Abstract
Pancreatic diseases, which include diabetes, pancreatitis, and pancreatic cancer, are often difficult to detect and/or stage, contributing to a reduced quality of life and lifespan for patients. Thus, there is need for a technology that can visualize tissue changes in the pancreas, improve understanding of disease progression, and facilitate earlier detection in the human population. Because of low spatial resolution, current clinical magnetic resonance imaging (MRI) at low field strength has yet to fully visualize the exocrine, endocrine, vascular, and stromal components of the pancreas. We used high field strength magnetic resonance microscopy (μMRI) to image mouse pancreas ex vivo without contrast agents at high spatial resolution. We analyzed the resulting high-resolution images using volume rendering to resolve components in the pancreas, including acini, islets, blood vessels, and extracellular matrix. Locations and dimensions of pancreatic components as seen in three-dimensional μMRI were compared with histological images, and good correspondence was found. Future longitudinal studies could expand on the use of in vivo μMRI in mouse models of pancreatic diseases. Capturing three-dimensional structural changes through μMRI could help to identify early cellular and tissue changes associated with pancreatic disease, serving as a mode of improved detection in the clinic for endocrine and exocrine pathologies.
Diagnosis of pancreatic diseases is often complicated by problems of detection. Blood screening (glucose tolerance test) is usually sufficient for diagnosis of late-onset or type 2 diabetes; nonetheless, borderline cases require long-term assessment, sometimes without resolution, and many cases of diabetes can go undetected for years. Furthermore, the potential increased risk for pancreatic cancer in cohorts of individuals with late-onset diabetes cannot be readily followed. Both pancreatitis and pancreatic cancer are difficult to diagnose, and discriminating between these two diseases can be extremely difficult without pathological evaluation of a tissue biopsy. Likewise, a method for tracking a high-risk familial pancreatitis population for the development of pancreatic cancer is not readily available. Reliable imaging methods would provide a powerful tool for visualizing tissue architecture and could substantially improve diagnosis of pathologies in the pancreas.
Magnetic resonance imaging (MRI) with high spatial resolution is an effective and well-established method for depicting normal tissue morphology and detecting pathological tissue changes. Current clinical imaging methods, however, cannot capture cellular architecture, glandular morphology, and compartmentalization of different cell types in the pancreas, because of low spatial resolution. In recent years, the advent of high field strength (>9.4 T) MRI systems has allowed imaging of mouse models of diseases using magnetic resonance microscopy (μMRI) (ie, imaging at microscopic spatial resolution, typically ∼50 μm).1 The greater sensitivity of high field strength imaging systems can be used to image the pancreas at high spatial resolution and to visualize tissue microstructure that is not visible at lower field strengths.2,3
The mouse pancreas has a number of features that make it an ideal organ for μMRI investigation of tissue microstructure, including i) physiologically different cell populations (acinar, ductal, and islet), ii) a clearly defined structure for each cellular component (acini, ducts, and islets), and iii) conserved stromal separation of these components with a distinct vasculature. In vivo MRI detection of morphological changes associated with pathology would reduce the need for risky biopsy procedures, which can lead to local inflammation and pancreatitis. In addition, artifacts and distortions due to tissue processing and sectioning would be avoided, allowing for maintenance of normal anatomical or pathological features in the pancreas. Before this technique can be applied in vivo, however, it is necessary to understand the μMRI phenotype of the exocrine and endocrine components of the mouse pancreas, which was the objective of the present study.
Previous MRI studies of mouse pancreas have used contrast agents to image orthotopic mouse models of pancreatic cancer and mouse models of diabetes; however, the ability to detect pathological cellular changes in the pancreatic tissue was limited by low spatial resolution at low field strengths.2,3 In the present study, we evaluated the use of high field strength (14.1 T) μMRI to capture the three-dimensional (3D) structure and morphology of cellular and anatomical components in normal mouse pancreas. μMRI images were obtained from ex vivo pancreas from chimeric mice (FVB6 F1) without the use of contrast agents. Contrast between different components of the pancreas was based on the endogenous differences in tissue relaxation times and proton density. High-resolution 3D μMRI data were reconstructed to provide volume images of the pancreatic components and to assess their spatial relationships. The present study represents the first 3D μMRI microimaging study of mouse pancreas. Our results indicate that volume images of the pancreas reconstructed from high-resolution μMRI images can reveal several important components of normal mouse pancreas, including parenchymal tissue (acini, islets, blood vessels) and connective tissue (stroma). Observing changes in these structures during the development of disease can be expected to shed light on important mechanisms that could contribute to the initiation and/or progression of pancreatic diseases.4 Clinically, changes in any of these cellular compartments can result in pathological states, including pancreatitis (acini), cancer (ductal), and diabetes (islets), and the ability to detect architectural changes will likely improve diagnostic capabilities for these diseases.
Materials and Methods
Mouse Strain
The mouse strain used for the present study was the FVB6 F1 strain, which is the first-generation chimera between FVB/N and C57/BL6. FVB6 F1 mice are a hybrid strain, the direct product of crossing FBV mice with B6 mice. A hybrid strain, although genetically similar across offspring, more realistically mimics the genetic heterogeneity observed in the human population. Mice were housed in microisolator cages with 12-hour light-dark cycles and a standard mouse chow ad libitum. All protocols were in compliance with the Animal Care and Use Committee of Northwestern University and with the guidelines established by the NIH for Care and Use of Laboratory Animals (7th edition, 1996).
Formalin Perfusion
Eleven-month-old mice were anesthetized with 100 mg/kg ketamine and 10 mg/kg xylazine delivered via a single intraperitoneal injection. The animals were perfused with heparinized saline for 5 minutes and then with 10% phosphate buffered formalin for 5 minutes. Immediately after perfusion, the pancreas was harvested by an abdominal midline laparotomy, along with portions of duodenum and spleen. The pancreas was removed from under the spleen and along the upper part of the intestine. The sample was stored in 10% phosphate-buffered formalin for further immersion fixation for a minimum of 2 weeks until prepared and mounted for MRI in fomblin (Sigma-Aldrich, St. Louis, MO).
Sample Preparation for μMRI
In all of the samples, a small portion of duodenum and of spleen was preserved on the sample, for orientation of the pancreas. The samples were gently stretched over a thin piece of plastic (2 cm × 4 cm) to maintain structural integrity and orientation. The pancreata were tied with sutures to the plastic to inhibit movement. These samples were placed in a 2-cm-diameter tube and submerged in fomblin. Fomblin is an inert liquid that has no proton magnetic resonance signal and is used to minimize magnetic susceptibility gradients and to keep the tissue from drying out during imaging.
μMRI Imaging
All imaging experiments were performed on a Bruker (Billerica, MA) Avance 14.1-T microimager using a 20-mm resonator tuned to proton frequency (600.133 MHz). High-resolution μMRI images were acquired using a 3D RARE (rapid acquisition with relaxation enhancement) pulse sequence. The following imaging parameters were used: TR 2500 ms, TE 7.5 ms, RARE factor 8, effective TE 30 ms, flip angle 90 degrees, field of view 20 mm × 16 mm × 3.55 mm, matrix size 320 × 256 × 100 for pixel size of 62 μm × 62 μm × 35 μm, antialiasing factor 1.0 in all three directions, fat suppression, and 8 averages, with a total scan time of 21 hours 20 minutes. For fat suppression, a frequency-selective Gaussian 90-degree pulse (bandwidth 2100 Hz) was applied at a constant frequency offset of −3.5 ppm with respect to water, which corresponds to the most intense signal in the fat spectrum. Transverse magnetization after application of the fat-suppression pulse was dephased using a spoiler gradient. The resulting image data set consisted of a series of 100 slices, which were either analyzed individually to identify pancreatic components or were used to generate volume-rendered 3D images.
Analysis of 3D μMRI Images
3D μMRI images were analyzed using direct volume rendering, a common data visualization technique that creates a two-dimensional (2D) image of a 3D data set by computing voxel intensity projections onto a plane. In this method, each pixel of the 2D image is computed by the weighted sum of the corresponding voxel intensities. Voxel weight defines voxel opacity and is computed as a transfer function (OPF) of voxel intensity. A typical OPF setting for displaying the desired magnetic resonance intensity interval [ILow, IHigh] is a linear function from 0 to MaxOp:
| (1) |
The voxel opacity function and color scheme can be manipulated (see Supplemental Figure S1 at http://ajp.amjpathol.org) to reveal 3D structures characterized by different magnetic resonance intensity amplitudes, structures that cannot be seen in the individual slice images used to generate the 3D image. For our analysis, we used VolView version 2.0 (Kitware, Clifton Park, NY), a freely available visualization framework that can directly load raw data files created by the Bruker system.
Histology
After μMRI, pancreata were removed from fomblin, rinsed in isopropanol, rinsed with PBS, and submerged into 10% phosphate-buffered formalin. After standard processing, pancreata were embedded in paraffin with maintained orientation, using a suture to landmark once the sample was unmounted from plastic (although processing does shrink and alter minor aspects of gross pathology). Samples were serially sectioned into slices 6 μm thick, with intermittent gaps of 50 or 100 μm. Slides were deparaffinized, hydrated, treated with H&E, and covered with glass coverslips. Overall phenotype was assessed by the presence of normal architecture (which included organization of acini and ducts, number and size of islets, presence of fibrosis and/or lipoatrophy) and any additional abnormalities via light microscopy. Histological images obtained by light microscopy were compared with μMRI images. Spleen and intestine were used as landmarks to identify the dorsal pancreas (adjacent to intestine, which correlates to the head of human pancreas) and ventral pancreas (adjacent to spleen, which correlates to the tail of human pancreas). Larger ducts are generally more visible near the dorsal end of the pancreas. These features served to orient pancreas samples, especially in comparison between μMRI and H&E images.
Results
Slice View of Pancreatic Structures in μMRI Images
High spatial resolution μMRI images of mouse pancreas acquired at 14.1 T demonstrated excellent contrast between various structures within the pancreas, based on their T2 and proton density differences (Figure 1). Regions of low, moderate, and high signal intensity were revealed in the T2-weighted spin-echo slice images (Figure 1A) and were identified by comparison with known anatomical and tissue features of the pancreas from the literature, as well as our own H&E histological evaluations (Figure 1B). Broad regions of low signal intensity correspond to acinar cells, which constitute ≥80% of the entire gland.5 Within these areas, distinct acinar lobules of cells with visibly dark nuclei can be seen. This assignment is consistent with H&E staining, which demonstrates dark eosinophilic regions (primarily protein-rich zymogen) along the apical ends and dark hematoxylin staining (nucleic acids) in nuclei. Several structures can be observed within the acinar parenchyma. Small elliptical structures with focal, moderate intensity distributed throughout the acinar cells are consistent with the arrangement of islets, the endocrine component. Regions of high signal intensity appear in the μMRI image as curves of varying thicknesses that cut through the organ, often connecting with one another, implying a network that is consistent with either ductal tree, vasculature, or stromal tissue in the pancreas. Further analysis of this hyperintense structural component was facilitated by visualizing the μMRI data as 3D volume images.
Figure 1.
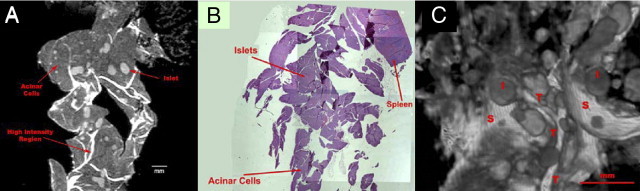
MRI and H&E histology of normal mouse pancreas. A: T2-weighted RARE 8 μMRI image (rapid acquisition with relaxation enhancement, factor 8) of a single longitudinal slice of formalin-fixed pancreas from an 11-month-old FVB F1 mouse. The image was obtained from a 3D data set (TR 2500 ms, effective TE 30 ms, slice thickness 35 μm, and in-plane resolution 62 μm). Islets (gray and circular), acinar cells (dark gray), and high-intensity regions (white) are labeled. B: H&E-stained histological image of the same mouse pancreas. All tissue is seen as purple. Spleen is present as a marker for the orientation of the pancreatic tissue. Islets are visible as round objects, slightly lighter purple, within the tissue. C: 3D volume-rendered image generated from 100 T2-weighted μMRI images such as that shown in A, using VolView version 2.0 software. Gross morphological distribution of islets (as spheres and ellipsoids), acinar cells (hypointense background), and novel structural features that appear to be tubes and sheets (hyperintense) are visualized in the volume image of the pancreatic tissue. Examples of islets (I) and novel structural features (tubes, T; sheets, S) are indicated on the image. Scale bars: 1 mm.
3D View of Pancreatic Structures in μMRI Images
The 3D volume-rendered image of mouse pancreas (Figure 1C) reveals the structural complexity of the gland. Broad, low-intensity regions corresponding to acinar cells can be observed (Figure 1A). Islets are visible in the 3D images as moderately intense spheres, which resemble the elliptical structures seen in the slice image. Islets are interspersed among a series of hyperintense tubes and a backdrop of a hyperintense sheet-like structure. To interpret these features in the 3D image, we analyzed regions based on spatial arrangement and signal intensity within the pancreas. Using small regions of the pancreas (Figure 2), structures were visualized with VolView version 2.0 software based on signal intensity and were rotated in space to determine their 3D characteristics. In so doing, we were able to distinguish islets, blood vessels, and extracellular matrix (ECM) among acinar tissue. Distinct features of these pancreatic components and their spatial relationship with each other as seen in μMRI images are discussed in sections that follow.
Figure 2.
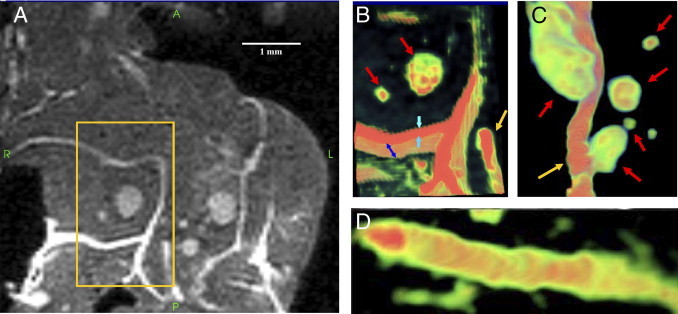
Volume-rendered (3D) μMRI image of normal mouse pancreas. A: Magnified view of the 2D μMRI image of a normal mouse pancreas (such as that shown in Figure 1A). B-D: 3D volume-rendered images of the boxed area in A. The color scale corresponds to the magnetic resonance signal intensity (green to red, low to high intensity). Cylindrical tube-like and planar sheet-like features can be identified in the high-intensity region of the volume image, in addition to islets, which appear ellipsoid. Note that these tubes and sheet-like features, both of which appear as high-intensity regions, cannot be clearly distinguished in the 2D μMRI image. B: A sheet (blue and cyan arrows) is seen underneath islets (red arrows). A tube appears in the bottom right corner (gold arrow). The sheet was measured to be ∼116 μm thick along the y axis (cyan arrows) and ∼298 μm in depth along the z axis (blue arrows). The total depth of this sheet through the tissue along the z axis (not shown) is ∼1400 μm. C: 3D volume-rendered image of the tube (gold arrow) shown in B in a different location with surrounding islets (red arrows). The diameter of the tube is ∼88 μm. The total length of the tube when tracked through the tissue (not shown) was measured to be ∼5850 μm. D: 3D volume-rendered image of a tube with a high-intensity lumen (red) and the lower-intensity wall (green). Note the clear distinction in morphology between the tube and the planar sheet.
μMRI of Islets
Islets appear as individual areas of intermediate and heterogeneous signal intensities and are easily identified by their spherical or elliptical appearance within acinar tissue of the pancreas (Figure 2). Although lymph nodes have a similar shape, they were ruled out as candidates based on the number and size of these spheres, their microstructural details, and pancreatic structures located nearby. Lymph nodes are fewer in number in the pancreas (three per pancreas) than are islets (≥500 per pancreas),6,7 and they tend to be much larger than islets and are generally located along the periphery of the gland (ie, not within the parenchyma)8,9 (based also on personal communication from Shannon J. Turley and Catharina Alam, authors of Refs. 8 and 9). See Supplemental Figure S2 at http://ajp.amjpathol.org.
Comparison of several normal mouse pancreata indicates that the number and shape of the islets vary widely from sample to sample. The sizes of these elliptical/spherical structures in μMRI images were compared with islets in H&E-stained tissue slides from the same tissue (Figure 3). Islets from several random sections of H&E-stained tissue were measured in two dimensions and the ellipsoids in μMRI were measured in three dimensions at their widest points (Table 1). Although tissue processing introduces additional disparity due to shrinkage, and although the H&E-stained islets could not necessarily be measured at its widest points, the sizes were within the same order of magnitude, and often matched two of the dimensions of the ellipsoids measured in the 3D μMRI images.
Figure 3.
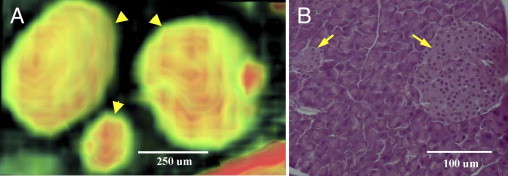
MRI and histology of islets in normal mouse pancreas. A: 3D volume-rendered μMRI image of islets in a normal mouse pancreas. The color gradient represents the magnetic resonance signal intensity gradient (green to red, low to high intensity). Note the varying intensity levels within each islet (arrowheads), which may reflect the presence of different cell types or microcirculation within pancreatic islets. B: H&E-stained histological image of islets (arrows) in a normal mouse pancreas. All tissue appears purple, with islets visible as round objects, slightly lighter purple, within the tissue. Acinar cells are visible surrounding the islets.
Table 1.
Dimensions of a Random Sample of Islets Present in Histological Slices and MR Volume-Rendered Images of a Normal Mouse Pancreas
| Islet | Measurements (μm), H&E |
Islet | Measurements (μm), μMRI |
|||
|---|---|---|---|---|---|---|
| x | y | x | y | z | ||
| 1⁎ | 150 | 150 | A⁎ | 390 | 540 | 510 |
| 2⁎ | 50 | 25 | B⁎ | 410 | 580 | 460 |
| 3 | 425 | 375 | C⁎ | 175 | 230 | 200 |
| 4 | 425 | 250 | D | 290 | 560 | 303 |
| 5 | 225 | 125 | E | 190 | 270 | 200 |
| 6 | 325 | 200 | F | 120 | 130 | 100 |
| 7 | 850 | 250 | G | 400 | 850 | 320 |
| 8 | 550 | 250 | H | 300 | 300 | 225 |
| 9 | 325 | 250 | I | 180 | 220 | 200 |
| 10 | 100 | 100 | J | 400 | 350 | 400 |
| 11 | 50 | 50 | K | 170 | 160 | 160 |
| 12 | 100 | 75 | L | 170 | 130 | 120 |
| 13 | 175 | 150 | M | 410 | 650 | 390 |
| 14 | 125 | 75 | N | 600 | 650 | 730 |
| O | 360 | 390 | 300 | |||
For each islet, measurements are from a single H&E histological preparation or a single μMRI image. Measurements are at the widest points along the x and y axes of islets in the histological image, and of the widest points along the x, y, and z axes of islets in the MR volume-rendered image.
μMRI of Blood Vessels and Ducts
Volume-rendered images revealed that the high-intensity tube-like networks that were observed in 2D images consisted of cylindrical and planar structures resembling tubes and sheets when observed in 3D (Figures 2 and 4). Based on current knowledge of the anatomy of mouse pancreas, the most prominent tube structures in the pancreas are blood vessels and ducts. Most pancreatic ducts tend to be small, especially in the splenic and gastric lobes of mouse pancreas. These smaller ducts converge to form larger ducts to transport digestive enzymes to the intestine. In the sample shown in Figure 2, B and C, the tube originates from the splenic end and moves straight down the tissue toward the duodenal end; it has an approximate diameter of 88 μm and total trackable length of 5850 μm. Measurement of the diameter of this tube at periodic intervals along its length indicates that the diameter of this tube changes by only ±10 μm. A tube near the duodenal end of the tissue in the lower left quadrant (Figure 4) has an approximate diameter of 120 μm and a trackable length of 2125 μm, and its diameter also does not change by more than 10 μm along its length. The uniformity of the tube diameter suggests that both tubes are blood vessels and not pancreatic ducts. A common feature of blood vessels in the pancreas is their proximity to islets, which are highly vascularized.10 In H&E sections of mouse pancreas, islets are often seen near blood vessels, a feature that is clearly visible in the volume-rendered μMRI images (Figures 2C and 4, B–D). In Figure 4, several islets of various sizes are seen at close proximity to a tube that we had earlier characterized as a blood vessel. It is likely that blood vessels remain visible because perfusion with formalin fixative preserved their 3D structural integrity, whereas fluid loss from ducts may have caused those smaller tubes to collapse. Thus, the constant diameter of the tubes, the presence of islets surrounding these tubes, and improved preservation of these tubes are consistent with blood vessels.
Figure 4.
3D volume-rendered image of a tube associated with multiple islets surrounded by acinar cells. Through manipulation of the opacity of the 3D volume image, different tissue components of the pancreas can be highlighted based on differences in magnetic resonance signal intensity, as reflected by the color scale (green to red, low to high intensity). Several notable structures can be observed: the high intensity of a lumen within a tube (A); the wall of the tube shown in A, with adjacent islets that have similar signal intensity (B); a tube and islets, with heterogeneous intensity visible within the islets (C); and islets and tubes as they appear in the tissue, surrounded by acinar cells (D). When their opacity is reduced, acinar cells appear translucent, allowing improved visualization of the islets and tube.
μMRI of ECM
The high-intensity planar structures in volume images resemble thin sheets that bend and wind throughout the gland (Figures 1 and 2). These structures appear to be consistent with mesenchymal tissue. H&E-stained pancreatic tissue rarely provides sections that include sheets of stromal tissue, and generally stroma is observed in H&E-stained sections of the pancreas only where diffuse and focal fibrosis exists in a pathological state, as evident in histology. Pancreatic ECM as seen in 3D reconstructed μMRI images is a complex network of planar structures with extensive branching in different directions throughout the tissue and connections with other sheets at different planes. The average thickness of the ECM in the normal mouse pancreas is 80 ± 20 μm, based on measurements at 15 different locations. The high intensity of the ECM in 2D images suggests a high concentration of mobile protons in this structure, which was confirmed by diffusion-weighted MRI. Whereas the surrounding tissue showed very little signal attenuation at b = 1000 s/mm2, nearly 100% signal attenuation was observed for ECM (see Supplemental Figure S3 at http://ajp.amjpathol.org).
Discussion
Here we report the first study to examine the microstructural features of intact mouse pancreas using μMRI. We used high spatial resolution imaging and 3D image reconstruction to visualize the exocrine and endocrine components of mouse pancreas. Understanding the 3D structure of the pancreas can be valuable in explaining the pathogenesis of pancreatic diseases, including diabetes, pancreatitis, and pancreatic cancer. Traditionally, the architecture of specific pancreatic components has been investigated using corrosion casts, histological methods, and electron microscopy.11,12 Although these methods provide much greater resolution than is currently possible with μMRI, they are not suitable for examining tissue in the intact state. Furthermore, unlike the μMRI method, none of the studies using previous methods revealed exocrine, endocrine, and vascular architecture simultaneously, as is essential to understanding the spatial relationship between these components in the pancreas. The existence of topographical associations and functional relationships between different pancreatic components emphasizes the need for an imaging approach that is capable of visualizing their 3D structure and morphology.13,14
Organization of the Exocrine Pancreas
The most striking feature of mouse pancreas in H&E-stained sections and in 2D high field strength μMRI images is the prominence of the acinar compartment. Acini make up approximately 80% to 85% of the pancreas and are responsible for prolific protein production. Each acinar cell can produce large amounts of zymogen granules, which are densely packed at the apical end of the acinar cell.15 Both the low nuclear/cytoplasmic ratio and the high protein content in the cytoplasm contribute to a highly eosinophilic cell. Although detection is limited to 50 μm, the relatively low signal intensity for acini in μMRI images of normal mouse pancreas can be attributed to lower water content, relative to other pancreatic components, and a shorter T2 relaxation time resulting from high protein and nucleic acid content. The ability to image normal acinar cells is important in understanding changes to these cells under pathological conditions. For example, chronic inflammatory diseases of the pancreas are characterized by the destruction of acinar cells and their replacement by connective tissue.16
Organization of the Endocrine Pancreas
Previous MRI studies of the pancreas have relied on contrast agents to visualize islets, because of the relatively poor native contrast between pancreatic islets and surrounding tissue.17 In our μMRI images, acquired without the use of a contrast agent, islets are clearly visible as moderately intense elliptical structures scattered within the parenchyma and distinct from the surrounding acinar tissue. The choice of TR and TE parameters, as well as the high spatial resolution used in our images, enables the visualization of islets and other pancreatic components based exclusively on differences in their spin-spin relaxation rates.
The 3D μMRI approach provides a novel methodology to visualize pancreatic islets and to quantify morphological features such as their size, distribution, and environment within the intact pancreas. 2D stereological techniques are limited in their ability to measure these features. An optical projection tomography method has been reported for quantitatively assessing the spatial distribution of insulin-labeled islet β-cells.18 In agreement with the optical projection tomography observations, our μMRI analysis found that each specimen displayed a wide range of islet sizes, which varied from sample to sample. However, whereas the optical projection tomography technique requires additional confocal imaging of the sample to calibrate pixel dimension, μMRI of the intact pancreas provides voxel dimensions directly from the image and does not require further calibration. Furthermore, unlike optical projection tomography, the μMRI method can also be used to characterize the environment of the islets. Obtaining information about the 3D structure of both endocrine and exocrine components of the pancreas simultaneously has great potential value, because of the systematic and complex physical, topographical, and functional associations between these components, which can reflect pathological changes.13,19
Organization of the Vasculature in the Pancreas
In general, blood vessels are typically observed in H&E histology as cross-sections, often containing red blood cells within the lumen. On occasion, longitudinal sections of these vessels can be observed in part as they traverse the gland. However, it is extremely unlikely that histological sectioning will follow these blood vessels consistently over long distances, given that they move above and below the plane of section. μMRI allows visualization of blood vessels without limitations due to the type and/or plane of section.
Most MRI done on the vasculature within the pancreas has been associated with islet cell transplantation or diabetes, used to detect blood vessel leakiness.20,21 Contrast-enhanced MRI using iron oxide nanoparticles was used to measure blood vessel leakiness in a mouse model of insulinoma22 and vascular volume fraction in targeted therapies of pancreatic adenocarcinoma.23 However, these studies did not include volume images of the vasculature and did not reveal the relationship of the vasculature to islets and other pancreatic components. Localization of areas with the greatest change in microvessel density, including changes in vascularity and blood vessel leakiness, is a vital observation for comparing diseased mouse pancreas to wild-type controls. In addition to illustrating the spatial relationship between islets and neighboring blood vessels, 3D volume μMRI could potentially visualize the microvasculature within pancreatic islets. The increased magnetic resonance signal intensity seen within the islets might represent the microcirculation of islets, previously seen only by scanning electron microscopy of vascular casts.10 This pattern of microcirculation affects the ability of cells within an islet to communicate among themselves,24 and therefore represents an additional source of useful information about islet function.
Organization of the ECM in the Pancreas
Formation of preinvasive lesions and their progression to cancer is often influenced by cellular interactions between tumor cells and the stromal microenvironment.25 The hyperintense appearance of ECM in T2-weighted spin-echo MRI images implies a long T2 moiety in pancreatic stroma, strongly suggesting the presence of hyaluronan, a glycosaminoglycan. Hyaluronan, which has a strong water-binding capacity, can maintain a high level of hydration leading to long T2-relaxation times and hence a hyperintense appearance in MRI images.26 Changes in ECM hyaluronan levels are known to occur in pancreatitis27 and in pancreatic cancer.28 Hyaluronan in the ECM provides a favorable microenvironment for cell proliferation and migration by maintaining the turgidity and hydration of tissue, in addition to activating intracellular signals through interaction with cell surface receptors. Increased expression of hyaluronan is sufficient to induce epithelial-mesenchymal transition and acquisition of transformed properties in phenotypically normal epithelial cell receptors. Associated changes in water content can be measured indirectly via alterations in tissue T2 relaxation time in MRI or directly by quantification of the water signal intensity using single voxel spectroscopy or chemical shift imaging, serving as a biomarker for those pathological conditions.
3D volume-rendered μMRI provides a means to explore stromal connective tissue and its association with neighboring parenchyma and vasculature. The ECM has been traditionally thought of as the structurally stable material that provides support for cells and tissues. The ECM is composed of a number of different macromolecules, whose structural integrity and functional composition are important in maintaining normal tissue architecture in development and in tissue-specific function,16 and abnormalities in ECM are of importance in both inherited and acquired diseases. Pancreatic diseases such as pancreatic cancer and chronic pancreatitis are characterized by profound alterations of ECM formation and composition.16 In fact, extensive remodeling of the ECM adjacent to parenchymal abnormalities is closely associated with tumor development and progression,29,30 which are likely to be reflected in the 3D architecture as observed by μMRI. ECM thickness, which may reflect its protein composition and its ability to tolerate mechanical stress,31 can be measured in volume-rendered μMRI images.
Clinical data confirm that the stromal compartment, which is uniquely dense in pancreatic intraepithelial neoplasia and invasive pancreatic ductal adenocarcinoma, acts as a barrier against therapy, as demonstrated in a mouse model of pancreatic cancer.32,33 The 3D-structural detail of the ECM with the adjacent microenvironment is not detectable by current imaging methods.34 Perentes et al35 reported in vivo imaging of ECM remodeling via multiphoton laser scattering microscopy. In that study, however, fluorescent labeling was necessary to show the ECM changes, whereas cellular expression of a labeling agent is not required for 3D volume-rendered μMRI microimaging. We believe that 3D volume-rendered images can demonstrate the critical anatomical changes of the stromal compartment in a variety of mouse models of pancreatic cancer at early and later stages of neoplastic and cancerous development. This capability could eventually lead to a similar approach suitable for clinical evaluation.
Organization of the Ductal Network in the Pancreas
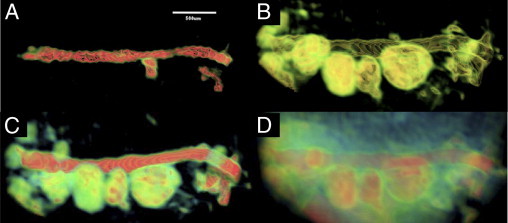
Both light and scanning electron microscopy have been used previously to examine specimens after retrograde injection of the pancreatic ductal system of normal rats with a silicone rubber compound.36 These studies revealed a ductal arrangement of the exocrine pancreas characterized by a complex curving and branching system of tubules. The inner surfaces of rat pancreatic ducts were investigated by using corrosion casts and scanning electron microscopy.11 Takahashi12 used scanning electron microscopy to study the finer details of acini and ducts after removing the interfering connective tissue by digestion with HCl. These studies provided great insight into the overall architecture of the pancreatic ductal tree with limited information regarding the interaction with acini; however, the methodologies were unable to provide details regarding the relationship of ducts to the endocrine, vascular, and stromal compartments and were not suitable for longitudinal studies in vivo. Our μMRI approach potentially enables evaluation of these interactions in a single animal over time, which is crucial for understanding the evolution of pathophysiology in animal models of pancreatic disease. We have obtained initial 3D volume μMRI images from FVB6 F1 mouse pancreas that illustrate a dense network of high-intensity structures which, in appearance, resemble the ductal network observed in corrosion casts of rodent pancreas (see Supplemental Figure S4 at http://ajp.amjpathol.org). In future work, we plan to focus on characterization of this structure in the normal mouse pancreas and its relationship to other pancreatic components.
Summary
The use of high field strength μMRI and 3D image reconstruction was critical to achieving the high spatial resolution needed to visualize pancreatic components and compartments in the excised, intact mouse pancreas. The present study provides an essential first step toward capturing the 3D structure and morphology of other internal organs. With further optimization, μMRI imaging of the pancreas and other internal organs can be performed in living mice to determine disease progression in longitudinal studies. Our ongoing imaging investigations are designed to evaluate the reliability and reproducibility of using 3D μMRI to detect architectural changes in exocrine, vascular, and stromal compartments of the pancreas during development of various diseases, including diabetes, pancreatitis, and preinvasive and invasive pancreatic cancer.
Acknowledgment
We thank Florie Bulaclac for administrative assistance.
Footnotes
Supported by the Jay Wertheimer Pancreatic Cancer Research Fund, with additional support from the Barnum and Zell Family Foundations at Northwestern University, the Nathan and Isabel Miller Family Foundation, and the IDP Foundation. This magnetic resonance approach was developed, in part, through a project funded by a Pancreatic Cancer Action Network–AACR Career Development Award (P.J.G.) in honor of Nancy Daly Riordan.
P.J.G. and P.N.V. contributed equally to this work.
Supplemental material for this article can be found at http://ajp.amjpathol.org or at doi: 10.1016/j.ajpath.2011.04.007.
Supplementary data
Image analysis of a 3D μMRI image of a mouse pancreas by volume rendering. A–C: Orthogonal slice images through the visualized volume. Pixels are displayed in grayscale, with white for high magnetic resonance intensity and black for low magnetic resonance intensity. Note the difference in magnetic resonance intensities associated with the tube (higher intensity: red arrows) and the islets (medium intensity: green arrows). D: A pseudo-color volume-rendered μMRI image showing a tube (red arrows) surrounded by islets (green arrows). E: Opacity function (OPF) [equation (1) under Materials and Methods] and color scale used to generate image D. ILow = 1700; IHigh = 65535; MaxOp = 1. The OPF function is superimposed on the magnetic resonance intensity histogram (shaded area). F–H: Volume rendering of the same μMRI image with different OPF functions allows selective visualization of pancreatic components. The OPF and color scale used to generate the images are shown at the bottom of each panel. OPF function parameters of ILow = 11200, IHigh = 14700, and MaxOp = 1 show the tube alone (F); OPF function parameters of ILow = 6000, IHigh = 9700, and MaxOp = 0.5 show only the islets and the outer layer (low magnetic resonance intensity) of the tube (G); and OPF function parameters of ILow = 6000, IHigh = 14700, and MaxOp = 1 show both islets and the tube (H).
Images of a lymph node in the mouse pancreas. A: H&E-stained histology image. B: 3D volume-rendered μMRI image shows that the lymph node is protruding from the pancreatic tissue. C: Detailed view of the same lymph node, 3D μMRI image. The OPF parameters were chosen to visualize the details within the lymph node. This resulted in removing the outside tissue intensities further. The lymph node diameter in orthogonal cross-sections was 1887, 1029, and 747 μm.
Diffusion-weighted images of normal FVB F1 mouse pancreas. The high-intensity region of the mouse pancreas consisting of sheet- and tube-like features in the 3D volume image has protons with high mobility, detected by diffusion-weighted imaging. Left: Spin-echo image of the pancreas with no diffusion weighting (b = 0 s/mm2) shows high-intensity regions over the entire pancreas. Right: Diffusion-weighted spin-echo image of the same section (b = 1000 s/mm2) shows nearly 100% signal attenuation only in the high-intensity regions. The tissue component surrounding the high-intensity region (ie, acinar cells) displays less signal attenuation in the diffusion-weighted image, suggesting the presence of protons with less mobility in that component. 2D images shown here were acquired using a diffusion-weighted spin-echo imaging sequence and the following parameters: TR/TE 2000 ms/29 ms, diffusion gradient duration 3 ms, delay between diffusion gradients 20 ms, b value 0 or 1000 s/mm2, slice thickness 500 μm, field of view 22 mm × 16 mm, and matrix size 256 × 256 for in-plane pixel size 86 μm × 62 μm.
3D volume-rendered image of the high signal intensity regions of normal FVB F1 mouse pancreas. The tree-like structure seen in the normal pancreas most likely represents networks of ducts, blood vessels, and stromal components. We plan to characterize the contribution of each component to this network in future investigations.
References
- 1.Lester D.S., Lyon R.C., McGregor G.N., Engelhardt R.T., Schmued L.C., Johnson G.A., Johannessen J.N. 3-Dimensional visualization of lesions in rat brain using magnetic resonance imaging microscopy. Neuroreport. 1999;10:737–741. doi: 10.1097/00001756-199903170-00014. [DOI] [PubMed] [Google Scholar]
- 2.Grimm J., Potthast A., Wunder A., Moore A. Magnetic resonance imaging of the pancreas and pancreatic tumors in a mouse orthotopic model of human cancer. Int J Cancer. 2003;106:806–811. doi: 10.1002/ijc.11281. [DOI] [PubMed] [Google Scholar]
- 3.Moore A., Grimm J., Han B., Santamaria P. Tracking the recruitment of diabetogenic CD8+ T-cells to the pancreas in real time. Diabetes. 2004;53:1459–1466. doi: 10.2337/diabetes.53.6.1459. [DOI] [PubMed] [Google Scholar]
- 4.Cabrera O., Berman D.M., Kenyon N.S., Ricordi C., Berggren P.O., Caicedo A. The unique cytoarchitecture of human pancreatic islets has implications for islet cell function. Proc Natl Acad Sci USA. 2006;103:2334–2339. doi: 10.1073/pnas.0510790103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song K.H., Ko S.H., Ahn Y.B., Yoo S.J., Chin H.M., Kaneto H., Yoon K.H., Cha B.Y., Lee K.W., Son H.Y. In vitro transdifferentiation of adult pancreatic acinar cells into insulin-expressing cells. Biochem Biophys Res Commun. 2004;316:1094–1100. doi: 10.1016/j.bbrc.2004.02.153. [DOI] [PubMed] [Google Scholar]
- 6.Jeong J.H., Kim S.H., Lee M., Kim W.J., Park T.G., Ko K.S., Kim S.W. Non-viral systemic delivery of Fas siRNA suppresses cyclophosphamide-induced diabetes in NOD mice. J Control Release. 2010;143:88–94. doi: 10.1016/j.jconrel.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haschek W.M., Wallig M.A., Rousseaux C.G. 2nd ed. Academic Press; San Diego, CA: 2009. Fundamentals of Toxicologic Pathology; p. 251. [Google Scholar]
- 8.Turley S.J., Lee J.W., Dutton-Swain N., Mathis D., Benoist C. Endocrine self and gut non-self intersect in the pancreatic lymph nodes. Proc Natl Acad Sci USA. 2005;102:17729–17733. doi: 10.1073/pnas.0509006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alam C., Valkonen S., Ohls S., Törnqvist K., Hänninen A. Enhanced trafficking to the pancreatic lymph nodes and auto-antigen presentation capacity distinguishes peritoneal B lymphocytes in non-obese diabetic mice. Diabetologia. 2010;53:346–355. doi: 10.1007/s00125-009-1599-x. [DOI] [PubMed] [Google Scholar]
- 10.Murakami T., Hitomi S., Ohtsuka A., Taguchi T., Fujita T. Pancreatic insulo-acinar portal systems in humans, rats, and some other mammals: scanning electron microscopy of vascular casts. Microsc Res Tech. 1997;37:478–488. doi: 10.1002/(SICI)1097-0029(19970601)37:5/6<478::AID-JEMT10>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 11.Ashizawa N., Watanabe M., Fukumoto S., Shimada Y. Scanning electron microscopic observations of three-dimensional structure of the rat pancreatic duct. Pancreas. 1991;6:542–550. doi: 10.1097/00006676-199109000-00007. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi H. Scanning electron microscopy of the rat exocrine pancreas. Arch Histol Jpn. 1984;47:387–404. doi: 10.1679/aohc.47.387. [DOI] [PubMed] [Google Scholar]
- 13.Bertelli E., Bendayan M. Association between endocrine pancreas and ductal system: More than an epiphenomenon of endocrine differentiation and development? J Histochem Cytochem. 2005;53:1071–1086. doi: 10.1369/jhc.5R6640.2005. [DOI] [PubMed] [Google Scholar]
- 14.Williams J.A., Goldfine I.D. The insulin-pancreatic acinar axis. Diabetes. 1985;34:980–986. doi: 10.2337/diab.34.10.980. [DOI] [PubMed] [Google Scholar]
- 15.Motta P.M., Macchiarelli G., Nottola S.A., Correr S. Histology of the exocrine pancreas. Microsc Res Tech. 1997;37:384–398. doi: 10.1002/(SICI)1097-0029(19970601)37:5/6<384::AID-JEMT3>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 16.Gress T.M., Menke A., Bachem M., Muller-Pillasch F., Ellenrieder V., Weidenbach H., Wagner M., Adler G. Role of extracellular matrix in pancreatic diseases. Digestion. 1998;59:625–637. doi: 10.1159/000007567. [DOI] [PubMed] [Google Scholar]
- 17.Medarova Z., Moore A. Non-invasive detection of transplanted pancreatic islets. Diabetes Obes Metab. 2008;10(Suppl 4):88–97. doi: 10.1111/j.1463-1326.2008.00942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alanentalo T., Asayesh A., Morrison H., Loren C.E., Holmberg D., Sharpe J., Ahlgren U. Tomographic molecular imaging and 3D quantification within adult mouse organs. Nat Methods. 2007;4:31–33. doi: 10.1038/nmeth985. [DOI] [PubMed] [Google Scholar]
- 19.Park I.S., Bendayan M. Characterization of the endocrine cells in the pancreatic-bile duct system of the rat. Anat Rec. 1992;232:247–256. doi: 10.1002/ar.1092320209. [DOI] [PubMed] [Google Scholar]
- 20.Turvey S.E., Swart E., Denis M.C., Mahmood U., Benoist C., Weissleder R., Mathis D. Noninvasive imaging of pancreatic inflammation and its reversal in type 1 diabetes. J Clin Invest. 2005;115:2454–2461. doi: 10.1172/JCI25048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medarova Z., Castillo G., Dai G., Bolotin E., Bogdanov A., Moore A. Noninvasive magnetic resonance imaging of microvascular changes in type 1 diabetes. Diabetes. 2007;56:2677–2682. doi: 10.2337/db07-0822. [DOI] [PubMed] [Google Scholar]
- 22.Sennino B., Raatschen H.J., Wendland M.F., Fu Y., You W.K., Shames D.M., McDonald D.M., Brasch R.C. Correlative dynamic contrast MRI and microscopic assessments of tumor vascularity in RIP-Tag2 transgenic mice. Magn Reson Med. 2009;62:616–625. doi: 10.1002/mrm.22040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guimaraes A.R., Rakhlin E., Weissleder R., Thayer S.P. Magnetic resonance imaging monitors physiological changes with antihedgehog therapy in pancreatic adenocarcinoma xenograft model. Pancreas. 2008;37:440–444. doi: 10.1097/MPA.0b013e31817c5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brunicardi F.C., Stagner J., Bonner-Weir S., Wayland H., Kleinman R., Livingston E., Guth P., Menger M., McCuskey R., Intaglietta M., Charles A., Ashley S., Cheung A., Ipp E., Gilman S., Howard T., Passaro E., Jr Microcirculation of the islets of Langerhans: Long Beach Veterans Administration Regional Medical Education Center Symposium. Diabetes. 1996;45:385–392. doi: 10.2337/diab.45.4.385. [DOI] [PubMed] [Google Scholar]
- 25.Bhowmick N.A., Moses H.L. Tumor-stroma interactions. Curr Opin Genet Dev. 2005;15:97–101. doi: 10.1016/j.gde.2004.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gensanne D., Josse G., Schmitt A.M., Lagarde J.M., Vincensini D. In vivo visualization of hyaluronic acid injection by high spatial resolution T2 parametric magnetic resonance images. Skin Res Technol. 2007;13:385–389. doi: 10.1111/j.1600-0846.2007.00241.x. [DOI] [PubMed] [Google Scholar]
- 27.Löhr M., Fischer B., Weber H., Emmrich J., Nizze H., Liebe S., Klopper G. Release of hyaluronan and laminin into pancreatic secretions. Digestion. 1999;60:48–55. doi: 10.1159/000007588. [DOI] [PubMed] [Google Scholar]
- 28.Theocharis A.D., Tsara M.E., Papageorgacopoulou N., Karavias D.D., Theocharis D.A. Pancreatic carcinoma is characterized by elevated content of hyaluronan and chondroitin sulfate with altered disaccharide composition. Biochim Biophys Acta. 2000;1502:201–206. doi: 10.1016/s0925-4439(00)00051-x. [DOI] [PubMed] [Google Scholar]
- 29.Kopfstein L., Christofori G. Metastasis: cell-autonomous mechanisms versus contributions by the tumor microenvironment. Cell Mol Life Sci. 2006;63:449–468. doi: 10.1007/s00018-005-5296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cretu A., Brooks P.C. Impact of the non-cellular tumor microenvironment on metastasis: potential therapeutic and imaging opportunities. J Cell Physiol. 2007;213:391–402. doi: 10.1002/jcp.21222. [DOI] [PubMed] [Google Scholar]
- 31.Black L.D., Allen P.G., Morris S.M., Stone P.J., Suki B. Mechanical and failure properties of extracellular matrix sheets as a function of structural protein composition. Biophys J. 2008;94:1916–1929. doi: 10.1529/biophysj.107.107144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scales S.J., de Sauvage F.J. Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol Sci. 2009;30:303–312. doi: 10.1016/j.tips.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 33.Olive K.P., Jacobetz M.A., Davidson C.J., Gopinathan A., McIntyre D., Honess D., Madhu B., Goldgraben M.A., Caldwell M.E., Allard D., Frese K.K., Denicola G., Feig C., Combs C., Winter S.P., Ireland-Zecchini H., Reichelt S., Howat W.J., Chang A., Dhara M., Wang L., Ruckert F., Grutzmann R., Pilarsky C., Izeradjene K., Hingorani S.R., Huang P., Davies S.E., Plunkett W., Egorin M., Hruban R.H., Whitebread N., McGovern K., Adams J., Iacobuzio-Donahue C., Griffiths J., Tuveson D.A. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohtani O. Three dimensional organization of the connective tissue fibers of the human pancreas: a scanning electron microscopic study of NaOH treated-tissues. Arch Histol Jpn. 1987;50:557–566. doi: 10.1679/aohc.50.557. [DOI] [PubMed] [Google Scholar]
- 35.Perentes J.Y., McKee T.D., Ley C.D., Mathiew H., Dawson M., Padera T.P., Munn L.L., Jain R.K., Boucher Y. In vivo imaging of extracellular matrix remodeling by tumor-associated fibroblasts. Nat Methods. 2009;6:143–145. doi: 10.1038/nmeth.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bockman D.E. Architecture of normal pancreas as revealed by retrograde injection. Cell Tissue Res. 1980;205:445–451. doi: 10.1007/BF00232285. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Image analysis of a 3D μMRI image of a mouse pancreas by volume rendering. A–C: Orthogonal slice images through the visualized volume. Pixels are displayed in grayscale, with white for high magnetic resonance intensity and black for low magnetic resonance intensity. Note the difference in magnetic resonance intensities associated with the tube (higher intensity: red arrows) and the islets (medium intensity: green arrows). D: A pseudo-color volume-rendered μMRI image showing a tube (red arrows) surrounded by islets (green arrows). E: Opacity function (OPF) [equation (1) under Materials and Methods] and color scale used to generate image D. ILow = 1700; IHigh = 65535; MaxOp = 1. The OPF function is superimposed on the magnetic resonance intensity histogram (shaded area). F–H: Volume rendering of the same μMRI image with different OPF functions allows selective visualization of pancreatic components. The OPF and color scale used to generate the images are shown at the bottom of each panel. OPF function parameters of ILow = 11200, IHigh = 14700, and MaxOp = 1 show the tube alone (F); OPF function parameters of ILow = 6000, IHigh = 9700, and MaxOp = 0.5 show only the islets and the outer layer (low magnetic resonance intensity) of the tube (G); and OPF function parameters of ILow = 6000, IHigh = 14700, and MaxOp = 1 show both islets and the tube (H).
Images of a lymph node in the mouse pancreas. A: H&E-stained histology image. B: 3D volume-rendered μMRI image shows that the lymph node is protruding from the pancreatic tissue. C: Detailed view of the same lymph node, 3D μMRI image. The OPF parameters were chosen to visualize the details within the lymph node. This resulted in removing the outside tissue intensities further. The lymph node diameter in orthogonal cross-sections was 1887, 1029, and 747 μm.
Diffusion-weighted images of normal FVB F1 mouse pancreas. The high-intensity region of the mouse pancreas consisting of sheet- and tube-like features in the 3D volume image has protons with high mobility, detected by diffusion-weighted imaging. Left: Spin-echo image of the pancreas with no diffusion weighting (b = 0 s/mm2) shows high-intensity regions over the entire pancreas. Right: Diffusion-weighted spin-echo image of the same section (b = 1000 s/mm2) shows nearly 100% signal attenuation only in the high-intensity regions. The tissue component surrounding the high-intensity region (ie, acinar cells) displays less signal attenuation in the diffusion-weighted image, suggesting the presence of protons with less mobility in that component. 2D images shown here were acquired using a diffusion-weighted spin-echo imaging sequence and the following parameters: TR/TE 2000 ms/29 ms, diffusion gradient duration 3 ms, delay between diffusion gradients 20 ms, b value 0 or 1000 s/mm2, slice thickness 500 μm, field of view 22 mm × 16 mm, and matrix size 256 × 256 for in-plane pixel size 86 μm × 62 μm.
3D volume-rendered image of the high signal intensity regions of normal FVB F1 mouse pancreas. The tree-like structure seen in the normal pancreas most likely represents networks of ducts, blood vessels, and stromal components. We plan to characterize the contribution of each component to this network in future investigations.


