H HSQC (data not shown) spectra of the Ile-to-Val mutants indicate no significant structural perturbation in the KSHV Pr core.
Fig. 2. KSHV Pr-DD2 titration.
(a) The distribution of isoleucine δ1-methyl groups on a full length KSHV Pr monomer structure, based upon 2PBK. The view is a 90 degree rotation about the horizontal axis, relative to that in Figure 1b. Isoleucines separate into five zones with respect to distance from Trp109 as indicated by colored balls. The 13C-1H HSQC titration spectra of the KSHV Pr M197D (b) and Δ196 (c) constructs with DD2, focusing on the isoleucine δ1-methyl region. The spectral overlays display apo (black) and > 5 molar equivalents DD2 (red). Ile44 (solid blue box), Ile105 (dotted blue box), and Ile71 (solid black box) are used as the binding probes. (d) The binding curves (M197D, solid black; Δ196, dashed red) represent the average apparent Kd values calculated for the three Ile probes, and are summarized as a bar chart (e). HSQC titration spectra and binding curves for the M197D-I201V construct are displayed in Figure S4.
As expected, Ile44 and Ile105, both located within 5 Å of the KSHV Pr dimer interface, display the largest chemical shift resonance perturbations upon truncation of the C-terminal residues (Fig. S2). Ile71 is located within 10 Å of the KSHV Pr dimer interface and exhibits moderate resonance perturbations. Conversely, isoleucine residues located greater than 10 Å from the dimer interface exhibit little or no resonance perturbations. In addition, overlap of the Ile71-le206 and Ile105-Ile201 resonances are observed in the Δ209 13C-1H HSQC spectrum, but not in the corresponding Δ196 spectrum (Fig. S2). Although the Δ191 and Δ196 spectra exhibit no differences (data not shown), the Δ191 construct displayed greater tendencies to aggregate in solution under the current NMR conditions. As a result, DD2 binding studies were performed using the Δ196 construct, in comparison with the full-length M197D obligate monomer.
DD2 binds to KSHV Pr in the presence and absence of the dynamic C-terminus
Addition of greater than five molar equivalents of DD2 to the ILV-labeled M197D obligate monomer induced significant chemical shift perturbations of the isoleucine residues located at the KSHV Pr dimer interface (Fig. 2b). Here, the methyl resonances of the isoleucine residues most proximal to the “hot spot” Trp109 (Ile44, Ile71, and Ile105) were the most affected. Not unexpectedly, resonances of the C-terminal isoleucines (Ile201, Ile206, and Ile222) are also perturbed, as these are the residues that would transiently interact with the KSHV Pr dimer interface in the absence of DD2. Because the Ile105 and Ile201 resonances of KSHV Pr M197D overlapped, the KSHV Pr M197D-I201V construct was also examined (Fig. S4). All isoleucine peaks in the M197D-I201V construct displayed similar resonances as those in the “wild-type” obligate monomer. Moreover, the absent Ile201 peak in the M197D-I201V construct allows for easier determination of the Ile105 chemical shift resonances. DD2 titration spectra acquired on the Δ196 truncation (Fig. 2c) display the same general patterns as observed for the full-length KSHV Pr monomer analogs.
Ile44, Ile71, and Ile105 were chosen as NMR reporter probes due to their proximity of less than 10 Å to the “hot spot” Trp109. The Hill equation was used to perform a nonlinear regression curvefit analysis of the δ1-methyl group chemical shift resonances as a function of total ligand concentration (Fig. 2d). The apparent Kd values from these three reporter residues were then averaged to obtain an estimate of DD2 binding. The resulting NMR-based binding curves indicate that DD2 binds with equal affinity to the M197D (Kd,app = 5.5 ± 3.5 μM) and M197D-I201V (Kd,app = 5.8 ± 2.1 μM) constructs (Fig. 2e). Notably, no binding events were observed for any of the KSHV Pr constructs using surface plasmon resonance or isothermal titration calorimetry (data not shown). DD2 appears to bind to the Δ196 construct with lower affinity (Kd,app = 13.0 ± 2.0 μM). However, this observation may be a reflection of the presence (M197D or M197D-I201V variants) or absence (Δ196 construct) of C-terminal residues transiently interacting with Trp109. Collectively, the NMR titration data clearly demonstrate that mutations or deletions of residues within the conformationally dynamic C-terminus have no significant effect towards DD2 binding affinity.
The crystal structure of the KSHV Pr Δ196-DD2 complex
In parallel to the NMR-based DD2 titration studies, a 2.0 Å resolution crystal structure of the KSHV Pr-DD2 complex was obtained (PDB accession code 3NJQ). To date, there have been no reported NMR or X-ray crystallographic structures of an allosterically inhibited HHV Pr monomer. In general, the majority of the published HHV Pr structures have either been in their respective apo or active-site inhibited dimeric forms, as illustrated by the apo (PDB accession code 1FL1)23 and covalently bound peptide phosphonate-inhibited (2PBK)22 dimeric KSHV Pr structures. Prior attempts to crystallize the full-length uninhibited KSHV Pr M197D obligate monomer proved fruitless, possibly due to the conformationally dynamic C-terminal residues disrupting the formation of a stable crystal lattice. Crystallization trials using the KSHV Pr M197D, Δ209, and Δ191 constructs yielded no appreciable crystals, either in the presence or absence of DD2. However, the Δ196 construct in the presence of excess molar equivalents of DD2 yielded small cube-like crystals that were used to acquire a 2.0 Å resolution X-ray diffraction data set (Table 1, Table S1). Notably, no crystals were observed for the apo Δ196 under similar reservoir conditions.
Table 1.
Summary of Crystallographic Information, 3NJQ
| Data collection and processing | ||
| Number of crystals used | 1 | |
| Wavelength (Å) | 1 | |
| Space group | I 222 | |
| Unit cell parameters | ||
| a, b, c (Å) | 69.3, 95.9, 119.2 | |
| α, β, γ (°) | 90, 90, 90 | |
| Matthews coefficient (A3/Da) | 2.32 | |
| Solvent content (%) | 46.5 | |
| Molecules per asymmetric unit | 2 | |
| Beamline | ALS 8.3.1 | |
| Diffraction Data | ||
| Resolution range (Å) | 50 – 2.0 | (2.1 – 2.0) |
| Unique reflections | 27221 | (1200) |
| R(I)sym (%)a | 6.3 | (31.1) |
| Completeness (%) | 99.9 | (70.1) |
| Redundancy | 4.1 | (2.0) |
| I/σ(I) | 30.5 | (3.3) |
| Refinement | ||
| Resolution range (Å) | 48 – 2.0 | |
| Reflections used in refinement (work/free) | 25855/1366 | |
| Final R values for all reflections (workb/freec) (%) | 20.3/24.8 | |
| Protein residues | 377 | |
| Inhibitor | 3 | |
| Water molecules | 106 | |
| RMS Deviations | ||
| Bonds (Å) | 0.019 | |
| Angles (°) | 1.98 | |
| Ramachandran parametersd | ||
| Residues in most favored regions | 90.0% | |
| Residues in additional allowed regions | 8.7% | |
| Residues in generously allowed regions | 0.6% | |
| Mean B-factor (Å2) | ||
| Protein | 46.4 | |
| Inhibitors | ||
| 3NJQ A 197 (DD2-A) | 31.0 | |
| 3NJQ B 198 (DD2-B) | 29.0 | |
| 3NJQ B 199 (DD2-C, bridging molecule) | 33.6 | |
| Water molecules | 27.0 | |
Numbers in parentheses represent the highest resolution shell.
Rsym = (Σ |(I-〈I〉)|/ΣI), where 〈I〉 is the average intensity of multiple measurements.
Rwork = (Σ |(Fobs – Fcalc)|/Σ|Fobs|)
Rfree = Rwork based on ~ 1000 (at least 10%) of reflections excluded from refinement
Calculated using Procheck51
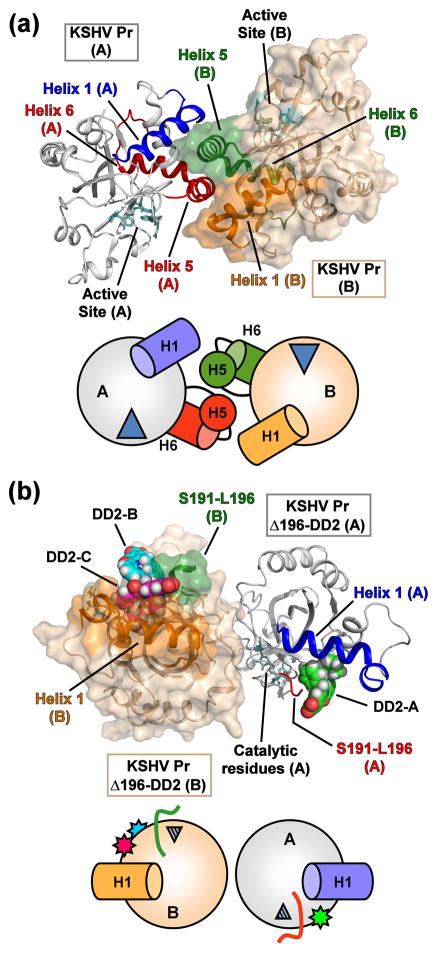
In contrast to the solution state, where all the KSHV Pr constructs are monomeric in the NMR spectra, the Δ196-DD2 complex crystallizes as an asymmetric pair of KSHV Pr Δ196 monomers containing three DD2 molecules (Fig. 3). Previously published structures of the full-length KSHV Pr dimers22,23 contain two symmetrical monomers centered about a C2 rotation axis. While the dimer interface of proteolytically active KSHV Pr consists of interfacial α-helices and a hydrophobic surface centered on Trp109, the Δ196-DD2 complex forms a dimer on the distal side of the molecule with respect to Trp109. The dimer interface of the Δ196-DD2 complex buries approximately 1800 Å2 total surface area, which mostly consists of hydrophilic residues, with the crystal structure containing a number of water molecules between the two monomers. Both monomers in the asymmetric unit are conformationally similar with respect to one another and to residues 1–196 of the previously published KSHV Pr dimer structures (1FL1 and 2PBK), with overall RMS deviations less than 1.0 Å for backbone and heavy atom overlays (Table S2). Comparing a monomeric unit of the KSHV Pr dimer and the Δ196-DD2 complex structures reveals two significant differences: the formation of an allosteric DD2 binding pocket and the conformational perturbation of the active site.
Fig. 3. Structural comparison of the “apo” and DD2-inhibited KSHV Pr monomers.
(a) The dimer structure of peptide-phosphonate inhibited KSHV Pr (2PBK). The catalytic residues (cyan) are located ~ 15 – 20 Å from the dimer interface. The interfacial helix 5 and the following helix 6 (monomer A, red; monomer B, green) are displayed. Helix 1 of monomer A (blue) and monomer B (orange) also form a portion of the dimer interface and are aligned in an anti-parallel orientation with respect to each other. (b) The structure of the KSHV Pr Δ196-DD2 complex (3NJQ) crystallizes as an asymmetric dimer, with dimerization occurring on the opposite face with respect to 2PBK. DD2 molecules bind to the hydrophobic surface normally occupied by helix 5. Monomer A of the complex contains one DD2 molecule (pose 1, green carbons), while monomer B contains two DD2 molecules (pose 2, magenta carbons; pose 3, cyan carbons). The truncated C-terminal residues of the Δ196 constructs (red, monomer A; green, monomer B) are also indicated. Helix 1 of monomer A (blue) and monomer B (orange) are oriented end-on-end with respect to each other. Below each structure are cartoon representations of the monomeric units, with the wedges representing the active site, and stars the DD2 molecules.
The “hot spot” Tryptophan 109 acts as a hinge to create the DD2 binding pocket
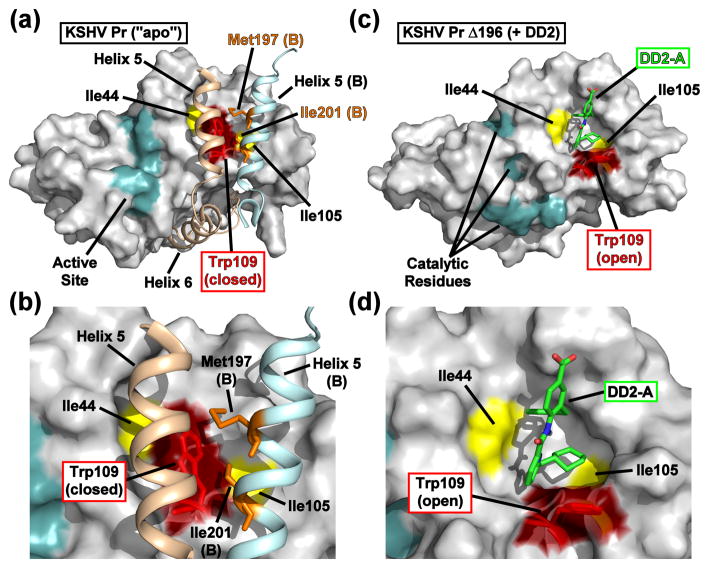
One of the major differences in the backbone conformation between the “apo” and DD2-bound states is apparent in the α1-α2 loop (residues 87–99) and helix 2 (residues 100–110) (Fig. 4 and Movie S2). In the case of the enzymatically active KSHV Pr dimer, the Trp109 indole ring adopts a “closed” conformation in each monomer, forming a relatively flat hydrophobic surface which interacts with the two interfacial helices. Dimerization is stabilized by the Met197 and Ile201 sidechains of the partner monomer, forming intermolecular hydrophobic interactions with Trp109 (Fig. 4a–b).21,22 Conversely, the Trp109 indole ring adopts an “open” conformation in each monomer of the Δ196-DD2 complex, creating a hydrophobic cavity that acts as the DD2 binding pocket (Fig. 4c–d).
Fig. 4. Comparison of the apo and DD2-bound KSHV Pr crystal structures.
(a) The dimer interface of KSHV Pr (2PBK) consists of two helices from each monomeric unit (helix 5, tan, monomer A; light blue, monomer B), which stabilize the active site (cyan) via the C-terminal helix 6 and occlude the Trp109 (red). (b) The Met197 and Ile201 sidechains from helix 5 of monomer B (orange) form hydrophobic interactions with Trp109 of monomer A. Both Δ196-DD2 monomer A (c) and monomer B (Fig. S5) exhibit independent DD2 binding pockets in which the Trp109 sidechain indole ring (red) adopts an “open” form (d). See also Movie S2.
The DD2 binding pocket of the KSHV Pr Δ196-DD2 complex
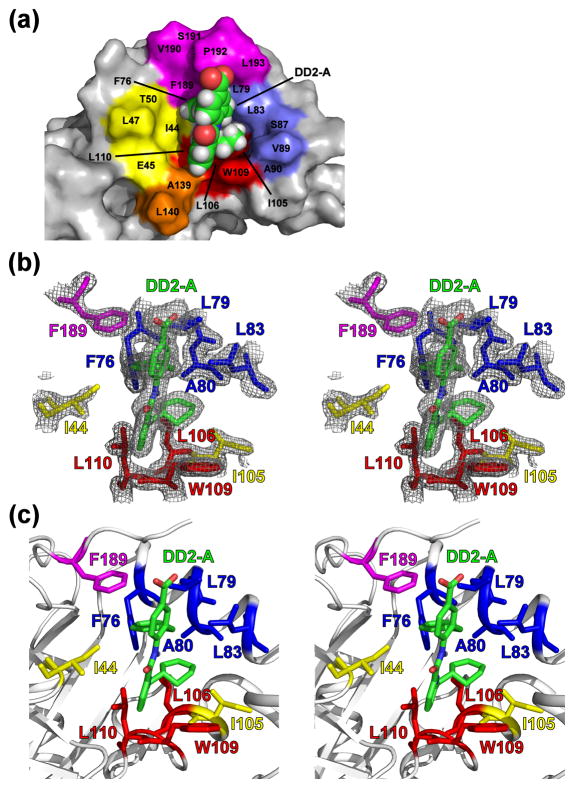
The DD2 binding pocket is dominated by aliphatic residues located in several secondary structure motifs. Residues within the β2-β3 loop (residues 44–52), helix 1 (residues 73–91), helix 2 (residues 100–110), the β6-β7 loop (residues 139–148), and the C-terminus (residues 189–196) are involved (Fig. 5, Fig. S5). With the exception of residues 189–196, the backbone conformation of the DD2 binding pockets are similar in the two asymmetric Δ196-DD2 monomers. Minor fluctuations in the orientation of the Trp109 sidechain indole ring and in the loop located between helix 1 and helix 2 suggest that the binding pocket may be able to sample other conformational states, even in the presence of the small-molecule inhibitor.
Fig. 5. The DD2 binding pocket.
(a) The hydrophobic DD2 binding pocket is composed of aliphatic residues from the β2-β3 loop (yellow), helix 1 and the α1-α2 loop (blue), helix 2 (red), the β6-β7 loop (orange), and the C-terminus (magenta). DD2 (green carbons) is shown as a space-filling model. (b) Stereoview of DD2 (green) within the Δ196 binding pocket of monomer A, in relation to the “hot spot” Trp109 (red) and the Ile44 and Ile105 reporter groups (yellow). Also displayed are buried aliphatic residues of helix 1 (blue), helix 2 (red), and the C-terminus (magenta) that compose the binding pocket. The mesh represents the 2Fo-Fc 1σ electron density map. (c) Figure 5b with the protein backbone ribbons displayed. Comparable views of monomer B are displayed in Figure S5.
Notably, the asymmetric dimer contains three DD2 poses. While monomer A (Fig. 5) contains only one DD2 molecule (DD2-A) in the binding pocket, monomer B (Fig. S5) displays two DD2 molecules (DD2-B and DD2-C). Inspection of the two monomers indicates that DD2-A and DD2-B are located within the DD2 binding pocket. Positioning of the DD2-A and DD2-B molecules within the binding pocket are offset by an approximate 80° rotation about the vertical axis of the “backbone” atoms (Fig 6). The differences in the rotation axis and backbone positioning are due to the DD2-C molecule displacing DD2-B. DD2-C is located outside the hydrophobic cavity and appears to be a consequence of a crystal contact, acting as a “bridge” molecule between adjoining unit cells. The average B-factor values observed for the three DD2 molecules (31.0 Å2, 29.0 Å2, 33.6 Å2), as well as residues within close proximity (≤ 5 Å) to the inhibitor (33.4 Å2), are lower than the overall mean B-factor of the complex (46.4 Å2). The specific individual B-factors are listed in Tables 1 and S3. In addition, each of the DD2 molecules exhibit occupancies of 1.0 in the 2Fo-Fc difference (Fig. 5 and S5) and Fo-Fc omit maps (Fig. S6). These results suggest that, upon DD2 binding, the sidechains of the hydrophobic binding pocket samples less conformational mobility relative to the rest of the structure.
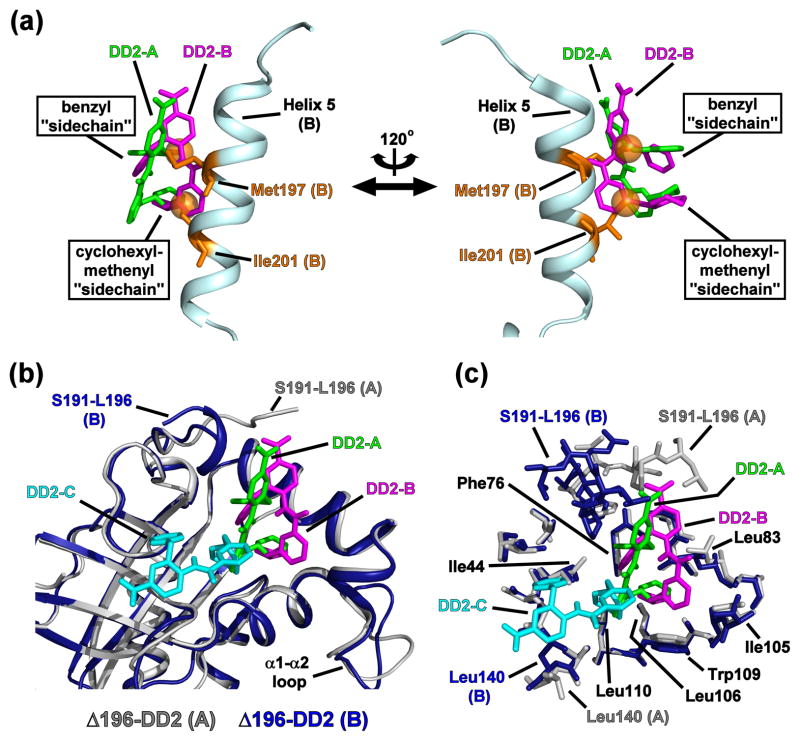
Fig. 6. The conformations of KSHV Pr-bound DD2 in situ.
(a) DD2-A (green) and DD2-B (magenta) within the Δ196-DD2 complex are overlaid with Helix 5 of the KSHV Pr partner monomer B (cyan). Orange balls represent the non-branched methyl groups of Met197 and Ile201. The DD2 “sidechains” match the relative positions observed for those of the native Met197 and Ile201, and are inserted into the hydrophobic pocket vacated by the Trp109 indole ring. The protein monomer structures of the Δ196-DD2 complexes and the apo KSHV Pr dimer are omitted for clarity. (b) An overlay of Δ196-DD2 monomers with their constituent DD2 molecules. Monomer A (gray) contains DD2-A (green), while monomer B (dark blue) contains DD2-B (magenta) and DD2-C (cyan). DD2-C is an artifact of crystal packing and is situated outside the DD2 binding pocket. Major conformational differences in the DD2 binding pocket are only observed for the C-terminal residues (Ser191 – Leu196). (c) An overlay of Δ196-DD2 monomers with their respective DD2 molecules displaying the sidechains of the residues constituting the DD2 binding pocket. The benzyl “sidechain” of DD2-A and DD2-B are in close proximity to Phe76, while the cyclohexyl ring interacts with Ile105, Leu106, and Leu110.
Overall RMS deviation values of the three conformers suggest that the individual DD2 structures are similar (Table S4). However, visual inspection of their structural overlays indicates that poses 1 and 2 are more closely related to each other than to DD2-C (Fig. S7). The DD2 benzyl and cyclohexyl-methylenyl “sidechain” groups adopt the same relative elevations and positions within the KSHV Pr binding pocket as the Met197 and Ile201 sidechains of the partner monomer in the active dimeric enzyme (Fig. 6a). This suggests that the DD2 “sidechains” mimic the i → i+4 positioning inherent with sidechains of residues that adopt a helical conformation. Moreover, the DD2 “sidechains” of both DD2-A and DD2-B are enveloped by the hydrophobic cavity formed upon the Trp109 indole ring adopting the “open” position (Fig. 5, Fig. S5). Notably, the DD2-A and DD2-B “sidechains” occupy the same conformational space (Fig. 6). In this case, the cyclohexyl rings are co-planar, but offset by one carbon atom, while the plane of the benzyl rings are rotated at an approximate 40° angle. Selected inter-atomic distances between the DD2 sidechain carbons and residues within the Δ196 binding pocket are reported in Table S5.
Further examination of the DD2 conformations may also explain the structure-activity relationships in the helical mimetic library used in the initial fluorescence-based activity screening. Of our original optimized mimetic library, only DD2 contained an amide group in the ortho position with respect to the nitrogen atom of the pyridinyl ring.21 The stacking positions of the cyclic DD2 “sidechain” groups prevent the amide proton and the pyridinyl nitrogen from interacting with the protease, either directly or via a water molecule (Fig. S7). Interatomic distances and “backbone” dihedrals measured from the crystallographic poses of DD2 suggest that the amide proton and pyridinyl nitrogen may form a weak intramolecular hydrogen bond (Table S4). Such interactions may help stabilize the conformation necessary for efficient binding to KSHV Pr. Further alterations to the DD2 “backbone”, such as the addition of polar functional groups, would be required to enhance solubility and, perhaps, increase efficacy towards the HHV proteases.
Loss of helix 6 disrupts the conformation of the KSHV Pr active site
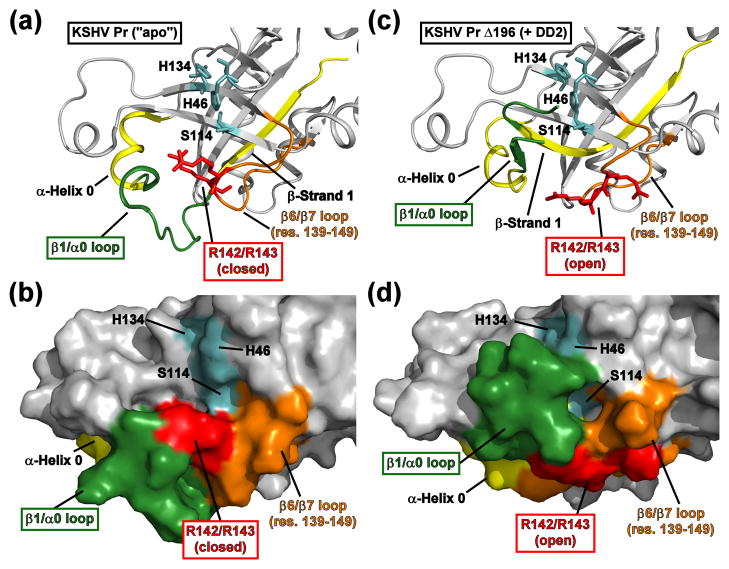
The loss of helix 6 results in a dramatic conformational perturbation of the KSHV Pr active site, located ~ 15 – 20 Å from the dimer interface (Fig. 7, Movie S3). The most noticeable change is observed for the conformation of β-strand 1 (residues 5–13), and is propagated through the β1-α0 loop (residues 14–27) and helix 0 (residues 27–33). In the case of the KSHV Pr dimer, β-strand 1 points outward from the structural core of the enzyme, allowing the β1-α0 loop and the following helices to adopt a conformation suitable for substrate binding and proteolytic cleavage (Fig. 7a–b). Although the β1-α0 loop plays a minimal role in substrate recognition, specifically the S3 pocket,22 its primary function is to help form one of the ridges of the substrate binding pocket. In the case of the Δ196-DD2 complex, the C-terminal portion of β-strand 1 adopts a nearly orthogonal conformation with respect to its corresponding position in the KSHV Pr dimer. This results in a rotation of the β1-α0 loop into a position that collapses the substrate binding pocket and occludes the catalytic triad (Fig. 7c–d).
Fig. 7. Structural perturbation of the KSHV Pr active site upon DD2 binding.
(a-b) The active site of the 2PBK represents an “apo” state of KSHV Pr. The catalytic triad (H46, H134, and S114, cyan) and the conserved oxyanion hole-stabilizing arginine residues (R142 and R143, red) are displayed as sticks. Also highlighted are the positions of β-strand 1 and α-helix 0 (yellow), the β1/α0 loop (dark green), and the β6/β7 loop (orange). Residues 197–230 are omitted for clarity. (c-d) The conformation of the “apo” state active site residues displays clear differences relative to the Δ196-DD2 complex (3NJQ). The Arg142 and Arg143 sidechains (red) adopt a “closed” conformation in the apo state, but an “open” conformation while in complex with DD2. In the DD2-bound state, the β1/α0 loop (dark green) occludes the catalytic triad (cyan) and disrupts the substrate binding pocket. See also Movie S3.
Another striking difference in this conformational rearrangement is the positioning of Arg142 and Arg143 (Fig. 7). These two arginine residues are conserved among all members of the HHV Pr family, and are known to have important roles in stabilizing the structure of the oxyanion hole.24 In the case of KSHV Pr, both the backbone amide and sidechain guanidino groups of Arg142 and Arg143 stabilize peptide substrate interactions within the active site via a series of intermolecular hydrogen bonds.22 The KSHV Pr dimer structure displays the Arg142 and Arg143 sidechains pointing inward towards the catalytic residues and forming a ridge of the substrate binding pocket (Fig. 7a–b). We term this the “closed” state in conjunction with the “closed” Trp109 indole ring. Alternatively, in the structure of the Δ196-DD2 complex, the sidechains of both arginine residues point outward towards the space originally occupied by helix 6 (Fig. 7c–d). We term this the “open” conformational state, analogous to the “open” Trp109 indole ring.
An examination of the average B-factor values indicates that the residues of the catalytic triad (33.6 Å2) are smaller than that of the overall structure (46.4 Å2), but those of the β1-α0 loop (56.6 Å2) are larger. In addition, the average B-factor values for residues of monomer A in the loop containing Arg142 and Arg143 are also significantly larger (59.1 Å2) than the overall average value. The individual B-factor values corresponding to these residues are listed in Table S6. This suggests a general allosteric mechanism of HHV Pr inhibition: that DD2 binding at the dimer interface enhances mobility of the oxyanion hole-stabilizing loop, allowing the active site to be conformationally destabilized. Notably, no significant differences in the average B-factor values are observed for the residues 138–149 of monomer B (35.2 Å2). However, this may reflect the presence of the bridging DD2 molecule in monomer B restricting conformational mobility of the Cys138-Val149 loop.
These observations point to the importance of a structured helix 5 in propagating conformational changes within KSHV Pr. Here, a DD2 molecule captures an inactive KSHV Pr conformation by binding to the hydrophobic patch in the area normally occupied by helix 5, disrupting interactions between the protease structural core and helices 5 and 6. If helix 5 was located at the dimer interface, helix 6 would be in a position to sterically force the sidechains of Arg142 and Arg143 inward towards the structural core of the enzyme, allowing the formation of the active site and oxyanion hole. The loss of helix 6 likely allows the loop containing Arg142 and Arg143 to sample multiple conformational states, effectively destabilizing both the oxyanion hole and the substrate binding pocket.
DD2 inhibits HCMV Protease by binding to the dimer interface
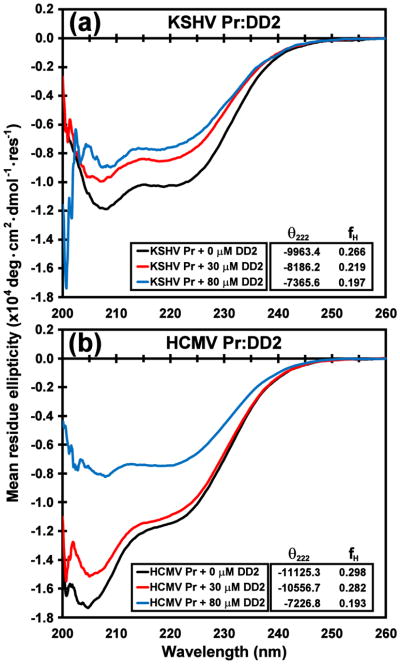
As all eight HHV proteases are structurally and functionally homologous,24 we speculated that DD2 acts upon human cytomegalovirus protease (HCMV Pr) in the same manner as on KSHV Pr. Using a fluorescence-based substrate cleavage kinetics assay previously described for KSHV Pr,21 the in vitro dissociation constant was re-determined for the HCMV Pr monomer-dimer equilibrium as 1.3 ± 0.1 μM. This value is nearly identical to that reported for KSHV Pr (Kd = 1.7 μM) under similar conditions,21,25 indicating that both HHV proteases are weakly associating dimers. Activity assays also indicate that DD2 exhibits four-fold weaker inhibition against HCMV Pr relative to KSHV Pr, with an IC50 value of 12.8 ± 1.1 μM. In addition, circular dichroism spectra of the wild-type HCMV Pr exhibit loss of helical content upon addition of excess molar equivalents DD2 (Fig. 8), suggesting that the C-terminal helices are unfolded.
Fig. 8. CD spectra of DD2 titrations with KSHV Pr and HCMV Pr.
The circular dichroism spectra of ~ 3 μM (a) KSHV Pr and (b) HCMV Pr in the presence of 0 μM (black), 30 μM (red), and 80 μM (blue) DD2. Estimated fractional helicity (fH) values derived from the mean residue ellipticity of the 222 nm band are listed in the insets, and indicate loss of helical content with increasing molar equivalents of DD2. Loss of helicity is a strong indication of HHV protease dimer disruption.
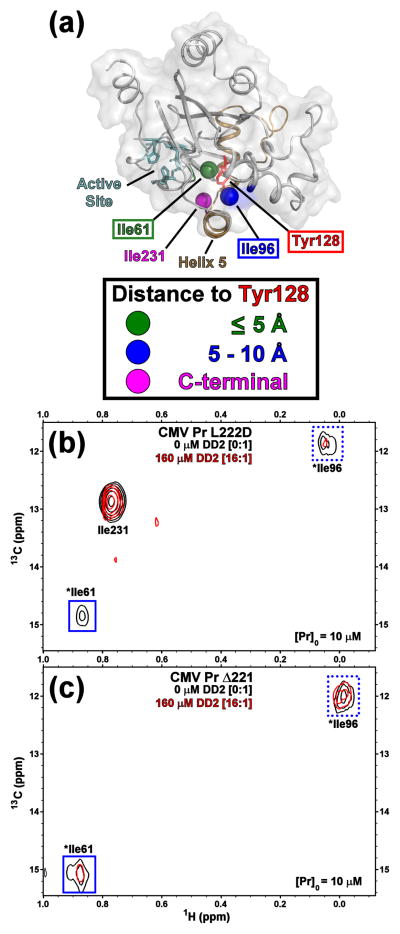
To further probe DD2 interactions with HCMV Pr, HSQC spectra of selective [13C-1H methyl] isoleucine labeled samples were acquired. Two HCMV Pr constructs homologous to the KSHV Pr monomers mentioned above were engineered. The first was an HCMV Pr obligate monomer (L222D), which exhibits an approximate 25-fold reduction in specific protease activity relative to the wild-type sequence (data not shown). The second was a HCMV Pr truncation (Δ221) that mimics the KSHV Pr Δ196 construct. In the case of the full-length HCMV Pr obligate monomer, two isoleucines (Ile61 and Ile96) are positioned at the dimer interface, while the third (Ile231) is located in the C-terminal region. Tyr128 was identified as the potential “hot spot” aromatic residue of HCMV Pr (Fig. 9a). 13C-1H HSQC spectra acquired on the selectively labeled HCMV Pr L222D full length obligate (Fig. 9b) and HCMV Pr Δ221 truncated monomers (Fig. 9c) indicates that DD2 binds to the dimer interface. Both Ile61 and Ile96 resonances are affected upon addition of DD2, with Ile61 exhibiting more extensive peak broadening. Importantly, as in the case of KSHV Pr, DD2 can bind to the C-terminal truncated HCMV Pr analog, but exhibits greater peak broadening and smaller chemical shift perturbations even at higher relative DD2 concentrations. The extensive peak broadening suggests a weaker binding affinity of DD2 to HCMV Pr, relative to KSHV Pr. Collectively, the fluorescence activity assay and the CD and NMR spectra suggest that DD2 binds to HCMV Pr in a structurally homologous pocket as KSHV Pr. The apparent weaker inhibition of HCMV Pr by DD2 may be the result of the smaller Tyr aromatic moiety creating a smaller hydrophobic pocket relative to KSHV Pr. Although the HCMV Pr Ty128 and the KSHV Pr Trp109 residues are out of register by one position in the primary sequences, their sidechains occupy the same general space on the surface of the dimer interface (Fig. 10). With further chemical optimization of DD2, these results provide a promising pathway for developing broad-spectrum allosteric inhibitors for all eight HHV proteases.
Fig. 9. HCMV Pr-DD2 titration data.
(a) The three isoleucine δ1-methyl groups in human CMV Pr are localized at the dimer interface, and color-coded with respect to distance to Tyr128, as indicated. Helix 5 (tan), the active site (cyan), and Tyr128 (red) are also displayed. Tyr128 is homologous to Trp109 of KSHV Pr. The 13C-1H HSQC spectra of selective [13C-1H methyl] isoleucine labeled CMV Pr L222D obligate monomer (b), and Δ221 truncation (c) in the presence of 0 (black) and 16 molar equivalents DD2 (red) indicates DD2 binds at the dimer interface. Both Ile61 and Ile96 are putatively assigned; Ile231 was assigned by the loss of the resonance in the Δ221 truncation.
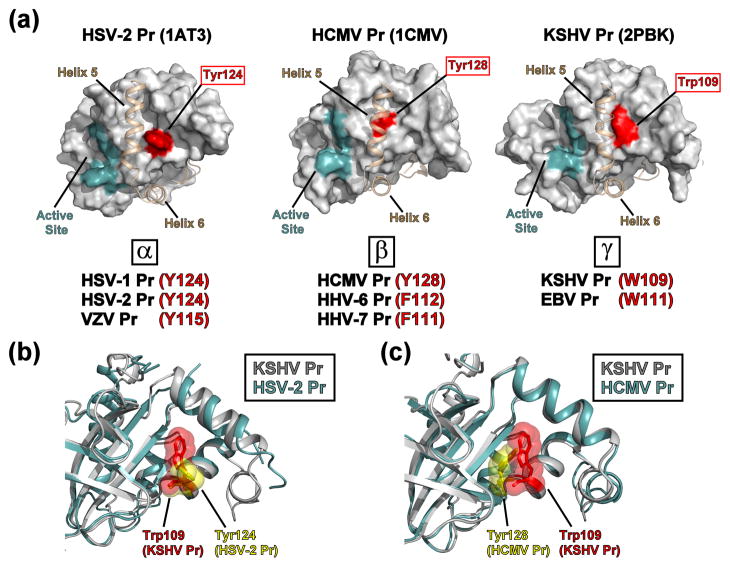
Fig. 10. Structural homology of the HHV protease “hot spots”.
(a) Representative X-ray crystallographic structures of the three structurally homologous HHV protease subfamilies (HSV-2 Pr, 1AT3; HCMV Pr, 1CMV; KSHV Pr, 2PBK), with the active site (cyan) and interfacial helix 5 and following helix 6 (tan) as indicated. “Hot spot” aromatic residues (red) are located at the center of the hydrophobic dimer interface and are potential target sites for small-molecule inhibitors that disrupt protein-protein interactions. α-subfamily HHV proteases: HSV-1 = herpes simplex virus-1/; HSV-2 = herpes simplex virus-2; VZV = Varcella Zoster virus. β-subfamily HHV proteases: HCMV = human cytomegalovirus; HHV-6 = human herpesvirus-6; HHV-7 = human herpesvirus-7. γ-subfamily HHV proteases: KSHV = Kaposi’s sarcoma-associated herpesvirus; EBV = Epstein-Barr virus. (b) Backbone overlay of KSHV Pr (2PBK, gray) and HSV-2 Pr (1AT3, cyan), focusing on the dimer interface “hot spot” region. The sidechains of Trp109 (KSHV Pr, red) and Tyr124 (HSV-2 Pr, yellow) are displayed in space-filling mode. Helices 5 and 6 are omitted for clarity. (c) Backbone overlay of KSHV Pr (2PBK, gray) and HCMV Pr (1CMV, cyan), focusing on the dimer interface “hot spot” region. The sidechains of Trp109 (KSHV Pr, red) and Tyr128 (HCMV Pr, yellow) are displayed in space-filling mode. Helices 5 and 6 are omitted for clarity.
Discussion
From a drug-discovery standpoint, focusing on a conformationally dynamic target and capturing an inactive state in order to influence regulatory pathways is an appealing concept.4,6 In particular, regulating PPIs via allosteric mechanisms has recently gained traction. Allosterically targeting the subunit interface of dimeric complexes, such as the interdigitated β-sheets of HIV protease, has become an important goal due to increased incidents of resistance towards active-site therapeutics. Recent reports have noted the development of peptidic HIV protease dimer disruptors,26,27 including an alkylated tripeptide which “sequesters” a monomer.28 Importantly, small molecule peptide mimetics have been shown to disrupt other PPIs. Perhaps the most well documented are the helical mimetics such as the Nutlins that inhibit p53/MDM229 interactions, and the multi-aryl compounds which disrupt calmodulin/smMLCK30 and Bcl-xL/Bak31,32 interactions. Multi-aryl and multi-cyclic compounds were also recently discovered to regulate CREB KID/CBP KIX domain33,34 and human survivin homodimer35 interactions.
We have validated a methodology in which small molecules are used to allosterically inhibit an active enzyme by targeting a conformationally dynamic region. In the case of KSHV Pr, this conformationally dynamic region also governs the PPIs which influence the inactive monomer/active dimer equilibrium. Taken together, the NMR and X-ray crystallography data indicate a “monomer trap” model of KSHV Pr inhibition:21 that DD2 binds to and captures a pre-existing inactive monomer, thereby shifting the monomer-dimer equilibrium towards the inactive conformational state. Since HHV proteases are known to play critical roles in the late lytic stage of the viral lifecycle,24 inhibiting protease function at this stage would be key to the development of novel therapeutics targeting herpesviruses. Although promising results are observed in vitro for two of the eight members of the HHV protease family examined thus far, modifications to DD2 or the discovery of new helical mimetic scaffolds with improved pharmacokinetic and pharmacodynamic properties are necessary for further development. In particular, DD2 did not display an inhibitory effect in preliminary viral infectivity studies (data not shown) using a stably KSHV-infected SLK219 endothelial cell line.36–38 These results are explained by poor solubility and cell permeability of DD2 (Table S7), and we are currently exploring different chemical modifications to the DD2 scaffold in order to address these issues.
As the first structure of an allosterically inhibited HHV Pr monomer to date, our KSHV Pr Δ196-DD2 complex provides valuable insights. First, conformational perturbations in the crystal structure of the inactive complex relative to the active KSHV Pr dimer suggest a reason why previous attempts at developing an active site inhibitor have failed. Specifically, the substrate binding pocket of the active site may be too shallow and too dynamic for a competitive inhibitor to bind, as evidenced by the lack of efficacious inhibitors that target the active site. Second, the Δ196-DD2 structure, focusing on the DD2 binding pocket, can be used as a template for in silico docking studies in future efforts to discover other allosteric KSHV Pr inhibitors, including those that exhibit more pharmacologically favorable characteristics than DD2. Third, and more importantly, although members of the HHV protease family have low sequence homology, they are structurally (Table S8) and functionally highly homologous. Inspection of the available HHV Pr structures (Fig. 10) reveals that all eight proteases have a potential hot spot aromatic residue at the dimer interface. This region could act as a target site for more potent allosteric, pan-specific inhibitors that trap the other HHV proteases in their respective inactive monomeric states. Allosteric inhibition as a method to regulate conformationally dynamic complexes such as protein-protein interactions remains a mostly unexploited pathway. By examining KSHV Pr-DD2 interactions, we have utilized this route as a potential solution towards developing therapeutics that regulate high value, but previously intractable targets.
Materials And Methods
Protease Truncation and Mutagenesis
The KSHV Pr M197D-S204G sequence14,16 was the template used to generate genes encoding the Δ209, Δ196, and Δ191 truncation constructs. Genes encoding all KSHV Pr M197D Ile → Val single point mutations were synthesized using the unoptimized sequences of KSHV Pr M197D or KSHV Pr Δ196 as templates. Briefly, codons within the gene sequence representing isoleucine (ATA, ATC, or ATT) were changed to those encoding valine (GTA, GTC, or GTT). All gene sequences were designed at UCSF, and synthesized/purchased from GeneArt, Inc. The wild-type HCMV Pr sequence39 was the template for site-directed mutagenesis for the HCMV Pr L222D construct, performed using the QuickChange Site Directed Mutagenesis Kit (Stratagene). The wild-type HCMV Pr sequence also was the template for generating the HCMV Pr ΔL221 truncation sequence using the pET11a expression vector (Novagen). All resulting genes were verified through DNA sequencing.
Primers (Integrated DNA Technologies, Inc.) encoding restriction sites were as follows:
KSHV Pr
| NcoI | |
| All Truncations: | |
| 5′-CCA TGG CAC AGG GCC TGT ACG TCG-3′ | |
| BamH1 | |
| Δ191: | 5′-GGA TCC TTA ACT GAC GAA ATT TGG TGT GGA AAG GTC CTC-3′ |
| Δ196: | 5′-GGA TCC TTA TAA GGT CTC CAG GGG ACT GAC GAA ATT TGG-3′ |
| Δ209: | 5′-GGA TCC TTA GCG GTC CCG TAT GAA GCT GCC ATC-3′ |
HCMV Pr
| L222D mutation | |
| 5′-GAC AGC TAC GGC CTG GAC GGC AAC AGC GTG GAC GC-3′ | |
| Δ221: | |
| Nde1: | 5′- GGT TCA TAT GCA TCA CCA TCA CCA TCA CAT GAC-3′ |
| BamH1: | 5′- AAT TGG ATC CTT ACA GGC CGT AGC TGT CTG AGC-3′ |
Protein Expression and Purification
All KSHV Pr protein samples were expressed in M9 minimal media (NMR) or Luria Broth (X-ray crystallography) and purified as previously described.15,21,22 Soluble HCMV Pr was expressed and purified as described previously40 with the following modifications. Following overnight expression at 16 °C cells were pelleted and lysed by sonication, 0.5-second pulse/1-second recovery for 5 minutes, followed by 3-second pulse/3-second recovery for 2 minutes. After centrifugation at 30,000 g for 45 minutes, soluble fractions were passed over a 1 ml HisTrap FF column (GE Healthcare LifeSciences) using the following buffers: Wash/binding (50 mM NaPi, 300 mM NaCl, 25 mM imidazole, 5 mM β-mercaptoethanol, pH 8.0); elution (50 mM NaPi, 300 mM NaCl, 300 mM imidazole, 5 mM β-mercaptoethanol). Eluted His-tagged proteins were buffer exchanged into 25 mM NaPi, 150 mM NaCl, 5 mM β-mercaptoethanol and further purified over a Superdex75 (26/60) size exclusion chromatography column (GE Healthcare LifeSciences).
Starting protein concentrations for all assays described below were measured using a Nanodrop 2000c UV spectrophotometer (Thermo Scientific). Selective [13C-1H methyl] isotopic labeling of the isoleucine, leucine, and valine residues was achieved by adding 100 mg/L [dimethyl-13C2]-α-ketoisovaleric acid (Cambridge Isotope Laboratories) and 50 mg/L [methyl-13C1]-α-ketobutyric acid (Sigma) to otherwise unlabeled M9 minimal media 1 hr prior to IPTG induction.41
Calculation of Protein Sample Concentrations
Starting protein concentrations for all assays described below were measured using a Nanodrop 2000c UV spectrophotometer (Thermo Scientific) at 280 nm using the following extinction coefficients: KSHV Pr M197D and KSHV Pr M197D-I201V, ε = 23,950 M−1 cm−1; KSHV Pr Δ209, Δ196, Δ191, and all Δ196 Ile → Val mutants, ε = 22,460 M−1 cm−1; HCMV Pr wild-type and HCMV Pr L222D, ε = 28,420 M−1 cm−1; HCMV Pr Δ221, ε = 23,950 M−1 cm−1.
Protease Kinetic Analysis
Kinetic proteolysis assays were performed on HCMV Pr variants using the same methodology as previously described for KSHV Pr studies.21 Monomer-dimer Kd (N=3) and IC50 (N=4) values reported herein represent the average and standard deviation.
Circular Dichroism Analysis
Protease was allowed to equilibrate with or without DD2 for 1 h at room temperature in buffer solution (25 mM sodium phosphate buffer, 75 mM NaCl, 1 mM β-mercaptoethanol, 0.8% dioxane, pH 8). Dioxane was used to solubilize the DD2 stocks in place of DMSO, which absorbs far UV CD wavelengths. Spectra were obtained using a 2 mm path length cuvette on a Jasco J-715 spectropolarimeter with a final protein concentration of ~3 μM. Data were acquired at a constant temperature of 25 °C with the following parameters: accumulations 3, scan rate 50 nm/min, data pitch 0.1 nm, response 4 s, bandwidth 20 nm, and standard sensitivity. Buffer with and without DD2 were used to acquire background spectra. Final CD data spectra are reported as mean residue ellipticity (deg · cm2 · dmol−1 · residue−1). Fractional helicity (fH) values were estimated from the 222 nm band:42
| (1) |
where θ222,max is the calculated mean residue ellipticity value for a theoretical 100% helical polypeptide of N residues, collected at temperature T (°C). At 25 °C: KSHV Pr (N = 229 residues), θ222,max = −37,338 deg cm2 dmol−1 residue−1; HCMV Pr (N = 262 residues), θ222,max = −37,390 deg cm2 dmol−1 residue−1.
NMR Data Acquisition and Analysis
All spectra were acquired on cryoprobe-equipped Bruker Avance 500 MHz or 800 MHz NMR spectrometers at 12 °C or 27 °C. Typical NMR samples used for the 13C-1H HSQC titration studies consisted of ~ 0.01 – 0.02 mM selective [13C-1H methyl] ILV labeled protein in 0.45 mL buffer. Sample preparation, including buffer conditions and DD2 titrations, and spectral acquisition parameters were as previously described.21 Each titration study contained at least one repeat acquisition point. All data were processed using NMRPipe43 and analyzed using Sparky44.
Calculation of NMR-based apparent Kd values
Chemical shift data for the δ1-methyl groups of Ile44, Ile71, and Ile105 were converted to frequency values. Chemical shift perturbation values versus resonances corresponding to the apo protein are expressed as combined 13C-1H frequency perturbations (Δωobs):
| (2) |
where ΔδH and ΔδC correspond to the 1H and 13C chemical shift perturbation versus the apo protein, respectively. specfreqH and specfreqC correspond to the spectral frequencies of 1H and 13C, respectively.
Titration data point from the 13C-1H HSQC spectra were fit to a modified Hill Equation 45 using Matlab (The Mathworks, Inc).
| (3) |
where Δωmax is the maximum frequency perturbation, corresponding to a fully saturated ligand-bound state. [L]T is the total ligand concentration, and N is the Hill coefficient. Estimated values for Δωmax, N, and Kd were calculated from a non-linear regression fitting of Equation 2, using a grid search methodology to minimize χ2 errors. Final estimated apparent Kd values and errors reported in this paper are averages and standard deviations calculated for Ile44, Ile71, and Ile105. Estimated experimental errors were based upon repeat data points acquired during the NMR titrations and were propagated throughout the calculations.
X-Ray Crystallography Data Acquisition and Structure Determination
KSHV Pr Δ196 stock solutions used for X-ray crystallography consisted of 25 mM Tris (pH 8.0), 150 mM KCl, 0.1 mM EDTA, and 2 mM DTT. Crystals were grown at 25 °C with the hanging drop vapor diffusion method, with DD2 in greater than five-fold molar excess with respect to Δ196. The reservoir solution consisted of 2 M (NH4)2SO4, 0.2 M NaCl, 0.05 M sodium cacodylate (pH 7.0), 0.1 M urea, and 0.1 M sodium acetate. Following one day of incubation, 1 μL of 14 M β-mercaptoethanol was added to the reservoir. After 14–21 days, crystals appeared as a small cube measuring 0.03 × 0.05 × 0.05 mm3. A 2.0 Å-resolution X-ray diffraction data set was collected at the Lawrence-Berkeley National Laboratory Advanced Light Source Beamline 8.3.1, using a crystal flash-cooled to 100 K in mother liquor with 20% glycerol as the cryoprotectant.
Diffraction images were processed using denzo and scalepack from the hkl-2000 suite.46 The resulting structure was solved by molecular replacement with phaser47 and using residues 1–196 of a monomeric unit of the phosphonate-inhibited KSHV Pr dimer (2PBK)22 as the template search model. The resulting structure model was a dimer in an asymmetric unit, and was subjected to multiple rounds of restrained refinement and isotrophic B-factor minimization with refmac48 and Coot49 (pre-refinement: R-factor = 45.6%, Rfree = 49.5%; post-refinement: R-factor = 20.3%, Rfree = 24.8%).
All structural figures and animations within this paper and the supplementary materials were created using Pymol 1.2 (Schrödinger, LLC). Morph calculations (Supplemental Movies 2–3) were performed using the Yale Morph Server (http://www.molmovdb.org/).
DD2 Solubility Measurements
Test compounds were serially diluted from 10,000 μM to 625 μM in DMSO and placed in columns 1–5 and 7–11 of a 96-well polypropylene plate (Costar 3365). Columns 6 and 12 were filled with DMSO as the background. From each well, 5 μL was transferred into the 96-well disposable UV-Star plate (Greiner Bio-One). Acetonitrile (97.5 μL) and pH 7.4 PBS buffer (97.5 μL) were added to each well, and the plate was agitated for 30 min using an IKA microtiter plate shaker. The UV spectra from 200–500 nm was measured for all wells and subtracted from the DMSO background. Correlations between concentrations and absorbance at 260, 280, and 300 nm were determined as slopes. Then, 5 μL from each well of the polypropylene plate was added to a MultiScreen Solubility Filter Plate (Millipore) and diluted with 195 μL of PBS. The plate was agitated for 2 hrs and filtered into a 96-well disposable UV-Star plate, and the UV absorbance at 260, 280, and 300 nm was measured. The aqueous solubility (Amax filtrate/slope) was determined for all three wavelengths, and values are given as the means with 95% confidence intervals.
DD2 Cell Permeability Measurements
All liquid-handling steps for the PAMPA assay were performed on a Biomek FX Laboratory Automation Workstation (Beckman-Coulter) and analyzed by pION’s (London, UK) PAMPA Evolution 96 Command Software. The PAMPA Evolution 96 Permeability Assay Kit includes the Acceptor Sink Buffer (ASB), Double-Sink Lipid Solution, and a PAMPA sandwich plate, preloaded with magnetic disks. For each experiment, 4 μL of lipid was transferred onto the support membrane in the acceptor well, followed by addition of 200 μL of ASB (pH 7.4). Then, 180 μL of diluted test compound (50 μM in system buffer at pH 7.4 starting from a 10 mM DMSO solution) was added to the donor wells. The PAMPA sandwich plate was assembled and placed on the Gut-Box and stirred for 30 min. The distribution of the compounds in the donor and acceptor buffers (100 μL aliquot) was determined by measuring the UV spectra from 200 to 500 nm using the SpectraMax reader (Molecular Devices). The permeability coefficient was determined using the maximum absorbance from 200 to 500 nm:50
| (4) |
where VD is the donor well volume (cm3); A is the filter area (cm2); CD(0) is the sample concentration in the donor well at time 0 (mole/cm3); CD(t) is the sample concentration in the donor well at time t (mole/cm3); t is the interval of time (sec); tLAG is the lag time needed to reach steady state conditions (sec); and R is the membrane retention (related to the membrane/water partition coefficient). Standards used were verapamil (Pe = 1505 × 10−6 cm/s) as a high permeability standard, carbamazepine (Pe = 150 × 10−6 cm/s) as medium permeability standard and ranitidine (Pe = 2.3 × 10−6 cm/s) as low permeability standard. The compounds were measured in triplicate, and values are given as the mean values with 95% confidence intervals.
Supplementary Material
Acknowledgments
This work was funded in part by grants from the National Institutes of Health (grant 1R01-A1067423) and the HIV Accessory and Regulatory Complex Center (grant P50-GM082250). This research was also supported by a grant from the National Institutes of Health, University of California San Francisco-Gladstone Institute of Virology & Immunology Center for AIDS Research (P30-A122763 to G.M.L.). We thank Prof. Don Ganem, Prof. R. Kip Guy, Dr. Leggy Arnold, and Mr. Ernest Lam for technical support. We also thank Prof. John D. Gross and Dr. Ana Lazic for helpful discussions of this manuscript.
Abbreviations
- CD
circular dichroism
- DD2
dimer disruptor 2
- HCMV Pr
human cytomegalovirus protease
- HHV Pr
human herpesvirus protease
- HSQC
heteronuclear single quantum coherence
- KSHV Pr
Kaposi’s sarcoma-associated herpesvirus protease
- PDB
Protein Data Bank
- PPIs
protein-protein interactions
Footnotes
Supplementary data associated with this article can be found in the online version, at doi:10.1016/j.jmb.2011.06.032.
PDB Accession Numbers
Structural coordinates for the KSHV Pr Δ196-DD2 complex have been deposited at the Brookhaven Protein Data Bank with accession number 3NJQ.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 2.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5:789–96. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zinzalla G, Thurston DE. Targeting protein-protein interactions for therapeutic intervention: a challenge for the future. Future Med Chem. 2009;1:65–93. doi: 10.4155/fmc.09.12. [DOI] [PubMed] [Google Scholar]
- 4.Metallo SJ. Intrinsically disordered proteins are potential drug targets. Curr Opin Chem Biol. 2010;14:481–488. doi: 10.1016/j.cbpa.2010.06.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kar G, Keskin O, Gursoy A, Nussinov R. Allostery and population shift in drug discovery. Curr Opin Pharmacol. 2010;10:715–22. doi: 10.1016/j.coph.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee GM, Craik CS. Trapping moving targets with small molecules. Science. 2009;324:213–5. doi: 10.1126/science.1169378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–17. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 8.Berg T. Small-molecule inhibitors of protein-protein interactions. Curr Opin Drug Discov Devel. 2008;11:666–74. [PubMed] [Google Scholar]
- 9.Yin H, Hamilton AD. Strategies for targeting protein-protein interactions with synthetic agents. Angew Chem Int Ed Engl. 2005;44:4130–63. doi: 10.1002/anie.200461786. [DOI] [PubMed] [Google Scholar]
- 10.Hershberger SJ, Lee SG, Chmielewski J. Scaffolds for blocking protein-protein interactions. Curr Top Med Chem. 2007;7:928–42. doi: 10.2174/156802607780906726. [DOI] [PubMed] [Google Scholar]
- 11.Ross NT, Katt WP, Hamilton AD. Synthetic mimetics of protein secondary structure domains. Philos Transact A Math Phys Eng Sci. 2010;368:989–1008. doi: 10.1098/rsta.2009.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–6. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 13.Ma B, Nussinov R. Trp/Met/Phe hot spots in protein-protein interactions: potential targets in drug design. Curr Top Med Chem. 2007;7:999–1005. doi: 10.2174/156802607780906717. [DOI] [PubMed] [Google Scholar]
- 14.Nomura AM, Marnett AB, Shimba N, Dotsch V, Craik CS. Induced structure of a helical switch as a mechanism to regulate enzymatic activity. Nat Struct Mol Biol. 2005;12:1019–20. doi: 10.1038/nsmb1006. [DOI] [PubMed] [Google Scholar]
- 15.Nomura AM, Marnett AB, Shimba N, Dotsch V, Craik CS. One functional switch mediates reversible and irreversible inactivation of a herpesvirus protease. Biochemistry. 2006;45:3572–9. doi: 10.1021/bi0523658. [DOI] [PubMed] [Google Scholar]
- 16.Shimba N, Nomura AM, Marnett AB, Craik CS. Herpesvirus protease inhibition by dimer disruption. J Virol. 2004;78:6657–65. doi: 10.1128/JVI.78.12.6657-6665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.del Sol A, Tsai CJ, Ma B, Nussinov R. The Origin of Allosteric Functional Modulation: Multiple Pre-existing Pathways. Structure. 2009;17:1042–1050. doi: 10.1016/j.str.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogilvie W, Bailey M, Poupart MA, Abraham A, Bhavsar A, Bonneau P, Bordeleau J, Bousquet Y, Chabot C, Duceppe JS, Fazal G, Goulet S, Grand-Maitre C, Guse I, Halmos T, Lavallee P, Leach M, Malenfant E, O’Meara J, Plante R, Plouffe C, Poirier M, Soucy F, Yoakim C, Deziel R. Peptidomimetic inhibitors of the human cytomegalovirus protease. J Med Chem. 1997;40:4113–35. doi: 10.1021/jm970104t. [DOI] [PubMed] [Google Scholar]
- 19.Borthwick AD, Crame AJ, Ertl PF, Exall AM, Haley TM, Hart GJ, Mason AM, Pennell AM, Singh OM, Weingarten GG, Woolven JM. Design and synthesis of pyrrolidine-5,5-trans-lactams (5-oxohexahydropyrrolo[3,2-b]pyrroles) as novel mechanism-based inhibitors of human cytomegalovirus protease. 2. Potency and chirality. J Med Chem. 2002;45:1–18. doi: 10.1021/jm0102203. [DOI] [PubMed] [Google Scholar]
- 20.Gopalsamy A, Lim K, Ellingboe JW, Mitsner B, Nikitenko A, Upeslacis J, Mansour TS, Olson MW, Bebernitz GA, Grinberg D, Feld B, Moy FJ, O’Connell J. Design and syntheses of 1,6-naphthalene derivatives as selective HCMV protease inhibitors. J Med Chem. 2004;47:1893–9. doi: 10.1021/jm030540h. [DOI] [PubMed] [Google Scholar]
- 21.Shahian T, Lee GM, Lazic A, Arnold LA, Velusamy P, Roels CM, Guy RK, Craik CS. Inhibition of a viral enzyme by a small-molecule dimer disruptor. Nat Chem Biol. 2009;5:640–6. doi: 10.1038/nchembio.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazic A, Goetz DH, Nomura AM, Marnett AB, Craik CS. Substrate Modulation of Enzyme Activity in the Herpesvirus Protease Family. J Mol Biol. 2007;373:913–923. doi: 10.1016/j.jmb.2007.07.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reiling KK, Pray TR, Craik CS, Stroud RM. Functional consequences of the Kaposi’s sarcoma-associated herpesvirus protease structure: regulation of activity and dimerization by conserved structural elements. Biochemistry. 2000;39:12796–803. doi: 10.1021/bi001019h. [DOI] [PubMed] [Google Scholar]
- 24.Tong L. Viral proteases. Chem Rev. 2002;102:4609–26. doi: 10.1021/cr010184f. [DOI] [PubMed] [Google Scholar]
- 25.Pray TR, Reiling KK, Demirjian BG, Craik CS. Conformational change coupling the dimerization and activation of KSHV protease. Biochemistry. 2002;41:1474–82. doi: 10.1021/bi011753g. [DOI] [PubMed] [Google Scholar]
- 26.Hwang YS, Chmielewski J. Development of low molecular weight HIV-1 protease dimerization inhibitors. J Med Chem. 2005;48:2239–42. doi: 10.1021/jm049581j. [DOI] [PubMed] [Google Scholar]
- 27.Frutos S, Rodriguez-Mias RA, Madurga S, Collinet B, Reboud-Ravaux M, Ludevid D, Giralt E. Disruption of the HIV-1 protease dimer with interface peptides: structural studies using NMR spectroscopy combined with [2-(13)C]-Trp selective labeling. Biopolymers. 2007;88:164–73. doi: 10.1002/bip.20685. [DOI] [PubMed] [Google Scholar]
- 28.Bannwarth L, Rose T, Dufau L, Vanderesse R, Dumond J, Jamart-Gregoire B, Pannecouque C, De Clercq E, Reboud-Ravaux M. Dimer disruption and monomer sequestration by alkyl tripeptides are successful strategies for inhibiting wild-type and multidrug-resistant mutated HIV-1 proteases. Biochemistry. 2009;48:379–87. doi: 10.1021/bi801422u. [DOI] [PubMed] [Google Scholar]
- 29.Hu CQ, Hu YZ. Small molecule inhibitors of the p53-MDM2. Curr Med Chem. 2008;15:1720–30. doi: 10.2174/092986708784872375. [DOI] [PubMed] [Google Scholar]
- 30.Orner BP, Ernst JT, Hamilton AD. Toward proteomimetics: terphenyl derivatives as structural and functional mimics of extended regions of an alpha-helix. J Am Chem Soc. 2001;123:5382–3. doi: 10.1021/ja0025548. [DOI] [PubMed] [Google Scholar]
- 31.Kutzki O, Park HS, Ernst JT, Orner BP, Yin H, Hamilton AD. Development of a potent Bcl-x(L) antagonist based on alpha-helix mimicry. J Am Chem Soc. 2002;124:11838–9. doi: 10.1021/ja026861k. [DOI] [PubMed] [Google Scholar]
- 32.Petros AM, Dinges J, Augeri DJ, Baumeister SA, Betebenner DA, Bures MG, Elmore SW, Hajduk PJ, Joseph MK, Landis SK, Nettesheim DG, Rosenberg SH, Shen W, Thomas S, Wang X, Zanze I, Zhang H, Fesik SW. Discovery of a potent inhibitor of the antiapoptotic protein Bcl-xL from NMR and parallel synthesis. J Med Chem. 2006;49:656–63. doi: 10.1021/jm0507532. [DOI] [PubMed] [Google Scholar]
- 33.Best JL, Amezcua CA, Mayr B, Flechner L, Murawsky CM, Emerson B, Zor T, Gardner KH, Montminy M. Identification of small-molecule antagonists that inhibit an activator: coactivator interaction. Proc Natl Acad Sci USA. 2004;101:17622–7. doi: 10.1073/pnas.0406374101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bates CA, Pomerantz WC, Mapp AK. Transcriptional tools: Small molecules for modulating CBP KIX-dependent transcriptional activators. Biopolymers. 2011;95:17–23. doi: 10.1002/bip.21548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wendt MD, Sun C, Kunzer A, Sauer D, Sarris K, Hoff E, Yu L, Nettesheim DG, Chen J, Jin S, Comess KM, Fan Y, Anderson SN, Isaac B, Olejniczak ET, Hajduk PJ, Rosenberg SH, Elmore SW. Discovery of a novel small molecule binding site of human survivin. Bioorg Med Chem Lett. 2007;17:3122–9. doi: 10.1016/j.bmcl.2007.03.042. [DOI] [PubMed] [Google Scholar]
- 36.Bechtel JT, Liang Y, Hvidding J, Ganem D. Host range of Kaposi’s sarcoma-associated herpesvirus in cultured cells. J Virol. 2003;77:6474–81. doi: 10.1128/JVI.77.11.6474-6481.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vieira J, O’Hearn PM. Use of the red fluorescent protein as a marker of Kaposi’s sarcoma-associated herpesvirus lytic gene expression. Virology. 2004;325:225–40. doi: 10.1016/j.virol.2004.03.049. [DOI] [PubMed] [Google Scholar]
- 38.Chandriani S, Ganem D. Array-based transcript profiling and limiting-dilution reverse transcription-PCR analysis identify additional latent genes in Kaposi’s sarcoma-associated herpesvirus. J Virol. 2010;84:5565–73. doi: 10.1128/JVI.02723-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loveland AN, Chan CK, Brignole EJ, Gibson W. Cleavage of human cytomegalovirus protease pUL80a at internal and cryptic sites is not essential but enhances infectivity. J Virol. 2005;79:12961–8. doi: 10.1128/JVI.79.20.12961-12968.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brignole EJ, Gibson W. Enzymatic activities of human cytomegalovirus maturational protease assemblin and its precursor (pPR, pUL80a) are comparable: [corrected] maximal activity of pPR requires self-interaction through its scaffolding domain. J Virol. 2007;81:4091–103. doi: 10.1128/JVI.02821-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goto NK, Gardner KH, Mueller GA, Willis RC, Kay LE. A robust and cost-effective method for the production of Val, Leu, Ile (delta 1) methyl-protonated 15N-, 13C-, 2H-labeled proteins. J Biomol NMR. 1999;13:369–74. doi: 10.1023/a:1008393201236. [DOI] [PubMed] [Google Scholar]
- 42.Wallimann P, Kennedy RJ, Kemp DS. Large Circular Dichroism Ellipticities for N-Templated Helical Polypeptides Are Inconsistent with Currently Accepted Helicity Algorithms. Angew Chem Int Ed Engl. 1999;38:1290–1292. doi: 10.1002/(SICI)1521-3773(19990503)38:9<1290::AID-ANIE1290>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 43.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 44.Goddard TD, Kneller DG. Sparky. Vol. 3. University of California; San Francisco: 1999. [Google Scholar]
- 45.Ramstad T, Hadden CE, Martin GE, Speaker SM, Teagarden DL, Thamann TJ. Determination by NMR of the binding constant for the molecular complex between alprostadil and alpha-cyclodextrin. Implications for a freeze-dried formulation. Int J Pharm. 2005;296:55–63. doi: 10.1016/j.ijpharm.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 46.Otwinowski Z, Minor W, Carter CW., Jr Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 47.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skubak P, Murshudov GN, Pannu NS. Direct incorporation of experimental phase information in model refinement. Acta Crystallogr D Biol Crystallogr. 2004;60:2196–201. doi: 10.1107/S0907444904019079. [DOI] [PubMed] [Google Scholar]
- 49.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Avdeef A. Physicochemical profiling (solubility, permeability and charge state) Curr Top Med Chem. 2001;1:277–351. doi: 10.2174/1568026013395100. [DOI] [PubMed] [Google Scholar]
- 51.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallog. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.