Abstract
It is advantageous to use biomaterials in tissue engineering that stimulate extracellular matrix (ECM) production by the cellular component. Connective tissue growth factor (CTGF) stimulates type I collagen (COL1A1) transcription, but is functionally limited as a free molecule. Using a matrix-binding domain (MBD) from microfibril-associated glycoprotein-1, the fusion protein MBD-CTGF was targeted to the ECM and tested for COL1A1 transcriptional activation. MBD-CTGF produced by the ECM-synthesizing fibroblasts, or provided exogenously, localized to the elastic fiber ECM. MBD-CTGF, but not CTGF alone, led to a two-fold enhancement of COL1A1 expression. This study introduces a targeting technology that can be used to elevate collagen transcription in engineered tissues and thereby improve tissue mechanics.
Keywords: biomaterials, extracellular matrix, fusion protein, growth factor, tissue engineering
Introduction
The primary principle of tissue engineering is schematized as a “triad” – interactions of cells, scaffold, and signals that lead to a final engineered tissue. Combining two of these components by targeting signals to the scaffold, such as the extracellular matrix (ECM) synthesized by seeded cells, could simplify the engineering process and improve the outcome. As yet, the ECM has been underutilized as a destination for targeted cargo, which is surprising since in vivo it serves as a reservoir of signaling molecules such as growth factors,[1] modulators of angiogenesis,[2] and proteins that alter cellular adhesion.[3]
A peptide sequence has been described that is necessary and sufficient for targeting microfibril-associated glycoprotein-1 (MAGP-1) to the 10–12 nm microfibril ECM.[4] This “matrix-binding domain” (MBD) has also been shown to bind to a 8-cysteine motif in the microfibril component fibrillin-2, suggesting how the MBD can target to microfibril-rich tissues.[5] Therefore, the MBD of MAGP-1 is a promising homing sequence for stimulatory growth factors in the context of engineering tissues with abundant microfibrils such as blood vessels, skin, and tendon.[6, 7]
Growth factors have shown great utility in the production of engineered tissues. In line with the notable roles of transforming growth factor-β (TGF-β) in dermal wound healing and stimulation of extracellular matrix production,[8, 9] TGF-β doping of fibrin scaffolds has been shown to promote production of two ECM components important for vascular tissue, collagen and elastin.[10–13] Retention of TGF-β in a fibrin scaffold has been examined,[14] but TGF-β can also have anti-proliferative and pro-contractile effects undesirable in a tissue engineering scaffold.[15, 16] Vascular endothelial growth factor and epidermal growth factor have also been explored as stimulatory signals for engineered tissue both in synthetic [17–20] and natural [21–23] polymeric scaffolds. Connective tissue growth factor (CTGF) presents a promising strategy for tissue engineering: it promotes cell migration, adhesion and survival as well as enhanced ECM production.[24]
CTGF (also called CCN2) is a 38 kD growth factor produced by a variety of cells including skin fibroblasts, mesangial cells, and osteoblasts.[25–27] CTGF is a ligand for the low density lipoprotein receptor-related protein; after binding to its receptor, CTGF is rapidly internalized and degraded.[28] Of primary importance to tissue engineering, CTGF is consistently associated with elevated type I collagen expression, both in fibrotic human diseases including scleroderma and renal fibrosis [29, 30] and in in vitro experiments on a variety of cell types.[30–33] Stimulated collagen transcription is crucial to engineering tissues since collagen fibers can confer the requisite tissue stiffness and strength, as well as cellular guidance cues. [23, 34, 35]
The central hypothesis of this study was that adding the MBD to CTGF would anchor it to the ECM without compromising its ability to stimulate collagen transcription. In these studies, neonatal human dermal fibroblasts were used for functional analysis of MBD-CTGF, since they have served as the cellular component of fibrin-based engineered heart valves and arteries.[35–37] Comparisons of MBD-CTGF to free CTGF produced by transfected fibroblasts demonstrated that the MBD was necessary for localization to the ECM and that the matrix-bound growth factor was able to stimulate COL1A1 expression by the fibroblasts.
Experimental Part
Antibodies
Mouse monoclonal anti-v5 IgG (Invitrogen), mouse monoclonal anti-human tropoelastin (BA4, Abcam), Alexa 488-conjugated donkey anti-mouse IgG, Alexa 594-conjugated goat anti-rabbit IgG (Molecular Probes), and horseradish peroxidase (HRP)-conjugated sheep anti-mouse IgG (Amersham) were commercially available. Rabbit polyclonal anti-mouse recombinant tropoelastin antibody (MRT 617) was described previously.[38]
Plasmids
All CTGF expression plasmids used the pcDNA 3.1 (Invitrogen) vector system to allow production of protein with a C-terminal v5-6xHis epitope/purification tag. To generate the CTGF-v5His plasmid, primers f2CTGF and rCTGF (Table I) were used to amplify the human connective tissue growth factor (CTGF) cDNA from template DNA (a generous gift from Leah Allen, Fibrogen) including sequence encoding residues Met1 through Ala349. This PCR product was cloned in frame 5′ to the coding sequence for v5-His in pcDNA3.1. To generate MBD-CTGF-v5His plasmid, primers fCTGF and rCTGF (Table I) were used along with template CTGF cDNA to amplify the sequence encoding residues Glu27 through Ala349. This PCR product was cloned in frame in between the coding sequence for MAGP-1 exons 7+8 and the v5-his epitope tag located in MAGP7+8v5His.[4] All plasmids were verified by DNA sequencing. The collagen reporter plasmid pColLuc was previously described;[39] briefly, it consists of the human COL1A1 promoter inserted upstream of a modified firefly luciferase gene, luc2, in the pGL4.14 vector (Promega).
Table I.
Primers used in this study
| fCTGF | 5′ TAA GGA TCC ACA GAA CTG CAG CGG GC 3′ |
| f2CTGF | 5′ TAA GGT ACC ATG AGG GCC TGG 3′ |
| rCTGF | 5′ TAA CTC GAG CCA TGT CTC CGT ACA TCT TCC 3′ |
“f” and “r” denote sense and antisense primers, respectively.
Cell Culture
Rat lung fibroblasts (RFL-6, ATCC CCL-192) were cultured in Ham’s F-12 nutrient mixture (Invitrogen) supplemented with 1.176 g/L sodium bicarbonate, 10 U/ml penicillin, 10 U/ml streptomycin, 2 mM L-glutamine, non-essential amino acids, and 20% Cosmic Calf serum (CCS, Hyclone). Chinese hamster ovary cells (CHO-K1) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with antibiotics, 2 mM L-glutamine, non-essential amino acids, and 10% CCS. Neonatal human dermal fibroblasts (NHDF, Clonetics) were cultured in DMEM/F12 (Invitrogen) supplemented with antibiotics and 10% fetal bovine serum (FBS, Hyclone). Selection media were supplemented with 500 μg/ml G418 (Invitrogen).
Transfection
Cells were transiently transfected with Lipofectamine 2000 or LTX (Invitrogen) according to the manufacturer’s protocol. Briefly, cells were plated in a 35 mm dish and allowed to reach confluence. Fresh antibiotic-free medium was given one day prior to transfection, then medium was switched to Opti-MEM (Gibco). For each transfected dish, 4 μg plasmid and 10 μL of Lipofectamine 2000, or 5 μL of Lipofectamine LTX, were used. 4–5 hours after transfection, medium was exchanged with fresh antibiotic-free culture medium to wash out transfection agents. To obtain stably transfected cells, cultures were split 48 hours post-transfection (1:2.25) and changed to selection media after 24 additional hours. Complete death of non-transfected cells occurred within five days.
Indirect Immunofluorescence
Cells were grown on a 22×22 mm coverslip in a 35 mm dish or on a Permanox LabTek chamber slide (Nalge Nunc). The cells were rinsed with PBS and fixed in cold (−20°C) methanol. After rinsing with PBS, the cells were incubated for ten minutes in block (PBS-TG: PBS pH 7.4, 0.1% Tween-20, 1% coldwater fish gelatin) followed by one hour in primary antibody solution (1:500 primary antibody in PBS-TG). Cells were washed three times in PBS-TG followed by one hour in secondary antibody solution (1:1000 secondary antibody in PBS-TG, Molecular Probes). Finally, the cells were washed in PBS-TG and PBS before mounting on a slide using Gel/Mount (BioMeda). Pictures were taken at 400X using a Zeiss fluorescence microscope and AxioVision software.
Bioluminescence Measurement
72 hours post-transfection, the medium was replaced with 1 mM VivoGlo (Promega) in Hank’s Buffered Saline (HBSS, Invitrogen). The bioluminescence of each well was then obtained by imaging the culture plate in a Xenogen system (IVIS, Caliper) using a 30 second exposure time, bin 4, followed by downstream quantification using ImageJ as described previously.[39]
Immunoblotting
Equal volumes of conditioned medium were suspended in reducing SDS-PAGE loading buffer, separated by 12.5% SDS-PAGE at 25 mA, transferred to Protran (Whatman), and blocked for 1 hour in PBS + 0.1% Tween-20 + 2.5% dry milk. Blots were incubated 1 hour with mouse anti-v5 primary antibody (1:1000 in block), washed in block, and incubated 1 hour with HRP-conjugated sheep anti-mouse IgG secondary antibody (1:2000, Amersham). Blots were washed in block and PBS and developed by enhanced chemiluminescence.
Results
The MAGP-1 MBD Directs Organization of Recombinant CTGF by Fibroblasts
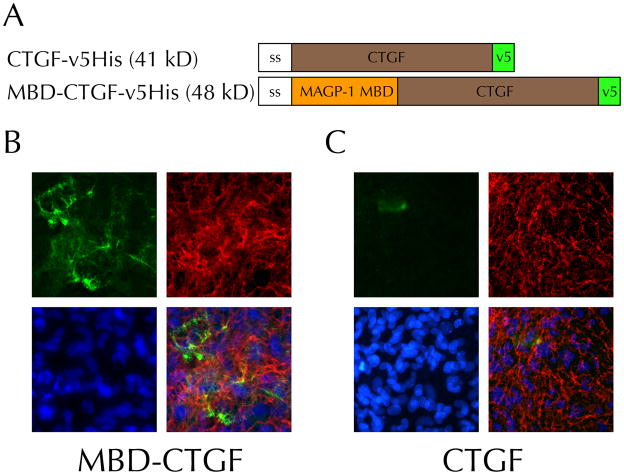
The MBD of human MAGP-1 directs localization of GFP to the ECM of RFL-6 cells in vitro.[4] Therefore, a fusion protein consisting of the MAGP-1 MBD and the growth factor CTGF was predicted to similarly localize to the ECM. All constructs had a N-terminal signal sequence and C-terminal v5-6xHis tag for efficient secretion, rapid identification, and easy purification (Figure 1A). Transiently transfected RFL-6 cells showed substantial matrix incorporation of MBD-tagged CTGF (Figure 1B). There were no qualitative differences between the immunofluorescence pattern obtained with the fusion protein and either the published pattern of MBD alone or the pattern of full-length MAGP-1 transfected in another well in the same assay ([4] and data not shown). Notably, this staining pattern overlapped with that for tropoelastin, another molecule that localizes to microfibrils in this system. The highest levels of MBD-CTGF matrix staining occurred in areas surrounding cells strongly expressing the transgene, suggesting a concentration effect on matrix incorporation. Other areas also showed matrix staining, but at an intermediate level, suggesting that protein released into the medium could be incorporated in trans following diffusion. In contrast to the transfections performed with MBD-CTGF, cells transfected to produce CTGF alone (henceforth referred to as “free CTGF”) did not localize CTGF to elastic fibers (Figure 1C). These cells instead showed a hazy pericellular staining pattern. Comparison of these two staining patterns indicates that the MAGP-1 MBD is necessary for matrix localization of CTGF in this system.
Figure 1. The MAGP-1 matrix binding domain (MBD) sequesters recombinant human connective tissue growth factor into the extracellular matrix of fibroblasts.
A) Schematic of constructs used in this study. Abbreviations: ss, signal sequence; MAGP-1 MBD, coding sequence for the matrix binding domain of MAGP-1; CTGF, coding sequence for CTGF; v5, v5 epitope followed by a 6× histidine repeat. Expected molecular weights are given in kilodaltons (kD).
B,C) MBD-CTGF fusion protein incorporates into the microfibrillar matrix but CTGF lacking the MBD is not. RFL-6 cells were transiently transfected with the MBD-CTGF- (B) or CTGF-v5His (C) plasmid. Indirect immunofluorescence for recombinant protein (v5, green) and tropoelastin (MRT617, red) antibodies is shown, as well as Hoechst 33342 nuclear staining (5 μg/ml, blue). Results are representative of four independent experiments.
MBD-CTGF Leads to Enhanced Expression of Type I Collagen by Fibroblasts
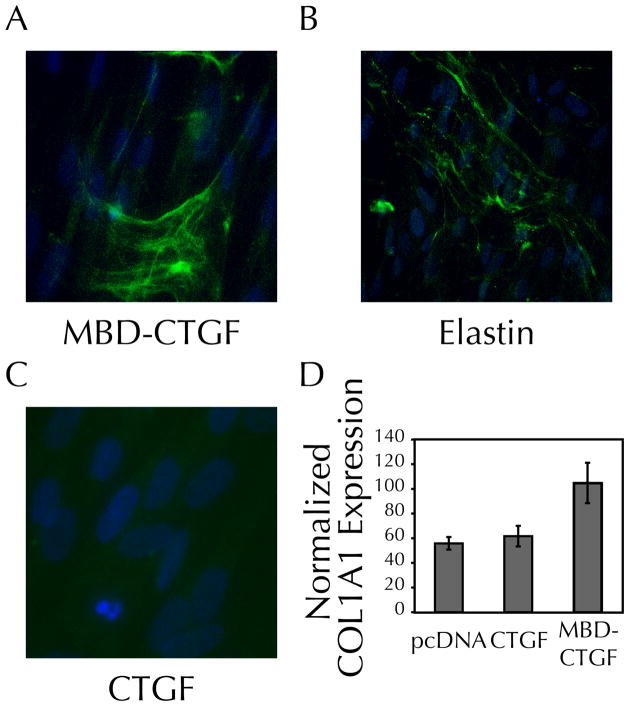
To determine if the matrix-binding CTGF had signaling capability that differed from free CTGF, expression of type I collagen (COL1A1) was measured in NHDF co-transfected with pColLuc, a reporter plasmid previously described to be useful in monitoring collagen production in tissue-engineered constructs.[39] Typically, CTGF signaling leads to enhanced expression of COL1A1.[32] NHDF transfected with MBD-CTGF show a CTGF staining pattern similar to that seen with RFL-6 cells (Figure 2A), and similar to the staining pattern for tropoelastin (Figure 2B). As was the case for RFL-6 cells, free CTGF does not show ECM localization (Figure 2C).
Figure 2. Expression of MBD-CTGF, but not CTGF alone, elevates transcription of type I collagen by fibroblasts.
A–C) NHDF transfected with a MBD-CTGF, but not a CTGF plasmid, demonstrate ECM incorporation of recombinant protein. 8 days post-transfection NHDF were analyzed by indirect immunofluorescence as in Figure 1B for recombinant protein (v5, green) and Hoechst 33342 (5 μg/ml, blue) for nuclear visualization. B) Tropoelastin staining (BA4, green) was similar to the pattern for MBD-CTGF.
D) Bioluminescence, measured 72 hours post-transfection, showed a two-fold increase in COL1A1 expression when fibroblasts were transfected with matrix-binding CTGF (p < 0.05 using the student’s t-test compared to either vector control or CTGF transfectants). Results are representative of four independent transfections, n=3–4 transfected wells per sample.
Bioluminescence was measured 72 hours post-transfection, to allow full expression of the transiently-transfected CTGF plasmid, its matrix incorporation and/or binding to cells, induction of downstream signaling, and expression of the luciferase transgene. Transfection with free CTGF failed to induce collagen expression above the levels of cells similarly transfected with the control vector pcDNA 3.1 (Figure 2D). However, transfection with the MBD-CTGF construct led to a nearly twofold increase in luciferase activity. Therefore, MBD-CTGF not only retains the ability to promote collagen expression, despite the addition of a peptide tag, but it also shows enhanced signaling function versus a similar transfection with free CTGF.
Fibroblasts Organize Exogenous Protein Produced by Co-cultured Epithelial Cells
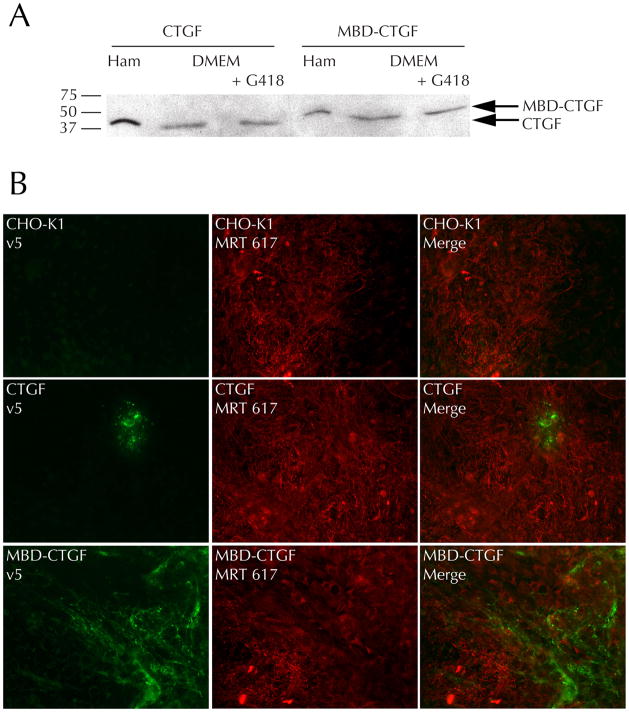
To test whether MBD-CTGF added exogenously to a fibroblast-generated ECM would result in its ECM localization, stable CHO-K1 cell lines were generated as a source of recombinant proteins. Each cell line produced a recombinant protein of the expected size and in a similar amount in both normal and selection medium (Figure 3A). The amount of protein produced was equal between MBD-CTGF and CTGF transfections. This suggests that the fibroblasts also produce equal amounts of CTGF and MBD-CTGF when transfected with equal amounts of DNA; quantification in that system is difficult since the protein partitions differently into media and cell/matrix fractions depending on matrix-binding ability.
Figure 3. MBD-CTGF secreted by CHO-K1 cells is incorporated into the ECM of co-cultured fibroblasts.
A) Conditioned media from confluent stable CHO-K1 transfectants grown in Ham’s F-12 (Ham) or DMEM with or without G418 for 72 hours was analyzed by immunoblotting for recombinant protein (v5 antibody). Arrows indicate the expected molecular weights of recombinant CTGF and MBD-CTGF.
B) Untransfected CHO-K1 cells or cells stably transfected with CTGF- or MBD-CTGF plasmids were co-cultured with RFL-6 cells in Ham’s F-12 medium. After 48 hours of culture cells were analyzed by indirect immunofluorescence as in Figure 1B. Results are representative of four independent experiments.
Stably transfected CHO-K1 cells were co-cultured with RFL-6 cells and allowed to grow past confluence. The fibroblasts’ ability to assemble ECM was unimpaired in co-culture, as judged by the formation of a normal tropoelastin matrix (Figure 3B). Co-culture with CHO-K1 cells expressing MBD-CTGF resulted in similar matrix incorporation of the recombinant protein compared to the case where the fibroblasts were expressing the transgene directly (Figure 1B), validating the hypothesis that MBD-CTGF can diffuse and incorporate in trans into the ECM. In agreement with earlier results, CHO-K1 expression of free CTGF did not result in significant matrix incorporation of the CTGF, although a pericellular pattern was once again apparent.
Discussion
This study shows that the MBD of MAGP-1 is sufficient to direct incorporation of functional recombinant CTGF into a microfibrillar ECM scaffold. MBD-CTGF stimulated collagen transcription twofold in the NHDF system, equivalent to previous studies using 50 ng/ml CTGF in the growth medium of hepatic stellate cells.[32] Without the MBD, recombinant CTGF failed to localize to the ECM or enhance collagen transcription. This lowered potency of the free CTGF could be due to diffusion into the culture medium and therefore a lowered local concentration near the cellular membrane where it can bind to its receptor. Alternately, free CTGF may be more rapidly endocytosed and degraded by the cell, terminating the signal before it leads to significant transcriptional activation. Likely matrix binding by MBD-CTGF is dependent on the presence of fibrillin in the ECM, since there is evidence for a direct interaction between the matrix-binding domain of MAGP-1 and fibrillin-2.[5]
While the molecular mechanism was not probed in this study, the elevated COL1A1 expression caused by MBD-CTGF is promising for tissue engineering. We previously showed that COL1A1 expression, monitored by the luciferase system, correlated strongly with collagen deposition by neonatal rat vascular smooth muscle cells in fibrin-based tissue constructs[39] and that collagen content strongly correlates with tissue stiffness and strength.[40] NHDF, which responded to MBD-CTGF with increased collagen expression in this study, are a promising cell source for tissue-engineered arteries[37] and heart valves,[41] therefore it is likely that MBD-CTGF treatment during culture could improve the mechanical properties of these tissues.
One intriguing consequence of generating matrix-bound CTGF is that it may synergize with latent TGF-β on fibrillin microfibrils. Human deficiencies in fibrillin, such as Marfan’s syndrome, can lead to altered TGF-β signaling, leading to the hypothesis that fibrillin is important for sequestering TGF-β and modulating its activity.[42–44] MAGP-1 itself has been hypothesized to modulate growth factor interactions with the microfibril.[45] Since some of the fibrogenic signals regulated by TGF-β require CTGF signaling [30], and owing to the usefulness of TGF-β in tissue engineering,[12, 16, 40] simultaneous co-localized activity of the two molecules could synergize, leading to enhanced collagen production. Positive feedback could emerge from this strategy, since binding of TGF-β to collagen can prolong its activity.[46]
While the current study employed transfection as the source of recombinant matrix-binding growth factor, it could also be used to dope the scaffold in other ways. Recombinant protein could be produced and purified in a bacterial or eukaryotic expression system then later added exogenously to the target cells or tissue. Purified recombinant CTGF has been effective at activating collagen transcription in previous studies[30–32] and bacterially produced proteins, including MAGP-1[47] and tropoelastin,[38] incorporate into the ECM when provided exogenously. Alternately, transfected cells could be co-entrapped into the tissue construct, providing a (potentially inducible) source of MBD-CTGF.
Conclusion
A new fusion protein was engineered in this study for stimulation of fibroblasts within a ECM scaffold. The protein was bifunctional, targeting to the elastic fiber ECM via the microfibril-associated glycoprotein-1 domain and activating type I collagen transcription via the CTGF domain. Notably, matrix localization of CTGF resulted in increased collagen transcription. Co-culture studies demonstrated that the fusion protein could also be delivered exogenously to a fibroblast scaffold. Based on the results of this study, the MBD-CTGF fusion protein could be used to dope engineered tissues during culture to increase collagen production by resident tissue cells.
Acknowledgments
This work was supported by funding from the National Institutes of Health (T32 HL007873 to J.S.W., HL084922 to R.P.M., HL083880 to R.T.T.) and from the National Science Foundation (Graduate Research Fellowship to J.S.W.
References
- 1.Dabovic B, Chen Y, Choi J, Vassallo M, Dietz HC, Ramirez F, von Melchner H, Davis EC, Rifkin DB. Journal of cellular physiology. 2009;219:14. doi: 10.1002/jcp.21643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nyberg P, Xie L, Kalluri R. Cancer research. 2005;65:3967. doi: 10.1158/0008-5472.CAN-04-2427. [DOI] [PubMed] [Google Scholar]
- 3.Kyriakides TR, Bornstein P. Thrombosis and haemostasis. 2003;90:986. doi: 10.1160/TH03-06-0399. [DOI] [PubMed] [Google Scholar]
- 4.Segade F, Trask BC, Broekelmann TJ, Pierce RA, Mecham RP. J Biol Chem. 2002;277:11050. doi: 10.1074/jbc.M110347200. [DOI] [PubMed] [Google Scholar]
- 5.Werneck CC, Trask BC, Broekelmann TJ, Trask TM, Ritty TM, Segade F, Mecham RP. J Biol Chem. 2004;279:23045. doi: 10.1074/jbc.M402656200. [DOI] [PubMed] [Google Scholar]
- 6.Boregowda R, Paul E, White J, Ritty TM. Matrix Biol. 2008;27:661. doi: 10.1016/j.matbio.2008.09.579. [DOI] [PubMed] [Google Scholar]
- 7.Ramirez F, Sakai LY. Cell and tissue research. 339:71. doi: 10.1007/s00441-009-0822-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cutroneo KR. J Cell Biochem. 2003;90:1. doi: 10.1002/jcb.10599. [DOI] [PubMed] [Google Scholar]
- 9.Steiling H, Werner S. Curr Opin Biotechnol. 2003;14:533. doi: 10.1016/j.copbio.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Choi WS, Mitsumoto A, Kochevar IE. Photochemistry and photobiology. 2009;85:1425. doi: 10.1111/j.1751-1097.2009.00611.x. [DOI] [PubMed] [Google Scholar]
- 11.Kuang PP, Zhang XH, Rich CB, Foster JA, Subramanian M, Goldstein RH. American journal of physiology. 2007;292:L944. doi: 10.1152/ajplung.00184.2006. [DOI] [PubMed] [Google Scholar]
- 12.Ross JJ, Tranquillo RT. Matrix Biol. 2003;22:477. doi: 10.1016/s0945-053x(03)00078-7. [DOI] [PubMed] [Google Scholar]
- 13.Long JL, Tranquillo RT. Matrix Biol. 2003;22:339. doi: 10.1016/s0945-053x(03)00052-0. [DOI] [PubMed] [Google Scholar]
- 14.Catelas I, Dwyer JF, Helgerson S. Tissue engineering. 2008;14:119. doi: 10.1089/ten.tec.2007.0262. [DOI] [PubMed] [Google Scholar]
- 15.Derynck R, Akhurst RJ, Balmain A. Nature genetics. 2001;29:117. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 16.Grouf JL, Throm AM, Balestrini JL, Bush KA, Billiar KL. Tissue Eng. 2007;13:799. doi: 10.1089/ten.2006.0206. [DOI] [PubMed] [Google Scholar]
- 17.Fan VH, Tamama K, Au A, Littrell R, Richardson LB, Wright JW, Wells A, Griffith LG. Stem cells (Dayton, Ohio) 2007;25:1241. doi: 10.1634/stemcells.2006-0320. [DOI] [PubMed] [Google Scholar]
- 18.Platt MO, Roman AJ, Wells A, Lauffenburger DA, Griffith LG. Journal of cellular physiology. 2009;221:306. doi: 10.1002/jcp.21854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leslie-Barbick JE, Moon JJ, West JL. Journal of biomaterials science. 2009;20:1763. doi: 10.1163/156856208X386381. [DOI] [PubMed] [Google Scholar]
- 20.Zisch AH, Lutolf MP, Ehrbar M, Raeber GP, Rizzi SC, Davies N, Schmokel H, Bezuidenhout D, Djonov V, Zilla P, Hubbell JA. Faseb J. 2003;17:2260. doi: 10.1096/fj.02-1041fje. [DOI] [PubMed] [Google Scholar]
- 21.Yang Y, Zhao Y, Chen B, Han Q, Sun W, Xiao Z, Dai J. Tissue Eng Part A. 2009;15:3589. doi: 10.1089/ten.TEA.2008.0648. [DOI] [PubMed] [Google Scholar]
- 22.Lee YB, Polio S, Lee W, Dai G, Menon L, Carroll RS, Yoo SS. Experimental neurology. [DOI] [PubMed] [Google Scholar]
- 23.Liu CZ, Xia ZD, Han ZW, Hulley PA, Triffitt JT, Czernuszka JT. Journal of biomedical materials research. 2008;85:519. doi: 10.1002/jbm.b.30975. [DOI] [PubMed] [Google Scholar]
- 24.Perbal B. Lancet. 2004;363:62. doi: 10.1016/S0140-6736(03)15172-0. [DOI] [PubMed] [Google Scholar]
- 25.Arnott JA, Nuglozeh E, Rico MC, Arango-Hisijara I, Odgren PR, Safadi FF, Popoff SN. Journal of cellular physiology. 2007;210:843. doi: 10.1002/jcp.20917. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Blom IE, Sa S, Goldschmeding R, Abraham DJ, Leask A. Kidney international. 2002;62:1149. doi: 10.1111/j.1523-1755.2002.kid567.x. [DOI] [PubMed] [Google Scholar]
- 27.Holmes A, Abraham DJ, Chen Y, Denton C, Shi-wen X, Black CM, Leask A. J Biol Chem. 2003;278:41728. doi: 10.1074/jbc.M305019200. [DOI] [PubMed] [Google Scholar]
- 28.Segarini PR, Nesbitt JE, Li D, Hays LG, Yates JR, 3rd, Carmichael DF. J Biol Chem. 2001;276:40659. doi: 10.1074/jbc.M105180200. [DOI] [PubMed] [Google Scholar]
- 29.Gore-Hyer E, Pannu J, Smith EA, Grotendorst G, Trojanowska M. Arthritis and rheumatism. 2003;48:798. doi: 10.1002/art.10953. [DOI] [PubMed] [Google Scholar]
- 30.Gore-Hyer E, Shegogue D, Markiewicz M, Lo S, Hazen-Martin D, Greene EL, Grotendorst G, Trojanowska M. Am J Physiol Renal Physiol. 2002;283:F707. doi: 10.1152/ajprenal.00007.2002. [DOI] [PubMed] [Google Scholar]
- 31.Frazier K, Williams S, Kothapalli D, Klapper H, Grotendorst GR. J Invest Dermatol. 1996;107:404. doi: 10.1111/1523-1747.ep12363389. [DOI] [PubMed] [Google Scholar]
- 32.Paradis V, Dargere D, Bonvoust F, Vidaud M, Segarini P, Bedossa P. Lab Invest. 2002;82:767. doi: 10.1097/01.lab.0000017365.18894.d3. [DOI] [PubMed] [Google Scholar]
- 33.Wang JF, Olson ME, Ball DK, Brigstock DR, Hart DA. Wound Repair Regen. 2003;11:220. doi: 10.1046/j.1524-475x.2003.11311.x. [DOI] [PubMed] [Google Scholar]
- 34.Wolf K, Muller R, Borgmann S, Brocker EB, Friedl P. Blood. 2003;102:3262. doi: 10.1182/blood-2002-12-3791. [DOI] [PubMed] [Google Scholar]
- 35.Robinson PS, Johnson SL, Evans MC, Barocas VH, Tranquillo RT. Tissue Eng Part A. 2008;14:83. doi: 10.1089/ten.a.2007.0148. [DOI] [PubMed] [Google Scholar]
- 36.Isenberg BC, Williams C, Tranquillo RT. Circulation research. 2006;98:25. doi: 10.1161/01.RES.0000196867.12470.84. [DOI] [PubMed] [Google Scholar]
- 37.Syedain ZH, Weinberg JS, Tranquillo RT. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6537. doi: 10.1073/pnas.0711217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozel BA, Ciliberto CH, Mecham RP. Matrix Biol. 2004;23:23. doi: 10.1016/j.matbio.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Weinbaum JS, Qi J, Tranquillo RT. Tissue Eng Part C: Methods. 2010;16:459. doi: 10.1089/ten.tec.2009.0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neidert MR, Lee ES, Oegema TR, Tranquillo RT. Biomaterials. 2002;23:3717. doi: 10.1016/s0142-9612(02)00106-0. [DOI] [PubMed] [Google Scholar]
- 41.Syedain ZH, Tranquillo RT. Biomaterials. 2009;30:4078. doi: 10.1016/j.biomaterials.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaudhry SS, Cain SA, Morgan A, Dallas SL, Shuttleworth CA, Kielty CM. J Cell Biol. 2007;176:355. doi: 10.1083/jcb.200608167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohn RD, van Erp C, Habashi JP, Soleimani AA, Klein EC, Lisi MT, Gamradt M, ap Rhys CM, Holm TM, Loeys BL, Ramirez F, Judge DP, Ward CW, Dietz HC. Nature medicine. 2007;13:204. doi: 10.1038/nm1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Habashi JP, Judge DP, Holm TM, Cohn RD, Loeys BL, Cooper TK, Myers L, Klein EC, Liu G, Calvi C, Podowski M, Neptune ER, Halushka MK, Bedja D, Gabrielson K, Rifkin DB, Carta L, Ramirez F, Huso DL, Dietz HC. Science (New York, NY) 2006;312:117. doi: 10.1126/science.1124287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weinbaum JS, Broekelmann TJ, Pierce RA, Werneck CC, Segade F, Craft CS, Knutsen RH, Mecham RP. J Biol Chem. 2008;283:25533. doi: 10.1074/jbc.M709962200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shibuya H, Okamoto O, Fujiwara S. Journal of dermatological science. 2006;41:187. doi: 10.1016/j.jdermsci.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 47.Weinberg JS. Biological and Biomedical Sciences (Molecular Cell Biology) Washington University; Saint Louis, Missouri: 2007. MAGP-1: An elastic fiber protein with matricellular function. Ph.D. [Google Scholar]