Abstract
In this study, we investigated the effects of ectopic estrogen receptor (ER)β1 expression in breast cancer cell lines and nude mice xenografts and observed that ERβ1 expression suppresses tumor growth and represses FOXM1 mRNA and protein expression in ERα-positive but not ERα-negative breast cancer cells. Furthermore, a significant inverse correlation exists between ERβ1 and FOXM1 expression at both protein and mRNA transcript levels in ERα-positive breast cancer patient samples. Ectopic ERβ1 expression resulted in decreased FOXM1 protein and mRNA expression only in ERα-positive but not ERα-negative breast carcinoma cell lines, suggesting that ERβ1 represses ERα-dependent FOXM1 transcription. Reporter gene assays showed that ERβ1 represses FOXM1 transcription through an estrogen-response element located within the proximal promoter region that is also targeted by ERα. The direct binding of ERβ1 to the FOXM1 promoter was confirmed by chromatin immunoprecipitation analysis, which also showed that ectopic expression of ERβ1 displaces ERα from the endogenous FOXM1 promoter. Forced expression of ERβ1 promoted growth suppression in MCF-7 cells, but the anti-proliferative effects of ERβ1 could be overridden by overexpression of FOXM1, indicating that FOXM1 is an important downstream target of ERβ1 signaling. Together, these findings define a key anti-proliferative role for ERβ1 in breast cancer development through negatively regulating FOXM1 expression.
Estrogens play a crucial role in the development and proliferation of normal tissues, as well as malignant mammary tissues,1 and their biological functions are mediated primarily through two estrogen receptors (ERs), ERα and ERβ, encoded by distinct genes, ESR1 and ESR2, respectively.2,3 ERα and ERβ bind to the natural estrogen 17β-estradiol (E2) with equal affinity, but they interact differentially with other natural and synthetic ligands.4 In response to estrogen signaling, ERα normally promotes the proliferation of breast epithelium and cancer cells, whereas ERβ has been shown to have an anti-proliferative and pro-apoptotic effect.2,3 In the presence of ligands, ERα and ERβ bind to the estrogen responsive element (ERE) located in gene promoter regions as either homodimers (ERα/ERα or ERβ/ERβ) or heterodimers (ERα/ERβ) to regulate the transcriptional activity of target genes. In addition, the effects of estrogens can also be mediated through plasma membrane-localized ERα and ERβ.3 It has also been reported that ERβ is able to regulate transcription independent of estrogen and in an ERE-independent manner.5–7 Although the biological function of ERβ varies in different organs, its deregulation has been comprehensively linked to breast and colon tumorigenesis.8
In humans, five common splice variants of ERβ (ERβ1 to ERβ5) have been identified.9 Among the five isoforms, ERβ1 [also called wild-type (WT) ERβ] and ERβ2 (also called ERβcx) are the most commonly expressed and frequently studied. Unlike ERβ1, ERβ2 does not bind estrogen and, therefore, can theoretically be a dominant-negative regulator of ERα. ERβ positivity in general has been shown to be associated with favorable prognosis, with patients having better response to endocrine therapy.10,11 Although the significance of each isoform is still unclear, both ERβ1 and ERβ2 have been shown to be good prognostic factors for endocrine therapy in breast cancer.12–15 As for ERα, despite its mitogenic function, its expression is generally associated with good prognosis in breast cancer, because approximately two-thirds of the patients positive for ERα respond to endocrine therapeutics, such as tamoxifen (OHT), fulvestrant (ICI 182780), and aromatase inhibitor.16–18
FOXM1, a member of forkhead box (FOX) family of transcription factors, is a critical regulator of cell-cycle progression,19,20 mitotic spindle integrity,21 angiogenesis,22 apoptosis,20,23 cell migration,22,23 metastasis,23 DNA damage repair,24,25 and tissue regeneration.26 FOXM1 is frequently overexpressed in a wide range of human cancer types, including colorectal,27 lung,28 prostate,29 liver,30 and breast31 carcinomas. In agreement, a microarray study also found FOXM1 expression to be elevated in multiple carcinomas, including prostate, lung, ovary, colon, pancreas, stomach, bladder, liver, kidney, and breast, compared with their normal counterparts.32
In addition to its involvement in breast cancer tumorigenesis,31 FOXM1 overexpression/dysregulation has also been implicated in the development of resistance to breast cancer drugs, including cisplatin,24 trastuzumab (Herceptin),31,33 and paclitaxel (Taxol).33 Consistently, high levels of FOXM1 expression are associated with poor prognosis in breast cancer.34 Previous work has shown that FOXM1 regulates ERα transcription in breast cancer cells.35 Conversely, ERα also controls FOXM1 expression at the transcription and gene promoter levels.36 In fact, FOXM1 is a key mediator of the mitogenic functions of ERα and estrogen signaling in breast cancer cells. As such, the deregulation of FOXM1 expression may contribute to insensitivity to breast cancer endocrine therapies.36 To explore the role of ERβ signaling in breast cancer development, we investigated the relation between FOXM1 and ERβ1 expression in breast cancer cell lines in vitro and in vitro and in clinical samples. In the present study, we found FOXM1 to be an ERβ1-regulated gene and ERβ1 represses FOXM1 expression through targeting ERα.
Materials and Methods
Cell Culture and Xenograft Model
The human breast carcinoma cell lines CAL51, MCF-7, MCF-7(ER-), MDA-MB-231, SKBR-3, T47D, ZR-75-1, and ZR-75-1(ER-) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2 mmol/L glutamine, and 100 U/mL penicillin/streptomycin in a humidified incubator at 37°C. The MCF-7(ER-) cells were established from prolonged culturing of adenovirally infected MCF-7 cells in estrogen-free conditions and were a kind gift from Laki Buluwela (London, UK). All experiments on the breast cancer cell lines were performed in full-serum conditions, unless indicated otherwise. Mice xenograft models have been described previously.37 Mice were housed at the Centre for Biotechnology, Karolinska Institute, Huddinge, Sweden. T47D-Tet-off-ERβ cells, stably transfected with the tetracycline-regulated ERβ expression plasmid, have previously been described.37 The T47D or T47D-ERβ cells were injected into the mammary fat pad of 5-week-old severe combined immunodeficient/beige mice (Taconic, Ry, Denmark). E2 pellets, 0.72 mg/pellet (Innovative Research of America, Sarasota, FL), were placed subcutaneously in the neck with a pellet trochar (Innovative Research of America). After 4, 8, 16, or 30 days, the mice were sacrificed, and the tumors were fixed in 4% paraformaldehyde and paraffin-embedded as described.37 Animal experiments were approved by the Swedish Board of Agriculture, reference number S 27-08, including approved animal welfare, experimental protocol, and animal toxicology.
Plasmids and Transfections
The pcDNA3-Flag-tagged human ERβ1 expression vector has previously been described.37 For transfections, cells were seeded to a confluence of ∼50% to 70% and incubated with a mix of transfection reagents containing FuGENE-6 (Roche, East Sussex, UK) and the plasmid DNA. CAL51 cells were transfected with Xfect (Clontech, Saint-Germain-en-Laye, France), and T47D and MCF-7-ER(-) cells were transfected with GenePulser II (Bio-Rad, Hemel Hempstead, UK). The optimized transfection efficiencies for these ER-positive and -negative breast cancer cells are usually between 30% and 80% (data not shown).
Luciferase Reporter Assay
The pGL3-FOXM1(Apa-I) WT and mERE4 reporter constructs have previously been described.36 Cells were transfected with pGL3-FOXM1 reporter constructs (WT or mERE4) and an internal transfection control plasmid expressing the Renilla-luciferase (pRL-TK; Promega, Southampton, UK) with the use of FuGENE-6 (Roche). For promoter analysis, 24 hours after transfection, cells were collected for firefly/Renilla luciferase assays with the use of the Dual-Glo Luciferase reporter assay system (Promega) according to the manufacturer's instructions. Luminescence was then measured with a plate reader (the 9904 TopCount; Perkin-Elmer, Beaconsfield, UK). The relative promoter activity was calculated from the ratio of the luciferase to Renilla luciferase activities.
Proliferation Assays
To determine cell proliferation, the sulforhodamine B (SRB) assay was performed as previously described23 with the use of the Sunrise microplate reader (Tecan UK, Reading, UK).
Western Blotting, ChIP Analysis, and Antibodies
Western blotting was performed on whole-cell extracts by lysing cells in buffer as previously described.38 Antibodies against cyclin B1 (H433), β-tubulin (H235), ERα (HC20), and FOXM1 (C-20) were obtained from Santa Cruz Biotechnology (Autogen Bioclear, Wiltshire, UK) and ERβ (ab3576) from Abcam (Cambridge, UK). Chromatin immunoprecipitation (ChIP) assays were performed as previously described.38,39 Anti-Flag antibody (F1804) was purchased from Sigma-Aldrich (Poole, UK).
Real-Time Quantitative PCR and Patient Samples
Frozen samples from patients who had undergone surgery at Charing Cross Hospital (London, UK) were used for RNA extraction. The ethical approval of this study was granted by the Riverside Research Ethics Committee, Hammersmith, London (reference number 05/Q0411/57). Total RNA (2 μg) isolated with the use of the RNeasy Mini kit (Qiagen, Crawley, UK) was reverse-transcribed with the Superscript III reverse transcriptase and random primers (Invitrogen, Paisley, UK), and the resulting first-strand cDNA was used as a template in the real-time PCR. All samples were performed in triplicates. The following gene-specific primer pairs were designed with the ABI Primer Express software version 3.0 (Applied Biosystems, Brackley, UK): FOXM1-sense, 5′-TGCAGCTAGGGATGTGAATCTTC-3′, and FOXM1-antisense, 5′-GGAGCCCAGTCCATCAGAACT-3′; ERα-sense, 5′-TGATCAGGTCCACCTTCTAGAATG-3′, and ERα-antisense, 5′-CGCCAGACGAGACCAATCAT-3′; ERβ-sense, 5′-CTGCTGGAGATGCTGAATGC-3′, and ERβ-antisense, 5′-CCGTGATGGAGGACTTGCA-3′; ERβ1-sense, 5′-ACTTGCTGAACGCCGTGACC-3′, and ERβ1-antisense, 5′-CAGATGTTCCATGCCCTTGTT-3′; L19-sense, 5′-GCGGAAGGGTACAGCCAAT-3′, and L19-antisense, 5′-GCAGCCGGCGCAAA-3′; and 18S-sense, 5′-CCTGCGGCTTAATTTGACTCA-3′, and 18S-antisense, 5′-AGCTATCAATCTGTCAATCCTGTCC-3′. Specificity of each primer was determined with Primer Express software (Applied Biosystems). Real-time PCR was performed with ABI PRISM 7700 Sequence Detection System with the use of SYBR Green Mastermix (Applied Biosystems). FOXM1, ERα, ERβ, L19, and 18S transcript levels were quantified with the standard curve method. L19 and 18S, non-regulated ribosomal housekeeping genes, were used as an internal control to normalize input cDNA.
IHC Analysis, TMAs, and Samples
With ethical approval from the Local Research Ethics Committee of the Leeds Teaching Hospitals NHS Trust (06/Q1206/180) formalin-fixed, paraffin-embedded tissue microarrays (TMAs) from 358 clinical samples collected from Leeds Teaching Hospitals were subjected to immunohistochemical (IHC) staining. IHC staining was performed on formalin-fixed, paraffin-embedded tissue sections with the use of the following antibodies: FOXM1 (C-20; Santa Cruz Biotechnology) and ERβ (ab3576; Abcam), ERα antibody (clone 6F11; Novocastra, Newcastle, UK), ERβ1 (PPG5/10; Serotec, Kidlington, UK), ERβ2 (57/3; Serotec), and ERβ5.40 ERα and each ERβ were assessed with the Allred score on the basis of proportion and intensity of nuclear staining with a cutoff value >3. FOXM1 was determined as positive when showing expression of moderate or strong intensity.
Statistical Analysis
Pearson's χ2 test and Fisher's exact test were used to test the relation between ERβ and FOXM1 expression. Correlations were assessed with Pearson's rank correlation test. SPSS version 16 (SPSS Inc., Chicago, IL) was used for all analyses. P ≤ 0.05 was considered significant.
Results
Expression of ERβ1 Represses Tumor Growth and FOXM1 Expression in Nude Mice Xenografts
To evaluate the role of ERβ1 on tumor growth in vivo, the estrogen-dependent T47D (control; ERβ-) and the derivatives ectopically expressing ERβ1, T47D-ERβ,37 breast carcinoma cells were transplanted into the mammary fat pads of athymic nude mice administrated with E2. After 4, 8, 16, and 30 days, four mice from each treatment group were sacrificed, and the xenografts were removed for examination. Tumor xenografts derived from T47D-ERβ cells were significantly smaller (P < 0.005) than the control group that received a transplant with the parental T47D cells (Figure 1A). The average volume of the T47D-ERβ tumors was 50 mm3 in diameter compared with 166 mm3 in the controls at 30 days after transplantation (Figure 1B). IHC staining of tumor sections showed that, although ERβ expression was undetectable, high levels of FOXM1 expression were found in the nuclei of the faster-growing control T47D-derived tumors. In contrast, the slower-growing T47D-ERβ tumors contained high levels of ERβ but low levels of FOXM1 staining (Figure 1C). The inverse relation between ERβ and FOXM1 expression in these mice xenografts suggested that ERβ1 might function through repressing FOXM1 expression to limit breast cancer growth.
Figure 1.
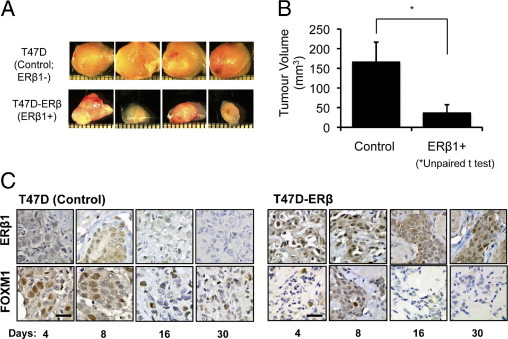
Expression of ERβ1 represses tumor growth and FOXM1 expression in nude mice xenografts. A: T47D (Control; ERβ1-) and the derivatives ectopically expressing ERβ1, T47D-ERβ (ERβ1+) breast carcinoma cells were transplanted into the mammary fat pads of athymic nude mice administrated with E2. After 4, 8, 16, and 30 days, four mice from each treatment group were sacrificed, and the xenografts were removed for examination. Photographs show tumors at 30 days after transplantation. B: Graph shows comparison of the average size between the control and T47D-ERβ tumors. T47D-ERβ tumors were significantly smaller (*P < 0.05) than the controls (6.5 mm and 11.5 mm in diameter, respectively) at 30 days after transplantation. C: IHC staining of tumor sections showed that, although the expression of ERβ1 was undetectable, high levels of FOXM1 expression were found in the nuclei of the controls. In contrast, the T47D-ERβ tumors expressed high levels of ERβ1 but low levels of FOXM1 staining. Scale bar = 30 μm.
Inverse Correlation between ERβ1 and FOXM1 Expression in Human Breast Cancer Samples
To test whether the inverse association between ERβ1 and FOXM1 expression also exists in human breast cancer, the expression patterns of individual ER isoforms, ERβ1, ERβ2, or ERβ5, and FOXM1 were examined in human breast cancer samples by IHC staining (Figure 2A; see also Supplemental Figure S1 at http://ajp.amjpathol.org). No significant correlation was observed between the expression levels of FOXM1 and ERβ2 or ERβ5; however, a potential but non-significant inverse correlation trend was detected between ERβ1 and FOXM1 expression (n = 245; P = 0.0717 t-test) when all samples were analyzed (Figure 2B). When IHC data were re-evaluated after excluding ERα-negative samples, a significant correlation between FOXM1 and ERβ1 expression (n = 173; P = 0.0197 t-test) was observed, consistent with the xenograft results (Figure 2C). As for ERβ2 and ERβ5, no association was observed with FOXM1 expression in the ERα-positive tumors. Similar results were obtained when the staining data were scored as plus or minus and analyzed by Fisher's test (see Supplemental Figure S2 at http://ajp.amjpathol.org). Notably, the clinical samples used in this study have previously been analyzed for correlations between the expression levels of ERβ isoforms and ERα. The results showed no significant associations, except for a positive correlation between ERβ2 and ERα expressions.40 This finding also indicates that the inverse relation between ERβ1 and FOXM1 is not because of the ability of ERβ1 to repress ERα expression.
Figure 2.
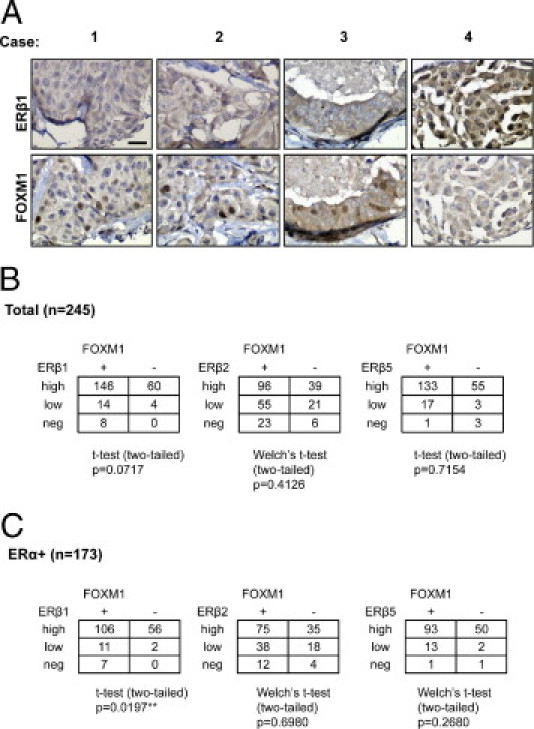
Inverse correlation between ERβ1 and FOXM1 expression in human breast cancer samples. A: Representative expression patterns of FOXM1 and ERβ1 in TMA. With the use of an ERβ1 antibody, IHC staining showed an inverse correlation/association between ERβ1 and FOXM1 expression in human breast cancer TMA. Scale bar = 30 μm. B: The expression of the individual ER variant, ERβ1, ERβ2, or ERβ5 was also investigated independently (see Supplemental Figure S1 at http://ajp.amjpathol.org). The staining of FOXM1, ERβ1, ERβ2, and ERβ5 (see Supplemental Figure S1 at http://ajp.amjpathol.org) was assessed with a scanscope (Scanscope Aperio Technologies, Inc., Vista, CA) connected to a personal computer. The staining intensity and percentage of staining in the cytoplasm and the nucleus were each scored independently in a semiquantitative fashion. For each case, a final score from the nucleus and the cytoplasm was obtained by multiplying the score of intensity with the score of the percentage, 8 being the maximum final score. To avoid subjectivity in evaluation, scoring was done by two independent persons. Allred scores of 0 to 2 are classified as negative (−), 3 to 5 as low positive (+), and 6 to 8 as high positive (++). Analysis of the staining results showed no significant correlation between the expression levels of FOXM1 and ERβ2 or ERβ5; however, a potential but not significant inverse correlation trend was detected between ERβ1 and FOXM1 expression (n = 245; P = 0.0717). C: Analysis of the staining results after the excluding ERα-negative patient samples showed a significant correlation between FOXM1 and ERβ1 expression (n = 173; P = 0.0197), further suggesting that ERβ1 represses FOXM1 expression.
Next, the expression levels of ERβ1 and FOXM1 mRNAs were analyzed by real-time quantitative PCR in a further 94 human breast cancer samples (Figure 3A). The results showed no significant relation between ERβ1 and FOXM1 mRNA transcript levels (n = 94; r = −0.149, P = 0.152 two-tailed Pearson's correlation) when all samples were analyzed, but the correlation became significant when the ERα-negative samples were removed from the analysis (n = 61; r = −0.256, P = 0.046 two-tailed; Figure 3B). These findings suggest that ERβ1 may negatively regulate FOXM1 expression at the transcriptional level, and this process depends on the presence of ERαexpression.
Figure 3.
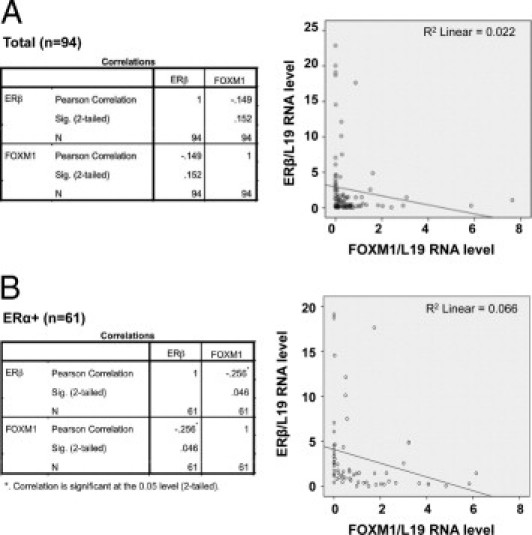
Inverse correlation between ERβ and FOXM1 mRNA expression in human breast cancer samples. Expression levels of ERβ and FOXM1 mRNA were analyzed by real-time quantitative PCR in 94 breast cancer patient samples with FOXM1, ERβ, and 18S primers. The FOXM1 and ERβ mRNA levels were normalized against 18S RNA levels. A: The results showed no significant relation between ERβ and FOXM1 mRNA transcript levels (n = 94; r = −0.149, P = 0.152 two-tailed) when all samples were studied by Pearson's correlation analysis. B: The correlation became significant when the only ERα-positive samples (n = 61) were analyzed (n = 61; r = −0.256, P = 0.046 two-tailed), showing a significant inverse correlation between ERβ and FOXM1 mRNA levels.
ERβ1 Represses FOXM1 Protein and mRNA Expression Only in ERα-Positive Breast Cancer Cells
To test the hypothesis that ERβ1 represses FOXM1 expression, we next used an ERα-positive T47D cell line expressing a Tet-Off-controlled ERβ1 construct (T47D-Tet-Off-ERβ),37 in which ERβ expression is inducible on doxycycline (Dox) withdrawal. Removal of Dox from the T47D-Tet-Off-ERβ cells in the presence of 10 nmol/L E2 resulted in an induction of ERβ1 expression and a corresponding decrease in FOXM1 expression (Figure 4A). In contrast, Dox withdrawal caused an increase in FOXM1 expression in the control T47D-Tet-Off cell line, probably because of the relief of the anti-proliferative effects of Dox (Figure 4A). Consistent with the Western blot results, real-time quantitative PCR analysis showed that Dox withdrawal resulted in a down-regulation of FOXM1 mRNA expression in the T47D-Tet-Off ERβ cells but an increase in FOXM1 mRNA levels in the control T47D-Tet-Off cells (Figure 4B).
Figure 4.
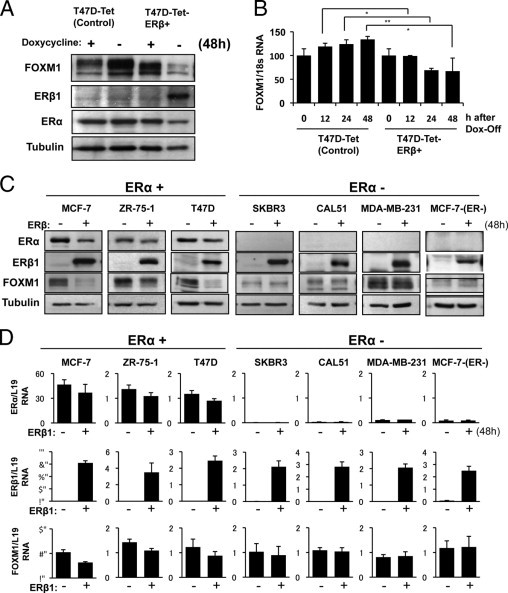
ERβ1 represses FOXM1 protein and mRNA expression only in ERα-positive breast cancer cells. Dox was removed from the control T47D-Tet cells and a T47D cell line expressing a Tet-Off controlled ERβ construct (T47D-Tet-ERβ), in which ERβ1 expression is inducible on Dox withdrawal. A: T47D-Tet and T47D-Tet-ERβ cells collected at 0 and 48 hours after Dox removal were used for Western blot analysis. Removal of Dox from the T47D-Tet-ERβ cells in the presence of 10 nmol/L E2 resulted in an induction of ERβ1 expression and a corresponding decrease in FOXM1 expression at 48 hours. In contrast, Dox withdrawal caused an increase in FOXM1 expression in the T47D-Tet-control cells. B: Real-time quantitative PCR analysis was performed on T47D-Tet and T47D-Tet-ERβ at 0, 12, 24, and 48 hours after Dox withdrawal. The results showed that Dox withdrawal caused a down-regulation of FOXM1 mRNA expression in the T47D-Tet-ERβ cells but an increase in FOXM1 mRNA levels in the T47D-Tet-control cells, probably as a result of the relief of the anti-proliferative effects of Dox. Statistical analyses were done using Student's t test. *P < 0.05, significant; **P < 0.01, very significant. C: Effects of ERβ1 ectopic expression on FOXM1 expression was examined in a panel of ERα-positive (MCF-7, ZR-75-1, and T47D) and ERα-negative [SKBR3, CAL51, MDA-MB-231 and MCF-7(ER-)] breast carcinoma cell lines. The breast cancer cells were transiently transfected with ERβ1 expression vector or an empty vector control and were collected at 48 hours for Western blot analysis for ERα, ERβ, FOXM1, and tubulin expression. D: The transfected cells were also analyzed for ERα, ERβ, FOXM1, and L19 RNA expression by real-time quantitative PCR. Both the Western blot and real-time quantitative PCR analysis results showed that ectopic expression of ERβ1 resulted in a down-regulation of FOXM1 expression in ERα-positive but not ERα-negative cell lines. Thus, these results suggest that the ability of ERβ1 to repress FOXM1 expression depends on the presence of ERα.
To confirm these results further and to explore the role of ERα in this regulatory mechanism, the effects of ERβ1 transfection on FOXM1 expression was studied in a panel of ERα-positive (MCF-7, ZR-75-1, T47D) and ERα-negative [SKBR3, CAL51, MDA-MB-231, MCF-7(ER-) and ZR-75-1(ER-)] breast carcinoma cell lines (Figure 4D; see also Supplemental Figure S3 at http://ajp.amjpathol.org). Western blot analysis showed that ectopic expression of ERβ1 resulted in a down-regulation of FOXM1 expression in ERα-positive cell lines, whereas ERβ1 overexpression had no effects on FOXM1 in the ERα-negative SKBR3, CAL51, and MDA-MB-231 cell lines. Consistent with this, ERβ1 overexpression did not alter the FOXM1 expression levels in clones of MCF-7 and ZR-75-1 (see Supplemental Figure S2 at http://ajp.amjpathol.org), which have lost ERα expression. It is notable that ERα expression was also down-regulated by ERβ1 overexpression, but the down-regulation is generally moderate and not sufficient to account for the considerable reduction in FOXM1 levels. The down-regulation of ERα expression by ERβ1 is probably partially because FOXM1 regulates ERα expression.35 Together with previous findings,36 these results suggest that the ability of ERβ1 to repress FOXM1 expression depends on the presence of ERα.
ERβ1 Represses FOXM1 Transcription by the ERE on the FOXM1 Promoter
ERα has previously been shown to regulate FOXM1 expression through an ERE located on proximal region (−45 bp from the transcription start site) of the FOXM1 promoter36 (Figure 5A). To examine whether ERβ1 suppresses FOXM1 expression through the ERE-targeted by ERα, co-transfection assays were performed in both MCF-7 and MCF-7(ER-) cells with either the WT or mutant (mERE4) FOXM1 promoter in the presence of different amounts of ERβ1 and E2 (Figure 5, B and C). The results showed that the WT, but not the mutant (mERE4), FOXM1 promoter was repressed by ERβ1 in the ERα-positive MCF-7 cells (Figure 5B). In contrast, both the WT and mutant (mERE4) FOXM1 promoters displayed low-basal activities and were not responsive to ERβ1 repression in the ERα-negative MCF-7(ER-) cells (Figure 5C). Together these results indicate that the ERE-like element located at −45 bp confers the responsiveness to ERβ1, confirming that FOXM1 is a target gene of ERβ1. Moreover, these results further highlight that ERβ1 requires ERα for its repression of FOXM1 expression.
Figure 5.
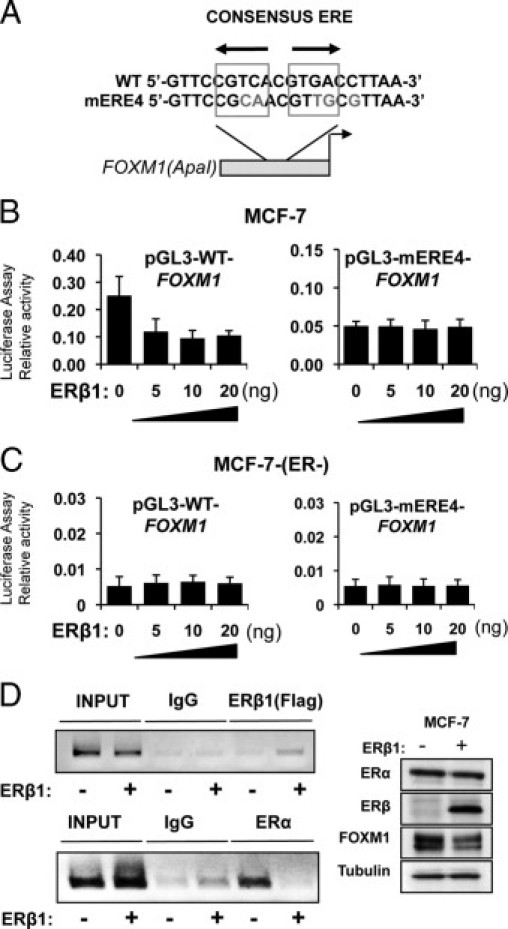
ERβ1 represses FOXM1 transcription by the ERE on the proximal promoter. A: Schematic representation of the ApaI FOXM1-luciferase reporter construct, showing the WT and the mutant ERE (mERE4) sequences. B: MCF-7 cells were transfected with pGL3-FOXM1(ApaI) WT or mERE4 and 0, 5, 10, or 20 ng of pcDNA3-Flag-ERβ1 expression vector in the presence of E2. The transfected cells were collected after 24 hours for firefly/Renilla luciferase assays with the use of the Dual-Glo Luciferase reporter assay system (Promega) according to the manufacturer's instructions. C: The ERα-negative, MCF-7(ER-) cells were transfected with pGL3-FOXM1(ApaI) WT or mERE4 and 0, 5, 10, or 20 ng of pcDNA3-Flag-ERβ1 expression vector in the presence of E2. The transfected cells were collected after 24 hours and analyzed for promoter activity as described. The WT, but not the mutant mERE4, FOXM1-luc activity was repressed by ERβ1 in the ERα-positive MCF-7 cells. In contrast, both the WT and mERE4 FOXM1-luc displayed low basal activities and were not responsive to ERβ1 repression in the ERα-negative MCF-7(ER-) cells. D: ChIP assays were performed to study the in vivo occupancy of the ERE region of the FOXM1 promoter. MCF-7 cells transfected with the control pcDNA3 vector or pcDNA3-Flag-ERβ1 were subjected to ChIP analysis with the ERα antibody and anti-Flag antibody, which recognized the transfected flag-tagged ERβ. The ChIP assays (inverted agarose gel images) showed that there was an increase in ERβ1 recruitment to the ERE region on ERβ1 ectopic expression. The occupancy of the ERE region by ERα was drastically reduced on ERβ1 expression, suggesting that ERβ1 displaces ERα from the ERE of FOXM1 promoter in vivo. Western blot analysis was also performed to show the expression levels of FOXM1, ERβ1, ERα, and tubulin in the transfected cells (right panel).
ERβ1 Displaces ERα from the ERE of FOXM1 Promoter in Vivo
To explore the mechanism by which ERβ1 represses FOXM1 expression, we studied the in vivo occupancy of the ERE site on the FOXM1 promoter by ERα and ERβ1 in MCF-7 cells in the absence or presence of ERβ1 expression by ChIP assays (Figure 5D). MCF-7 cells collected 24 hours after transfection with pcDNA3 as a control or pcDNA3-Flag-ERβ1 (Figure 5D) were subjected to ChIP analysis with the use of an ERα antibody and an anti-Flag antibody, which recognized the transfected Flag-tagged ERβ1. The ChIP assays showed that there was an increase in ERβ1 recruitment to the ERE region on ERβ1 ectopic expression. Concomitantly, occupancy of ERE region by ERα was drastically reduced in MCF-7 cells on ectopic ERβ1 expression, indicating that ERβ1 expression caused the disassociation of ERα from ERE region of the FOXM1 promoter (Figure 5D).
FOXM1 Is an Important Downstream Target of ERβ1
To show that FOXM1 is a functionally important downstream target of ERβ1, the parental MCF-7 cells and MCF-7 cells overexpressing WT FOXM1 (MCF-7-FOXM1) were transiently transfected with pcDNA3 or pcDNA3-Flag-ERβ1, and the rates of cell proliferation were monitored by SRB assays over 72 hours. Western blot analysis showed that overexpression of ERβ1 repressed FOXM1 expression in the MCF-7 cells but had little effects on FOXM1 in the MCF-7-FOXM1 cells at 48 hours (Figure 6A). Cell proliferation SRB assays showed that, although the MCF-7 cells transiently transfected with ERβ1 grew slower than the control cells, ERβ1 overexpression had no detectable effects on the proliferation of MCF-7-FOXM1 cells (Figure 6B), suggesting that FOXM1 is a critical downstream target of ERβ1 in the control of cell proliferation. The results also support the notion that ERβ1 negatively regulates FOXM1 expression at the gene promoter level, because FOXM1 expression was driven by the exogenous viral cytomegalovirus promoter, and, consequently, its transcription was not affected by ERβ1.
Figure 6.
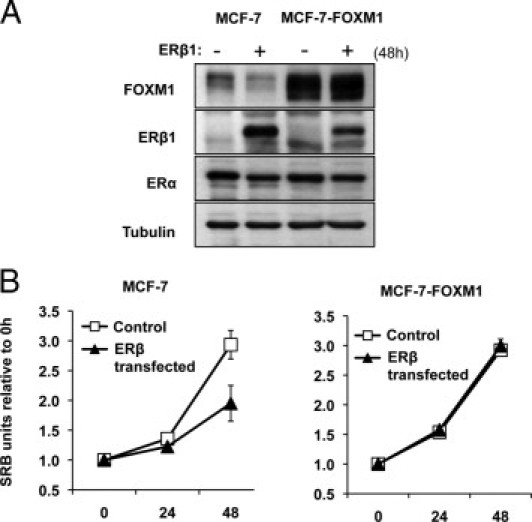
FOXM1 is a key downstream target of ERβ1 in breast cancer cells. Parental MCF-7 cells and MCF-7 cells overexpressing WT FOXM1 (MCF-7-FOXM1) were transiently transfected with pcDNA3 or pcDNA3-Flag-ERβ1 and used for Western blot analysis and SRB assays. A: Transfected cells were collected at 48 hours and analyzed for FOXM1, ERβ1, ERα, and tubulin expression by Western blot analysis. B: SRB assays were performed on these cells at 0, 24, 48, and 72 hours. Aliquots of the transfected cells were split into 96-well plates, and their proliferation was analyzed at the times indicated by SRB assays. Cell proliferation assays showed that. although the MCF-7 cells transiently transfected with ERβ1 grew slower than the control cells, ERβ1 overexpression had no detectable effects on the proliferation of MCF-7-FOXM1 cells.
Discussion
In humans, ERβ has at least five major isoforms (ERβ1 to ERβ 5) with distinct functions and tissue distributions.9 The importance of ERβ expression in breast cancer is well documented, with a number of studies showing that patients with ERβ-positive breast cancer treated with adjuvant tamoxifen have a better survival rate.41 Consistently, ERβ expression has been shown to be frequently associated with lower-grade tumors and negative axillary node status.42 In contrast, other studies have found that breast tumors co-expressing ERα and ERβ are often node positive and of higher grades,43 and breast tumors with increased ERβ expression are linked to tamoxifen resistance.44 These findings highlight the discrepancies in the knowledge on ERβ and its isoforms and emphasize that the clinical implications of ERβ expression, their mechanisms of action, and downstream targets in breast cancer still remain enigmatic.
The forkhead transcription factor FOXM1 is fundamental to breast cancer initiation and progression. Accordingly, FOXM1 expression increases during breast cancer tumorigenesis,31 and deregulated FOXM1 expression has been linked to resistance to chemotherapeutic agents, including gefitinib, lapatinib, and cisplatin, in breast cancer.24,31,45 Recently, FOXM1 has been shown to have a role in sensitivity and resistance of breast cancer endocrine therapy.36 Here, we studied the effects of ERβ1 ectopic expression in breast cancer cell lines and nude mice xenografts and observed that ERβ1 expression suppresses tumor cell proliferation and represses FOXM1 expression at mRNA and protein levels in ERα-positive but not ERα-negative breast cancer cells. This notion is further supported by the finding of a significant inverse correlation between ERβ1 and FOXM1 expression at both protein and mRNA transcript levels in ERα-positive breast cancer patient samples. Notably, there is no correlation between FOXM1 and ERβ2 or ERβ5 expression in total or ERα-positive breast cancer cases, although we cannot exclude that this might relate to the modest sample size. The finding that the repression of FOXM1 expression by ERβ1 in breast cancer depends on ERα suggests that ERβ1 represses FOXM1 expression through ERα. Consistent with this idea, the activity of WT and ERE-mutant FOXM1 promoters was lower in the ERα-negative MCF-7-(ER-) cells compared with the ERα-positive MCF-7 cells. Although ERβ is able to act in an ERE-independent manner,7 our data evidently show that ERβ1 acts through the ERE located on the FOXM1 promoter. Indeed, the in vitro promoter analysis showed that ERβ1 represses FOXM1 promoter activity through a proximal ERE site, which has previously been shown to be responsible for ERα induction in breast cancer cells.36 In addition, ChIP assays showed that ERβ1, when ectopically expressed, displaces ERα from the ERE region of the FOXM1 promoter in vivo, indicating that ERβ1 functions by competing with ERα for ERE binding. ERβ/ERβ homodimers have been suggested to have lower transcriptional activity than ERα/ERα or ERα/ERβ,46 and this might provide one mechanistic explanation as to how ERβ antagonizes ERα transcriptional output in the regulation of FOXM1 expression. The data also showed that ERβ1 functions primarily through antagonizing the action of ERα, because ERβ1 overexpression had no effects on FOXM1 expression and FOXM1 promoter activity in ERα-negative breast cancer cells.
The findings that ERβ1 is able to repress FOXM1 through antagonizing ERα is expected to have fundamental implications for the treatment of ERα-positive breast cancer. A number of highly selective ERβ ligands have already been generated, and some are currently under clinical evaluation for breast cancer treatment.4,47 However, loss of ERβ expression is a common event during breast and ovarian cancer tumorigenesis as well as progression,43 and this loss of ERβ expression has been linked to DNA methylation.48–50 In fact, treatment of ovarian and breast carcinoma cells with the demethylating agent 5-aza-2′ deoxycytidine has been shown to result in re-expression of the ERβ gene.49–51 Thus, treatment of ERβ-negative breast cancer with demethylating agents can be a viable strategy to reactivate ERβ expression to antagonize the ERα signaling in breast cancer. Because most patients with ERα-positive breast cancer are treated with and respond to endocrine therapy,16–18 these demethylating agents could also be used in combination with anti-estrogens or aromatase inhibitors to antagonize ERα signaling and, thus, to increase the efficacy of endocrine therapy in breast cancer. Equally, ERβ1 can also be an important prognostic biomarker as well as predictive factor for endocrine treatment sensitivity in breast cancer. Consistent with the hypothesis that ERβ1 is an important target for breast cancer treatment and marker for prognosis, we have obtained preliminary data showing that re-expression of ERβ1 can enhance the anti-proliferative effects of tamoxifen in the ERα-positive breast cancer cell line MCF-7 (see Supplemental Figure S4 at http://ajp.amjpathol.org).
In summary, our study shows that the most common ERβ isoform, ERβ1, negatively regulates the expression of the oncogenic forkhead transcription factor FOXM1 in breast cancer cells. This is consistent with the observations that ERβ1 is associated with the suppression of breast cancer cell proliferation and survival. We also observed that ERβ1 represses FOXM1 expression primarily through competing with ERα on binding to the ERE located on the proximal promoter. The inverse relation between ERβ1 and FOXM1 expression was confirmed in human breast cancer samples. In addition, our data showed that suppression of FOXM1 expression is the key mechanism mediating the anti-proliferative actions of ERβ1, and ectopic expression of FOXM1 can override the anti-proliferative effects of ERβ1. In summary, our findings provide insights into the role and mechanism of action of ERβ1 and identify FOXM1 as a crucial downstream target of ERβ1 in breast cancer. The indirect regulation of FOXM1 by ERβ1 by ERα could determine the responsiveness to breast cancer endocrine therapy, and this estrogen-signaling axis can therefore be important for breast cancer treatment and prognosis, especially because some two-thirds of breast carcinomas co-express ERα and ERβ.52
Footnotes
Supported by Breast Cancer Campaign (J.M. and E.W.-F.L.) and Cancer Research UK (Y.O., K.-K.H., R.C.C., and E.W.-F.L.).
Supplemental material for this article can be found at http://ajp.amjpathol.org or at doi: 10.1016/j.ajpath.2011.05.052.
Supplementary data
Representative staining of TMAs with antibodies against ERα, ERβ1, ERβ2, ERβ5, and FOXM1. Scale bar = 30 μm.
Statistical analysis of FOXM1, ERβ1, ERβ2, and ERβ5 staining by Fisher's test. The staining of FOXM1, ERβ1, ERβ2, and ERβ5 was assessed with a scanscope (Scanscope Aperio Technologies, Inc, Vista, CA) connected to a personal computer. The staining intensity and percentage of staining in the cytoplasm and the nucleus were each scored independently in a semiquantitative fashion. For each case, a final score from the nucleus and the cytoplasm was obtained by multiplying the score of intensity with the score of the percentage, 8 being the maximum final score. To avoid subjectivity in evaluation, scoring was done by two independent persons. Scores of 0 to 3 are classified as negative (-) and 4 to 8 as positive (+). A: There was no significant correlation between the expression levels of FOXM1 and ERβ2 or ERβ5. A potential but non-significant inverse correlation trend was detected between ERβ1 and FOXM1 expression (n = 245; P = 0.051). B: When IHC data were re-evaluated after excluding ERα-negative samples, a significant correlation was observed between FOXM1 and ERβ1 expression (n = 173; P = 0.039) but not between FOXM1 and ERβ2 or ERβ5.
ERβ1 has no effects on FOXM1 expression in the ERα-negative ZR75-1(ER-) cells. Effects of ERβ1 ectopic expression on FOXM1 expression was examined in the ERα-negative ZR75-1(ER-) breast carcinoma cell line. The breast cancer cells were transiently transfected with ERβ1 expression vector or an empty vector control and were collected at 48 hours for Western blot analysis for ERα, ERβ, FOXM1, and tubulin expression (left panel) and for and real-time quantitative PCR analysis (right panels).
ERβ1 can combine with tamoxifen to inhibit the proliferation of the ERα-positive MCF-7 breast cancer cells and has no effects on FOXM1 expression in the ERα-negative ZR75-1(ER-) cells. MCF-7 breast cancer cells transfected with ERβ1 expression vector or an empty vector control pcDNA3 were cultured with 0, 1, or 5 μmol/L tamoxifen in the presence of E2. Aliquots of the transfected cells were collected for Western blot analysis, ERβ, FOXM1, and tubulin expression (top panel) and used or SRB proliferation analysis (lower panels).
References
- 1.Musgrove E.A., Sutherland R.L. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9:631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 2.Koehler K.F., Helguero L.A., Haldosen L.A., Warner M., Gustafsson J.A. Reflections on the discovery and significance of estrogen receptor beta. Endocr Rev. 2005;26:465–478. doi: 10.1210/er.2004-0027. [DOI] [PubMed] [Google Scholar]
- 3.Speirs V., Walker R.A. New perspectives into the biological and clinical relevance of oestrogen receptors in the human breast. J Pathol. 2007;211:499–506. doi: 10.1002/path.2130. [DOI] [PubMed] [Google Scholar]
- 4.Nilsson S., Gustafsson J.A. Estrogen receptors: therapies targeted to receptor subtypes. Clin Pharmacol Ther. 2011;89:44–55. doi: 10.1038/clpt.2010.226. [DOI] [PubMed] [Google Scholar]
- 5.Lim W., Cho J., Kwon H.Y., Park Y., Rhyu M.R., Lee Y. Hypoxia-inducible factor 1 alpha activates and is inhibited by unoccupied estrogen receptor beta. FEBS Lett. 2009;583:1314–1318. doi: 10.1016/j.febslet.2009.03.028. [DOI] [PubMed] [Google Scholar]
- 6.Poola I., Fuqua S.A., De Witty R.L., Abraham J., Marshallack J.J., Liu A. Estrogen receptor alpha-negative breast cancer tissues express significant levels of estrogen-independent transcription factors: ERbeta1 and ERbeta5: potential molecular targets for chemoprevention. Clin Cancer Res. 2005;11:7579–7585. doi: 10.1158/1078-0432.CCR-05-0728. [DOI] [PubMed] [Google Scholar]
- 7.Zhang G., Liu X., Farkas A.M., Parwani A.V., Lathrop K.L., Lenzner D., Land S.R., Srinivas H. Estrogen receptor beta functions through nongenomic mechanisms in lung cancer cells. Mol Endocrinol. 2009;23:146–156. doi: 10.1210/me.2008-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Younes M., Honma N. Estrogen receptor beta. Arch Pathol Lab Med. 2011;135:63–66. doi: 10.5858/2010-0448-RAR.1. [DOI] [PubMed] [Google Scholar]
- 9.Moore J.T., McKee D.D., Slentz-Kesler K., Moore L.B., Jones S.A., Horne E.L., Su J.L., Kliewer S.A., Lehmann J.M., Willson T.M. Cloning and characterization of human estrogen receptor beta isoforms. Biochem Biophys Res Commun. 1998;247:75–78. doi: 10.1006/bbrc.1998.8738. [DOI] [PubMed] [Google Scholar]
- 10.Hopp T.A., Weiss H.L., Parra I.S., Cui Y., Osborne C.K., Fuqua S.A. Low levels of estrogen receptor beta protein predict resistance to tamoxifen therapy in breast cancer. Clin Cancer Res. 2004;10:7490–7499. doi: 10.1158/1078-0432.CCR-04-1114. [DOI] [PubMed] [Google Scholar]
- 11.Murphy L.C., Watson P.H. Is oestrogen receptor-beta a predictor of endocrine therapy responsiveness in human breast cancer? Endocr Relat Cancer. 2006;13:327–334. doi: 10.1677/erc.1.01141. [DOI] [PubMed] [Google Scholar]
- 12.Honma N., Saji S., Kurabayashi R., Aida J., Arai T., Horii R., Akiyama F., Iwase T., Harada N., Younes M., Toi M., Takubo K., Sakamoto G. Oestrogen receptor-beta1 but not oestrogen receptor-betacx is of prognostic value in apocrine carcinoma of the breast. APMIS. 2008;116:923–930. doi: 10.1111/j.1600-0463.2008.01122.x. [DOI] [PubMed] [Google Scholar]
- 13.Murphy L.C., Peng B., Lewis A., Davie J.R., Leygue E., Kemp A., Ung K., Vendetti M., Shiu R. Inducible upregulation of oestrogen receptor-beta1 affects oestrogen and tamoxifen responsiveness in MCF7 human breast cancer cells. J Mol Endocrinol. 2005;34:553–566. doi: 10.1677/jme.1.01688. [DOI] [PubMed] [Google Scholar]
- 14.Palmieri C., Lam E.W., Mansi J., MacDonald C., Shousha S., Madden P., Omoto Y., Sunters A., Warner M., Gustafsson J.A., Coombes R.C. The expression of ER beta cx in human breast cancer and the relationship to endocrine therapy and survival. Clin Cancer Res. 2004;10:2421–2428. doi: 10.1158/1078-0432.ccr-03-0215. [DOI] [PubMed] [Google Scholar]
- 15.Zhao C., Toresson G., Xu L., Koehler K.F., Gustafsson J.A., Dahlman-Wright K. Mouse estrogen receptor beta isoforms exhibit differences in ligand selectivity and coactivator recruitment. Biochemistry. 2005;44:7936–7944. doi: 10.1021/bi047691m. [DOI] [PubMed] [Google Scholar]
- 16.Gapinski P.V., Donegan W.L. Estrogen receptors and breast cancer: prognostic and therapeutic implications. Surgery. 1980;88:386–393. [PubMed] [Google Scholar]
- 17.Osborne C.K., McGuire W.L. The use of steroid hormone receptors in the treatment of human breast cancer: a review. Bull Cancer. 1979;66:203–209. [PubMed] [Google Scholar]
- 18.Elkak A.E., Mokbel K. Pure antiestrogens and breast cancer. Curr Med Res Opin. 2001;17:282–289. [PubMed] [Google Scholar]
- 19.Myatt S.S., Lam E.W. Targeting FOXM1 (letter to the editor) Nat Rev Cancer. 2008;8:242. doi: 10.1038/nrc2223-c2. [DOI] [PubMed] [Google Scholar]
- 20.Myatt S.S., Lam E.W. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 21.Laoukili J., Kooistra M.R., Bras A., Kauw J., Kerkhoven R.M., Morrison A., Clevers H., Medema R.H. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 22.Li Q., Zhang N., Jia Z., Le X., Dai B., Wei D., Huang S., Tan D., Xie K. Critical role and regulation of transcription factor FoxM1 in human gastric cancer angiogenesis and progression. Cancer Res. 2009;69:3501–3509. doi: 10.1158/0008-5472.CAN-08-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwok J.M., Myatt S.S., Marson C.M., Coombes R.C., Constantinidou D., Lam E.W. Thiostrepton selectively targets breast cancer cells through inhibition of forkhead box M1 expression. Mol Cancer Ther. 2008;7:2022–2032. doi: 10.1158/1535-7163.MCT-08-0188. [DOI] [PubMed] [Google Scholar]
- 24.Kwok J.M., Peck B., Monteiro L.J., Schwenen H.D., Millour J., Coombes R.C., Myatt S.S., Lam E.W. FOXM1 confers acquired cisplatin resistance in breast cancer cells. Mol Cancer Res. 2010;8:24–34. doi: 10.1158/1541-7786.MCR-09-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan Y., Raychaudhuri P., Costa R.H. Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Mol Cell Biol. 2007;27:1007–1016. doi: 10.1128/MCB.01068-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H., Ackermann A.M., Gusarova G.A., Lowe D., Feng X., Kopsombut U.G., Costa R.H., Gannon M. The FoxM1 transcription factor is required to maintain pancreatic beta-cell mass. Mol Endocrinol. 2006;20:1853–1866. doi: 10.1210/me.2006-0056. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida Y., Wang I.C., Yoder H.M., Davidson N.O., Costa R.H. The forkhead box M1 transcription factor contributes to the development and growth of mouse colorectal cancer. Gastroenterology. 2007;132:1420–1431. doi: 10.1053/j.gastro.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 28.Kim I.M., Ackerson T., Ramakrishna S., Tretiakova M., Wang I.C., Kalin T.V., Major M.L., Gusarova G.A., Yoder H.M., Costa R.H., Kalinichenko V.V. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 2006;66:2153–2161. doi: 10.1158/0008-5472.CAN-05-3003. [DOI] [PubMed] [Google Scholar]
- 29.Kalin T.V., Wang I.C., Ackerson T.J., Major M.L., Detrisac C.J., Kalinichenko V.V., Lyubimov A., Costa R.H. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 2006;66:1712–1720. doi: 10.1158/0008-5472.CAN-05-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalinichenko V.V., Major M.L., Wang X., Petrovic V., Kuechle J., Yoder H.M., Dennewitz M.B., Shin B., Datta A., Raychaudhuri P., Costa R.H. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–850. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Francis R.E., Myatt S.S., Krol J., Hartman J., Peck B., McGovern U.B., Wang J., Guest S.K., Filipovic A., Gojis O., Palmieri C., Peston D., Shousha S., Yu Q., Sicinski P., Coombes R.C., Lam E.W. FoxM1 is a downstream target and marker of HER2 overexpression in breast cancer. Int J Oncol. 2009;35:57–68. doi: 10.3892/ijo_00000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pilarsky C., Wenzig M., Specht T., Saeger H.D., Grutzmann R. Identification and validation of commonly overexpressed genes in solid tumors by comparison of microarray data. Neoplasia. 2004;6:744–750. doi: 10.1593/neo.04277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carr J.R., Park H.J., Wang Z., Kiefer M.M., Raychaudhuri P. FoxM1 mediates resistance to herceptin and paclitaxel. Cancer Res. 2010;70:5054–5063. doi: 10.1158/0008-5472.CAN-10-0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin K.J., Patrick D.R., Bissell M.J., Fournier M.V. Prognostic breast cancer signature identified from 3D culture model accurately predicts clinical outcome across independent datasets. PLoS ONE. 2008;3:e2994. doi: 10.1371/journal.pone.0002994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madureira P.A., Varshochi R., Constantinidou D., Francis R.E., Coombes R.C., Yao K.M., Lam E.W. The Forkhead box M1 protein regulates the transcription of the estrogen receptor alpha in breast cancer cells. J Biol Chem. 2006;281:25167–25176. doi: 10.1074/jbc.M603906200. [DOI] [PubMed] [Google Scholar]
- 36.Millour J., Constantinidou D., Stavropoulou A.V., Wilson M.S., Myatt S.S., Kwok J.M., Sivanandan K., Coombes R.C., Medema R.H., Hartman J., Lykkesfeldt A.E., Lam E.W. FOXM1 is a transcriptional target of ERalpha and has a critical role in breast cancer endocrine sensitivity and resistance. Oncogene. 2010;29:2983–2995. doi: 10.1038/onc.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hartman J., Lindberg K., Morani A., Inzunza J., Strom A., Gustafsson J.A. Estrogen receptor beta inhibits angiogenesis and growth of T47D breast cancer xenografts. Cancer Res. 2006;66:11207–11213. doi: 10.1158/0008-5472.CAN-06-0017. [DOI] [PubMed] [Google Scholar]
- 38.Essafi A., Fernandez de Mattos S., Hassen Y.A., Soeiro I., Mufti G.J., Thomas N.S., Medema R.H., Lam E.W. Direct transcriptional regulation of Bim by FoxO3a mediates STI571-induced apoptosis in Bcr-Abl-expressing cells. Oncogene. 2005;24:2317–2329. doi: 10.1038/sj.onc.1208421. [DOI] [PubMed] [Google Scholar]
- 39.Essafi A., Gomes A.R., Pomeranz K.M., Zwolinska A.K., Varshochi R., McGovern U.B., Lam E.W. Studying the subcellular localization and DNA-binding activity of FoxO transcription factors, downstream effectors of PI3K/Akt. Methods Mol Biol. 2009;462:201–211. doi: 10.1007/978-1-60327-115-8_13. [DOI] [PubMed] [Google Scholar]
- 40.Shaaban A.M., Green A.R., Karthik S., Alizadeh Y., Hughes T.A., Harkins L., Ellis I.O., Robertson J.F., Paish E.C., Saunders P.T., Groome N.P., Speirs V. Nuclear and cytoplasmic expression of ERbeta1: ERbeta2, and ERbeta5 identifies distinct prognostic outcome for breast cancer patients. Clin Cancer Res. 2008;14:5228–5235. doi: 10.1158/1078-0432.CCR-07-4528. [DOI] [PubMed] [Google Scholar]
- 41.Mann S., Laucirica R., Carlson N., Younes P.S., Ali N., Younes A., Li Y., Younes M. Estrogen receptor beta expression in invasive breast cancer. Hum Pathol. 2001;32:113–118. doi: 10.1053/hupa.2001.21506. [DOI] [PubMed] [Google Scholar]
- 42.Jarvinen T.A., Pelto-Huikko M., Holli K., Isola J. Estrogen receptor beta is coexpressed with ERalpha and PR and associated with nodal status, grade, and proliferation rate in breast cancer. Am J Pathol. 2000;156:29–35. doi: 10.1016/s0002-9440(10)64702-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leygue E., Dotzlaw H., Watson P.H., Murphy L.C. Altered estrogen receptor alpha and beta messenger RNA expression during human breast tumorigenesis. Cancer Res. 1998;58:3197–3201. [PubMed] [Google Scholar]
- 44.Speirs V., Malone C., Walton D.S., Kerin M.J., Atkin S.L. Increased expression of estrogen receptor beta mRNA in tamoxifen-resistant breast cancer patients. Cancer Res. 1999;59:5421–5424. [PubMed] [Google Scholar]
- 45.McGovern U.B., Francis R.E., Peck B., Guest S.K., Wang J., Myatt S.S., Krol J., Kwok J.M., Polychronis A., Coombes R.C., Lam E.W. Gefitinib (Iressa) represses FOXM1 expression via FOXO3a in breast cancer. Mol Cancer Ther. 2009;8:582–591. doi: 10.1158/1535-7163.MCT-08-0805. [DOI] [PubMed] [Google Scholar]
- 46.Lindberg M.K., Moverare S., Skrtic S., Gao H., Dahlman-Wright K., Gustafsson J.A., Ohlsson C. Estrogen receptor (ER)-beta reduces ERalpha-regulated gene transcription, supporting a “ying yang” relationship between ERalpha and ERbeta in mice. Mol Endocrinol. 2003;17:203–208. doi: 10.1210/me.2002-0206. [DOI] [PubMed] [Google Scholar]
- 47.Mohler M.L., Narayanan R., Coss C.C., Hu K., He Y., Wu Z., Hong S.S., Hwang D.J., Miller D.D., Dalton J.T. Estrogen receptor beta selective nonsteroidal estrogens: seeking clinical indications. Expert Opin Ther Pat. 2010;20:507–534. doi: 10.1517/13543771003657164. [DOI] [PubMed] [Google Scholar]
- 48.Skliris G.P., Munot K., Bell S.M., Carder P.J., Lane S., Horgan K., Lansdown M.R., Parkes A.T., Hanby A.M., Markham A.F., Speirs V. Reduced expression of oestrogen receptor beta in invasive breast cancer and its re-expression using DNA methyl transferase inhibitors in a cell line model. J Pathol. 2003;201:213–220. doi: 10.1002/path.1436. [DOI] [PubMed] [Google Scholar]
- 49.Zhao C., Lam E.W., Sunters A., Enmark E., De Bella M.T., Coombes R.C., Gustafsson J.A., Dahlman-Wright K. Expression of estrogen receptor beta isoforms in normal breast epithelial cells and breast cancer: regulation by methylation. Oncogene. 2003;22:7600–7606. doi: 10.1038/sj.onc.1207100. [DOI] [PubMed] [Google Scholar]
- 50.Rody A., Holtrich U., Solbach C., Kourtis K., von Minckwitz G., Engels K., Kissler S., Gatje R., Karn T., Kaufmann M. Methylation of estrogen receptor beta promoter correlates with loss of ER-beta expression in mammary carcinoma and is an early indication marker in premalignant lesions. Endocr Relat Cancer. 2005;12:903–916. doi: 10.1677/erc.1.01088. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki F., Akahira J., Miura I., Suzuki T., Ito K., Hayashi S., Sasano H., Yaegashi N. Loss of estrogen receptor beta isoform expression and its correlation with aberrant DNA methylation of the 5′-untranslated region in human epithelial ovarian carcinoma. Cancer Sci. 2008;99:2365–2372. doi: 10.1111/j.1349-7006.2008.00988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skliris G.P., Leygue E., Watson P.H., Murphy L.C. Estrogen receptor alpha negative breast cancer patients: estrogen receptor beta as a therapeutic target. J Steroid Biochem Mol Biol. 2008;109:1–10. doi: 10.1016/j.jsbmb.2007.12.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative staining of TMAs with antibodies against ERα, ERβ1, ERβ2, ERβ5, and FOXM1. Scale bar = 30 μm.
Statistical analysis of FOXM1, ERβ1, ERβ2, and ERβ5 staining by Fisher's test. The staining of FOXM1, ERβ1, ERβ2, and ERβ5 was assessed with a scanscope (Scanscope Aperio Technologies, Inc, Vista, CA) connected to a personal computer. The staining intensity and percentage of staining in the cytoplasm and the nucleus were each scored independently in a semiquantitative fashion. For each case, a final score from the nucleus and the cytoplasm was obtained by multiplying the score of intensity with the score of the percentage, 8 being the maximum final score. To avoid subjectivity in evaluation, scoring was done by two independent persons. Scores of 0 to 3 are classified as negative (-) and 4 to 8 as positive (+). A: There was no significant correlation between the expression levels of FOXM1 and ERβ2 or ERβ5. A potential but non-significant inverse correlation trend was detected between ERβ1 and FOXM1 expression (n = 245; P = 0.051). B: When IHC data were re-evaluated after excluding ERα-negative samples, a significant correlation was observed between FOXM1 and ERβ1 expression (n = 173; P = 0.039) but not between FOXM1 and ERβ2 or ERβ5.
ERβ1 has no effects on FOXM1 expression in the ERα-negative ZR75-1(ER-) cells. Effects of ERβ1 ectopic expression on FOXM1 expression was examined in the ERα-negative ZR75-1(ER-) breast carcinoma cell line. The breast cancer cells were transiently transfected with ERβ1 expression vector or an empty vector control and were collected at 48 hours for Western blot analysis for ERα, ERβ, FOXM1, and tubulin expression (left panel) and for and real-time quantitative PCR analysis (right panels).
ERβ1 can combine with tamoxifen to inhibit the proliferation of the ERα-positive MCF-7 breast cancer cells and has no effects on FOXM1 expression in the ERα-negative ZR75-1(ER-) cells. MCF-7 breast cancer cells transfected with ERβ1 expression vector or an empty vector control pcDNA3 were cultured with 0, 1, or 5 μmol/L tamoxifen in the presence of E2. Aliquots of the transfected cells were collected for Western blot analysis, ERβ, FOXM1, and tubulin expression (top panel) and used or SRB proliferation analysis (lower panels).


