Abstract
Background
Quantitative real-time PCR (qPCR) is a widely used technique for gene expression analysis. Its reliability is highly dependent upon selection of the appropriate reference genes for accurate gene expression normalization. In this study, we investigated the expression stability of 10 commonly used reference genes in a mouse myocardial infarction model.
Methods & Results
The expression stability of the 10 reference genes (Actb, B2m, Eef1a1, Gapdh, Hprt, Polr2a, Ppia, Rpl13a, Tbp, Tpt1) was analyzed using the geNorm software. Overall, the combination of Hprt, Rpl13a and Tpt1 was the most stable reference gene set in our experiments. Gapdh, Polr2a and Actb consistently showed the highest gene expression variability and the expression levels of Gapdh, Polr2a, Actb, B2m and Eef1a1 were found to be selectively up- or downregulated after myocardial infarction. We normalized the expression of Nppb and Vcam1, using different reference gene strategies and demonstrated that their induction after myocardial infarction was most clearly revealed with the optimal reference gene combination. However, the use of suboptimal reference gene combinations resulted in detrimental effects on gene expression levels and variability with a gradual loss of the expression differences and a significant reduction in statistical power.
Conclusions
Hprt, Rpl13a and Tpt1 are a set of stably expressed reference genes for accurate gene expression normalization in myocardial infarction studies in mice. We found that Gapdh, Polr2a and Actb display high expression variability in mouse myocardial infarction tissues and that loss of statistical power and increase in sample size are the evident consequences of choosing suboptimal combinations of reference genes. We furthermore caution against the use of Gapdh, Polr2a, Actb, B2m and Eef1a1 for gene expression normalization in myocardial infarction studies because of selective up- or downregulation after myocardial infarction, which could potentially lead to biased study outcomes.
Introduction
Quantitative real-time PCR (qPCR) is the method of choice for gene expression research and analysis of biological pathways for several reasons: the technique is fast, extremely sensitive, highly reproducible and can be integrated in high-throughput systems. Therefore, it is routinely applied in many research areas. Despite the advantages and its apparent simplicity, qPCR expression analysis also has a number of caveats, for example the need for a proper endogenous control to normalize relative gene expression data. Although many studies have warned against the use of a single reference gene for gene expression normalization [1], [2], the expression of reference genes, such as glyceraldehyde-3-phosphate dehydrogenase (Gapdh) or beta-actin (Actb), or the amount of 18S ribosomal RNA (rRNA) are still frequently used as a single endogenous control [3]. However, the use of a single unvalidated reference gene may give rise to biased study results, especially when study conditions are changed or experimental variability is increased [4]. The increase in reference gene variability becomes even more problematic if genes with relatively small expression differences are studied.
In the setting of myocardial infarction and congestive heart failure (CHF), this instability in reference gene expression has already been described. For instance, Brattelid et al. [5] reported that the reduced expression of Gapdh compared to 18S rRNA significantly influenced the outcome and interpretation of mRNA expression levels. In another in vitro reference gene study, the expression levels of either beta-2 microglobulin (B2m), Gapdh, Actb, or 18S rRNA were found to be highly dependent upon the experimental conditions [4]. This implicated that the choice for a given reference gene for gene expression normalization could potentially bias relative mRNA expression results and alter the study outcome. These problems on the validity of single reference genes for qPCR gene expression normalization led Vandesompele et al. to propose an innovative strategy by using the geometric mean of an optimal number of stably expressed reference genes (geNorm algorithm) [2]. This approach has subsequently been used by numerous researchers for the optimization of reference gene normalization strategies in distinct research applications.
Thus, stable reference gene expression is a prerequisite for conducting accurate and valid gene expression studies. This is particularly true when reference gene variability is expected to be high, as in the case of myocardial infarction or CHF studies. This study aimed to determine an optimal combination of stably expressed reference genes for use in mouse myocardial infarction studies using the geNorm algorithm. In addition, the effects of using suboptimal combinations of reference genes for gene expression analysis on statistical parameters such as significance, power and sample size were studied.
Methods
Ethics Statement
National and European principles of laboratory animal care were followed. The Animal Care and Use Committee of the University of Antwerp approved all animal experimental procedures (Permit Number: 2008-03).
Selection of candidate reference genes
The selection of the reference genes was based on a literature search on the subject of reference gene studies in myocardial infarction. Five commonly used reference genes were included: Actb, B2m, hypoxanthine phosphoribosyltransferase 1 (Hprt), Gapdh, TATA box binding protein (Tbp). Furthermore, five less known reference gene candidates were selected based on their promising stability characteristics, as determined by previous reference gene studies: eukaryotic translation elongation factor 1 alpha 1 (Eef1a1) [6], polymerase (RNA) II (DNA directed) polypeptide A (Polr2a) [7], peptidylprolyl isomerase A (cyclophilin A) (Ppia) [8], ribosomal protein L13a (Rpl13a) [6], tumor protein, translationally-controlled 1 (Tpt1) [6]. Care was taken not to include genes of the same functional pathway, as such avoiding problems of co-regulation and false positive reference gene selection.
Myocardial infarction mouse model and sample collection
The tissue specimens analyzed in this study were taken from a large mouse myocardial infarction tissue sample set. Myocardial infarction was induced by ligation of the left anterior coronary artery (LAD). In brief, 3- to 6-month-old male mice of C57BL/6 background were were anesthetized (tribromoethanol 0.25mg/g, intraperitoneally), intubated using a 22G intravenous catheter and mechanically ventilated with a small rodent ventilator (MiniVent type 845, Harvard Apparatus, ventilation at 10 µl/g, 180 breaths/min, 2 cm H2O positive end-expiratory pressure). A left parasternal thoracotomy was performed transecting ribs 4 and 5. After adequate exposure of the heart, the pericardium was cleaved and the LAD was ligated approximately 2 mm below the left atrial appendage using 7-0 polypropylene sutures (Pronova® BV-1, Ethicon, Johnson & Johnson). Ten age-matched C57BL/6 mice were sham-operated, i.e., without tightening of the LAD ligature. White discoloration of the myocardium, elevation of the ST segment on electrocardiographical monitoring and visual identification of the ligated artery in the infarction zone evidenced successful LAD ligation. Subsequently, the operation wound was closed in layers, mice were weaned from ventilation, extubated and placed under a heat source until full recovery. The tissue sampling was performed one week post-myocardial infarction. Infarcted left ventricle (ILV, n = 10, group 1) was harvested by macrodissection, immediately flash-frozen in liquid nitrogen and stored at −80°C for later use. Likewise, tissue specimens from non-infarcted left ventricular (NILV) free wall zones in myocardial infarction (NILV, n = 10, group 2) and sham-operated mice (sham, n = 10, group 3) were collected and stored. Group characteristics are summarized in Table S1.
Tissue homogenization, RNA extraction and quality, cDNA synthesis
The tissues were homogenized with an OmniTH tissue homogenizer (Mettler-Toledo) on ice. RNA was isolated using AllPrep DNA/RNA/Protein mini kit (Qiagen) and turbo DNA free kit (Ambion) was used to remove contaminating DNA leftovers, both following the manufacturer's instructions. The RNA concentration and purity were analyzed using a Nanodrop spectrophotometer (Nanodrop technologies), measuring spectral absorption at 260 and 280 nm. The assessment of the RNA integrity was done using Agilent 2100 Bioanalyzer. cDNA was synthesized using the transcriptor first strand cDNA synthesis kit (Roche) according to the manufacturer's instructions, using a combined oligo(dT)/random hexamer primer strategy for reverse transcription (RT). RT was performed on 0.5 µg RNA starting material in a final reaction volume of 20 µl. The RT reaction sequence consisted of incubation at 25°C for 10 minutes, followed by 30 minutes at 55°C. The transcriptor reverse transcriptase enzyme was inactivated by heating to 85°C for 5 minutes. cDNA samples were placed on ice and stored at −20°C until further use.
qPCR
Taqman® gene expression assays (Applied Biosystems) were used for qPCR analysis on a LightCycler® 480 instrument (Roche) (table 1). All primers were designed to be intron-spanning to prevent the replication of residual contaminating DNA. To minimize the influence of PCR inhibitors in qPCR applications, all cDNA samples were diluted by a factor 50. qPCR was performed on 5 µl cDNA using the LightCycler® 480 Probes Master (Roche) in a final reaction volume of 20 µl in LightCycler® 480 white 96 Multiwell Plates (Roche). All samples were run in duplicate and no template controls (NTC) were included in all runs to exclude possible DNA contamination. Reactions were carried out as follows: after an initial denaturation-activation step at 95°C for 10 min, amplifications consisted of 45 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 30 s and measurement of fluorescence at 72°C for 1 s. The quantification cycle (Cq) was measured using the baseline-independent second derivative maximum method [9]. The assay efficiency was measured by serial dilution (5-point 4-fold dilution series, using triplicates) of cDNA of pooled samples based on the slope of the standard dilution curve (E = 10(1/-slope)-1) (table 2).
Table 1. Gene abbreviations and respective protein function.
| Gene | Full name | Protein function |
| Actb | beta-actin | Cytoskeletal structure protein |
| B2m | beta-2-microglobulin | Beta–chain of MHC class I molecules |
| Eef1a1 | eukaryotic translation elongation factor 1 alpha 1 | Aminoacyl-tRNA binding to ribosomes during protein biosynthesis |
| Gapdh | glyceraldehyde-3-phosphate dehydrogenase | Oxido-reductase in glycolysis and gluconeogenesis |
| Hprt | hypoxanthine phosphoribosyltransferase 1 | Purine synthesis through the purine salvage pathway |
| Polr2a | polymerase (RNA) II (DNA directed) polypeptide A | DNA-dependent RNA polymerase |
| Ppia | peptidylprolyl isomerase A (cyclophilin A) | Protein folding |
| Rpl13a | ribosomal protein L13a | Structural component of the large 60S ribosomal subunit |
| Tbp | TATA box binding protein | RNA polymerase II transcription factor |
| Tpt1 | tumor protein, translationally-controlled 1 | Involved in calcium binding and microtubule stabilization |
| Nppb | natriuretic peptide B | Cardiac hormone, key role in natriuresis and vasorelaxation |
| Vcam1 | vascular cell adhesion molecule 1 | Adhesion molecule expressed on inflamed endothelium |
Table 2. qPCR assay information.
| Gene | AB ID | GenBank ID | Amplicon (bp) | E(%) |
| Actb | Mm01205647_g1 | NM_007393.3 | 72 | 95 |
| B2m | Mm00437762_m1 | NM_009735.3 | 77 | 92 |
| Eef1a1 | Mm01966109_u1 | NM_010106.2 | 150 | 88 |
| Gapdh | Mm99999915_g1 | NM_008084.2 | 107 | 94 |
| Hprt | Mm00446968_m1 | NM_013556.2 | 65 | 97 |
| Polr2a | Mm00839493_m1 | NM_009089.2 | 85 | 92 |
| Ppia | Mm02342430_g1 | NM_008907.1 | 148 | 102 |
| Rpl13a | Mm01612986_gH | NM_009438.4 | 122 | 93 |
| Tbp | Mm00446973_m1 | NM_013684.3 | 73 | 94 |
| Tpt1 | Mm03009502_g1 | NM_009429.2 | 150 | 91 |
| Nppb | Mm01255770_g1 | NM_008726.4 | 68 | 94 |
| Vcam1 | Mm00449197_m1 | NM_011693.3 | 97 | 93 |
AB ID: Applied biosystems Taqman® gene expression assay ID; bp: number of base pairs; E: Assay efficiency. The assay efficiency was measured by serial dilution of cDNA of pooled samples based on the slope of the standard dilution curve (E = 10(1/-slope)-1) and expressed as percentage (%).
Reference gene expression stability and statistical analysis
The geNorm software [2] for Microsoft Excel® was used for the analysis of the relative gene expression data. To calculate the expression stability of a given gene (gene stability measure M), the program uses an algorithm based on the mean of the pairwise variation of a given reference gene compared to all other control genes. The higher the value of M, the higher the expression variability of the corresponding reference gene. The least stable gene, i.e., the gene with the highest M value, is excluded from the subsequent analysis and all M values are then recalculated until the two most stable genes remain. A stability plot shows the stepwise exclusion of the most unstable reference gene. To determine the optimal number of reference genes in a particular experimental setting, a pairwise variation coefficient Vn/n+1 is calculated between two sequential normalization factors NFn and NFn+1. Values below a given threshold, signify sufficient reference gene set stability and hence validity for gene normalization. As a general rule, values of ≤0.5 for M and of ≤0.15 for Vn/n+1 are indicative of a stably expressed reference gene or gene set, respectively [2]. It is recommended to use at least the 3 most stable reference genes. Additionally, using the BestKeeper software tool [10], we performed a coefficient of variance analysis in which the coefficient of variance is expressed as a percentage of the average Cq level. To highlight the effects of the different normalization strategies on statistical parameters, such as significance, power and sample size, the expression of natriuretic peptide B (Nppb) and vascular cell adhesion molecule 1 (Vcam1) was measured and normalized using different normalization strategies. A non-parametric Mann-Whitney U test was used because of the non-normality of the relative expression data. A two-sided p-value of <0.05 indicated statistical significance. Statistical analysis was performed in PASW® statistics 18 (IBM Corp.). After logarithmic (log2) transformation, the statistical power of a two-sided Student's t-test and the sample size necessary to obtain a power of at least 0.8 were calculated using the distribution parameters of the normalized relative expression data [11].
MIQE guidelines
To increase the reliability and integrity of study results and to promote the effort for experimental consistency and transparency between research laboratories, we adhere to the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines [12]. A MIQE checklist is published as Table S2.
Results
Thirty samples of left ventricular tissue were harvested. To yield representative results for gene expression normalization in myocardial infarction experiments in mice, both ILV (n = 10, group 1) and NILV (n = 10, group 2) specimens from infarcted hearts, as well as tissue samples of the left ventricular free wall of sham-operated animals (n = 10, group 3) were included in the analysis (supplementary table 1). RNA quantity and quality assessment confirmed good RNA yields and adequate RNA purity and integrity with a mean A260/A280 ratio of 2.03±0.10 and mean RNA integrity number of 8.06±1.30 respectively.
For each sample, qPCR Taqman® assays were performed for 10 candidate reference genes (table 1). Dilution series were run for all candidate reference genes to quantify qPCR amplification efficiency (table 2). For all genes, samples were assayed within the same PCR-run (sample maximization experimental design) to avoid the introduction of inter-run variation. All samples were analyzed in duplicate. All NTCs were PCR-negative. Relative expression quantities were calculated using the E−ΔCq method. These values were imported in geNorm. Analysis provided a gene expression stability measure (M) for each reference gene that permitted ranking from least stable (highest M value) to most stably expressed (lowest M value) (figure 1). In addition, the pairwise variation value Vn/n+1 and the effects of stepwise inclusion of the next best reference gene on Vn/n+1 were calculated (figure 2).
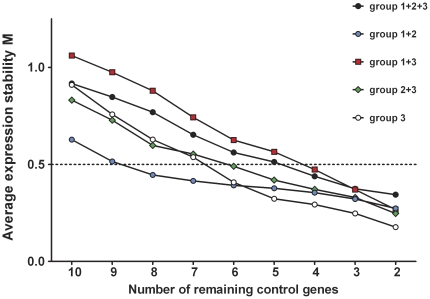
Figure 1. Average expression stability values of reference genes during stepwise exclusion.
The average expression stability values for mouse myocardial infarction tissue are depicted for all (n = 30) and different subsets of samples. Pairwise variation decreases from left to right, due to stepwise exclusion of the least stable reference gene. The least stable gene is found on the right; the best pairwise combination is depicted on the left. To retrieve the corresponding gene names, we refer to table 3. Group 1: infarcted left ventricle; group 2: non-infarcted left ventricle: group 3: sham left ventricular heart tissue.
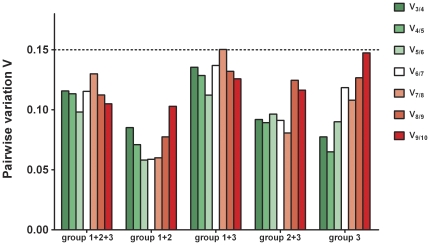
Figure 2. Pairwise variation analysis between sequential normalization factors.
The pairwise variation coefficient, calculated between sequential normalization factors, is used to determine of the optimal number of control genes. For instance, V3/4 indicates the change in stability when using four instead of three reference genes to calculate the normalization factor. A pairwise variation coefficient of ≤0.15 is regarded as an appropriate cut-off. Therefore, a combination of three reference genes is appropriate to use in each sample subset. To the right, the increasing values are due to the inclusion of unstable reference genes, such as Actb, Polr2a and Gapdh into the analysis. Group 1: infarcted left ventricle; group 2: non-infarcted left ventricle: group 3: sham left ventricular heart tissue.
Initially, we performed a geNorm analysis on the overall sample set of 30 infarcted and non-infarcted heart specimens (group 1+2+3), because this is a commonly used experimental setup in mouse myocardial infarction studies. For this experimental setup, Tpt1, Hprt, Rpl13a and Ppia showed M values below the theoretical threshold of 0.5, indicating adequate gene stability (figure 1). M values increased moderately for other reference genes, except for Actb, Polr2a and Gapdh, which showed higher gene expression variability as evidenced by an increase in the slope of the M value curve. Gapdh, although frequently used for gene expression normalization, yielded the highest M value in this data set, which is indicative of the largest overall variability. In figure 2, the optimal number of reference genes for use in gene normalization can be deduced from the value of the pairwise variation between sequential normalization factors. As recommended by Vandesompele et al. [2] 0.15 is generally considered as an appropriate cut-off, although this should be regarded as a reference rather than a strict value. Below this threshold the addition of extra reference genes does not result in a significant improvement of the expression stability of the reference gene set and is therefor not recommended. In our study, the pairwise variation V3/4 between the normalization factors with the combination of the three and four most stable reference genes, yielded a value of 0.117, which was already below the theoretical threshold of 0.15. This means that the addition of Ppia to the gene set composed of Tpt1, Hprt and Rpl13a is not needed and only results in a relatively small gain in stability and decrease in pairwise variation. For this reason and because an optimal balance needs to be found between the absolute gain in statistical power by reducing expression variability and the extra costs and efforts when measuring more reference genes, the set of the three best reference genes (NF3) is suitable as reference gene set in our study.
Subsequently, analysis of the tissue subsets revealed that the average expression stability values could be dependent upon the specific tissues used in the experimental setup (figure 1). For analysis of ILV and NILV specimens (group 1+2), all reference genes, except Gapdh and B2m showed reference gene stability below the theoretical threshold of 0.5. Tpt1, Eef1a1, Ppia, Rpl13a, Hprt and B2m performed best in the sham subset (group 3) and when combining ILV or NILV specimens with sham tissues (group 1+3 or 2+3), M values for Tpt1, Hprt, Rpl13a and Ppia or Rpl13a, Hprt, Tpt1, Ppia, Eef1a1 and B2m respectively, were found to meet the aforementioned stability criteria (table 3). In all the tissue subsets the combination of the three best performing genes was found to be sufficient for gene normalization with pairwise variation values of 0.15 or less (figure 2). Interestingly, the Tpt1, Hprt, Rpl13a and Ppia genes consistently showed M values below 0.5 in all of the five experimental setups described, while Eef1a1 displayed acceptable stability three times, B2m twice and Tbp, Actb and Polr2a only once. On the other hand, Gapdh invariantly disclosed suboptimal gene expression stability in all analyzed subsets. The suboptimal performance of Gapdh, Polr2a and Actb, and to a lesser extent of B2m, Eef1a1 and Tbp, was further confirmed by a coefficient of variance analysis using the BestKeeper software (table 4) and by heatmap analysis of the relative gene expression of all reference genes compared accross all samples (figure 3). Also, comparison of the reference gene expression over the three experimental groups showed a manifest (more than twofold) upregulation of Polr2a, Actb, B2m and Eef1a1 gene expression in ILV or NILV tissues compared to sham controls and a distinct (twofold or more) downregulation of Gapdh in ILV specimens compared to sham controls or NILV, respectively. Except for the expression of Gapdh in ILV versus sham tissues, all these differential expression patterns, were determined to be statistically significant, even after controlling for false discovery rate (Benjamini-Hochberg correction for multiple comparisons [13]). Selective up- or downregulation of reference genes under specific experimental conditions might, in addition to affecting statistical parameters such as power and sample size, give rise to biased study results when one of these genes would be used as reference for gene expression normalization.
Table 3. Reference genes ranked by their expression stability for different sample combinations.
| 1 + 2 + 3 | 1 + 2 | 1 + 3 | 2 + 3 | 3 |
| Gapdh | Gapdh | Gapdh | Polr2a | Polr2a |
| Polr2a | B2m | Polr2a | Actb | Actb |
| Actb | Eef1a1 | Actb | Gapdh | Gapdh |
| B2m | Actb | B2m | Tbp | Tbp |
| Eef1a1 | Polr2a | Eef1a1 | B2m | B2m |
| Tbp | Hprt | Tbp | Eef1a1 | Hprt |
| Ppia | Tpt1 | Ppia | Ppia | Rpl13a |
| Rpl13a | Tbp | Rpl13a | Tpt1 | Ppia |
| Hprt – Tpt1 | Ppia – Rpl13a | Hprt – Tpt1 | Hprt – Rpl13a | Eef1a1 – Tpt1 |
The reference genes are ranked using the expression stability value (M) obtained from the geNorm analysis. M increases from top to bottom. M≤0.5 (bold) indicates stable gene expression. Group 1: infarcted left ventricle; group 2: non-infarcted left ventricle; group 3: sham left ventricular heart tissue.
Table 4. Mean, standard deviation (SD) and coefficient of variance (CV) of quantification cycle (Cq) values.
| Gene | Cq | SD | CV (%) |
| Gapdh | 24.06 | 1.25 | 5.19 |
| Polr2a | 30.85 | 1.64 | 5.30 |
| Actb | 24.46 | 1.52 | 6.23 |
| B2m | 26.24 | 1.14 | 4.36 |
| Eef1a1 | 24.97 | 1.00 | 4.02 |
| Tbp | 32.88 | 1.04 | 3.15 |
| Ppia | 27.23 | 0.82 | 3.02 |
| Rpl13a | 26.35 | 0.67 | 2.53 |
| Hprt | 29.99 | 0.79 | 2.62 |
| Tpt1 | 25.06 | 0.66 | 2.62 |
CV: coefficient of variance is expressed as the percentage of the Cq standard deviation to the mean Cq.
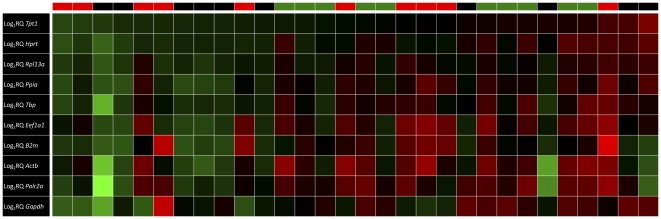
Figure 3. Heatmap of expression variability of individual reference genes.
The relative gene expression (RQ) of all samples (n = 30) for the 10 reference genes is depicted (color-coded, expression values are log2 transformed). Tpt1, Hprt, Rpl13a and Ppia show low variability in gene expression. Gapdh, Polr2a, Actb, B2m and Eef1a1 show high within-group expression variability and more extreme - sometimes opposing - expression values for individual samples compared to stably expressed genes. Tbp demonstrates intermediate expression variability. Red: ILV; green: NILV; black: sham.
Finally, to show the effect of suboptimal reference gene selection, we normalized Nppb and Vcam1 expression using different normalization strategies. Both these genes are known to be upregulated in the setting of myocardial infarction [14], [15]. When Nppb or Vcam1 gene expression was normalized against the combination of the three worst performing reference genes (i.e., Gapdh, Polr2a and Actb) or against a combination of all 10 reference genes (NF10), the relative expression differences were blunted and statistical significance was suppressed. However, normalization against the most optimal combination of reference genes (i.e., Tpt1, Hprt and Rpl13a) yielded a higher relative expression difference and a reduction in sample variance, which resulted in a dramatic improvement of the statistical power and significance (table 5).
Table 5. Effect of normalization strategy on expression levels and statistical significance.
| Normalization strategy | mean ± SD | mean ± SD | ∂ | p-value | power | |
| Nppb | 3 most stable genes | 0.65 ± 0.65 | -0.43 ± 0.61 | 1.09 | 0.003 | 0.954 |
| All 10 reference genes | 0.49 ± 0.63 | 0.04 ± 0.85 | 0.46 | 0.075 | 0.252 | |
| 3 least stable genes | 0.23 ± 0.62 | 0.35 ± 1.54 | -0.12 | 0.684 | 0.055 | |
| Vcam1 | 3 most stable genes | 0.79 ± 0.46 | -0.97 ± 0.92 | 1.76 | <0.001 | 0.999 |
| All 10 reference genes | 0.41 ± 0.44 | -0.51 ± 0.61 | 0.92 | 0.002 | 0.953 | |
| 3 least stable genes | 0.43 ± 0.61 | -0.19 ± 0.68 | 0.62 | 0.089 | 0.527 |
SD: standard deviation; 3 least stable genes: Gapdh, Polr2a, Actb; 3 most stable genes: Tpt1, Hprt and Rpl13a. The mean ± SD are calculated on log2 transformed expression data because of non-normality. Two-tailed Mann-Whitney U test was used for calculating p-values. Statistical power was calculated for a Student's t-test using statistical parameters of log2 transformed expression data. Nppb: group 2 vs. group 3; Vcam1: group 1 vs. group 3.
The calculation of the statistical significance and power for the different normalization strategies unambiguously demonstrated that application of suboptimal reference genes (table 6) or gene combinations (table 7), can dramatically affect both these factors. It is clear from this example that inclusion of more reference genes does not necessarily improve the statistical parameters. For example, the combination of the three most stable genes (NF3) required a sample size of 7 or 5 for Nppb or Vcam1, respectively, in order to obtain a power of at least 0.8. However, normalization using six or more reference genes led to a gradual increase in sample size. In addition, expression analysis using individual reference genes yielded considerable variability in optimal sample size (Nppb 5 - 296; Vcam1 4 - >1000) and tenfold or more reduction in statistical power (Nppb 0.998 - 0.078; Vcam1 1 - 0.050).
Table 6. Effect of normalization strategy on statistical parameters calculated for Nppb and Vcam1 expression using single reference genes.
| Tpt1 | Hprt | Rpl13a | Ppia | Tbp | Eef1a1 | B2m | Actb | Polr2a | Gapdh | ||
| Nppb | p-value | <0.001 | 0.023 | 0.005 | 0.023 | 0.123 | 0.280 | 0.631 | 0.436 | 0.912 | 0.005 |
| power | 0.998 | 0.715 | 0.933 | 0.647 | 0.153 | 0.231 | 0.078 | 0.255 | 0.107 | 0.488 | |
| n | 5 | 13 | 7 | 14 | 82 | 48 | 296 | 42 | 137 | 20 | |
| Vcam1 | p-value | <0.001 | <0.001 | 0.001 | 0.029 | 0.004 | 0.631 | 0.579 | 0.105 | 0.796 | 0.001 |
| power | 0.998 | 1 | 0.992 | 0.619 | 0.937 | 0.136 | 0.059 | 0.345 | 0.050 | 0.997 | |
| n | 5 | 4 | 6 | 15 | 7 | 96 | 943 | 21 | >1000 | 5 |
Two-tailed Mann-Whitney U test was used for calculating p-values. The statistical power was calculated for a Student's t-test using statistical parameters of log2 transformed expression data. Sample size (n) is number per group needed to obtain a power of at least 0.8. Nppb: group 2 vs. group 3; Vcam1: group 1 vs. group 3.
Table 7. Effect of normalization strategy on statistical parameters calculated for Nppb and Vcam1 expression using combinations of multiple reference genes.
| NF3 | NF4 | NF5 | NF6 | NF7 | NF8 | NF9 | NF10 | ||
| Nppb | p-value | 0.003 | 0.009 | 0.007 | 0.009 | 0.035 | 0.029 | 0.043 | 0.075 |
| power | 0.954 | 0.926 | 0.848 | 0.748 | 0.7 | 0.652 | 0.417 | 0.252 | |
| n | 7 | 8 | 9 | 12 | 13 | 14 | 24 | 43 | |
| Vcam1 | p-value | <0.001 | 0.001 | 0.001 | 0.001 | 0.003 | 0.007 | 0.007 | 0.002 |
| power | 0.999 | 0.991 | 0.993 | 0.962 | 0.891 | 0.835 | 0.857 | 0.953 | |
| n | 5 | 6 | 6 | 7 | 9 | 10 | 9 | 7 |
Two-tailed Mann-Whitney U test was used for calculating p-values. The statistical power was calculated for a Student's t-test using statistical parameters of log2 transformed expression data. Sample size (n) is number per group needed to obtain a power of at least 0.8. Nppb: group 2 vs. group 3; Vcam1: group 1 vs. group 3.
Discussion
The importance of adequate reference gene selection for the normalization of qPCR data cannot be underestimated. The inappropriate choice of reference genes frequently results in loss of accuracy, statistical significance and power [16], in particular in case of genes with small expression differences. Over the last decade, several new strategies for data normalization have been proposed and nowadays optimization of reference genes should be recommended as a crucial first step in every gene expression experiment.
Using the geNorm algorithm, we were able to identify a set of three reference genes, Hprt, Rpl13a and Tpt1, which can be used for accurate gene expression normalization in qPCR experiments on mouse myocardial infarction tissue. The application of different combinations of starting material yielded slightly different optimal reference gene sets. However Gapdh, which is frequently used for gene expression normalization in many cardiovascular studies, together with Polr2a and Actb, showed the largest gene variability and the worst performance as reference genes. This finding could be problematic for a substantial number of gene expression studies that utilize Gapdh for data normalization. On the other hand, it is not that surprising to find that Gapdh, which is an enzyme of the glycolysis pathway, displays high variability in the setting of myocardial infarction, where in the absence of oxygen, anaerobic pathways are activated to fulfill energy demands. Recently, Gapdh has also been reported to play a role in the mitochondria during apoptosis, inducing mitochondrial membrane permeabilization, which leads to loss of the inner mitochondrial transmembrane potential [17]. These findings question the unvalidated use of Gapdh as a reliable internal reference in ischemic conditions, not only in qPCR experiments, but also for normalization in other molecular techniques, such as western blotting.
The three most stably expressed genes in our experimental setting, Hprt, Rpl13a and Tpt1, encode proteins with independent physiological functions. Tpt1, also known as histamine-releasing factor (HRF), has a high degree of tissue conservation between species [18] and is expressed in a multitude of different tissues under extensive transcriptional control [19]. It was found to promote cell growth and has anti-apoptotic properties [20]. Rpl13a encodes one of about 50 proteins that are part of the large 60S ribosomal subunit, implicating Rpl13a in protein synthesis. The Hprt gene is translated into the enzyme hypoxanthine-guanine phosphoribosyltransferase, which has a pivotal role in purine nucleotide generation, providing the transfer of a phosphoribosyl group to hypoxanthine or guanine.
Over the last years, only few studies have dealt with reference gene normalization in the setting of myocardial infarction. A first study by Perez et al. [8] described the most suitable control genes in human myocardial tissues. The authors proposed a combination of Ppia (encodes a protein folding enzyme of the peptidyl-prolyl cis-trans isomerase family), Rplp (a ribosomal subunit protein analogous to Rpl13a) and Gapdh for accurate normalization. In another study by Pilbrow et al. [6], a microarray screen of publicly available gene expression data was used to select 10 genes of high abundance and consistent gene expression. The subsequent geNorm analysis indicated Srp14, Tpt1 and Eef1a1 as the most stable genes and ranked Gapdh as the least stable gene. The striking difference in Gapdh stability between these two studies can be explained by the difference in tissue sampling. The tissues analyzed in the study by Perez et al. [8] were derived from the myocardium of human heart donors not suffering from ischemic heart disease, whereas in the study by Pilbrow et al. [6] specimens from failing hearts together with non-failing donor hearts were analyzed. In our analysis we also noticed a better performance of Gapdh in NILV or sham tissues, which supports the observation that Gapdh still performed relatively well in the study of Perez et al. [8] On the other hand, the occurrence of contradictory findings between studies with a different study design shows the importance of adequate reference gene analysis in each specific experimental setup, even for different combinations of tissue samples. A recent study investigated human and rodent reference genes in the setting of heart failure and identified Gapdh as stable in mouse, but not in rat or human heart tissues [7]. These authors used a data set of only three sham and three heart failure specimens in their analysis. Also, heart failure tissue was harvested at a distance from the infarcted area excluding the infarction border zone and only included the remaining viable septum and left ventricular free wall. As we observed that, in particular in infarcted tissues, Gapdh expression behaves very unpredictably compared to other reference genes, the different outcome of this study is probably due to differences in tissue sampling and – importantly – the limited sample size. Our data in the setting of myocardial infarction are in line with the heart failure data of Pilbrow et al. [6] who also found indications for avoiding Gapdh as reference gene for routine gene expression normalization in human myocardial studies. However, we have to bear in mind that, because of interspecies differences in gene expression, claims on optimal reference gene combinations are generally not applicable to other species and have to be validated accordingly.
More in general, we stress that our conclusions are valid and refer to myocardial tissues in mice, and more particularly to tissues of a C57BL/6 mouse genetic background. This should be taken into consideration, because strain differences were recently found to result in altered expression stability of reference genes [21]. Secondly, since all tissue specimens were taken one week post-infarction, it is possible that other optimal reference gene combinations might be more appropriate for other timepoints. Furthermore, we limited our analysis to 10 reference genes, which we compiled on the basis of previous reference gene studies in myocardial infarction. We do not exclude the possibility that more optimal reference gene combinations can be found when other stably expressed genes are included in the analysis. However, as evidenced by the low value for the average expression stability value M, our reference gene set outperforms all other reported reference gene sets analyzed so far with the geNorm algorithm in the setting of myocardial infarction. Finally, our findings only apply to the setting of myocardial infarction in mice, and, therefore, do not preclude Gapdh from being an adequate reference gene in other conditions, tissues or species.
In conclusion, we identified and validated a stably expressed reference gene set for use in mouse myocardial infarction studies. Optimal reference gene normalization greatly improves statistical significance, power and can dramatically reduce sample size. Our results indicate in particular that Gapdh, which is commonly used for gene expression normalization in myocardial infarction studies, has rather high expression variability in myocardial infarction tissues in mice. We furthermore caution against the use of Gapdh, Polr2a, Actb, B2m and Eef1a1 for gene expression normalization in myocardial infarction studies because of selective up- or downregulation after myocardial infarction. Therefore, inclusion of Gapdh or other suboptimal reference genes will potentially lead to biased gene expression results. Given the risk of inducing reference gene instability when altering experimental conditions, we recommend the validation of a stable set of reference genes as an initial and essential step in all qPCR experiments.
Supporting Information
Group characteristics. The data are shown as mean ± SD. HW: heart weight; BW: body weight; TL: tibia length; LVEDD: left ventricular end-diastolic diameter; AWT: anterior wall thickness. * p<0.05 vs. sham; **p<0.01 vs sham (student's t-test). Transthoracic echocardiography (AplioXV®, 13MHz linear probe, Toshiba) was performed on anesthetized mice one week post-myocardial infarction. Left ventricular end-diastolic diameters (LVEDD) and anterior wall thickness (AWT) were measured at the mid-papillary muscle level.
(DOCX)
MIQE checklist.
(XLS)
Acknowledgments
The authors would like to acknowledge the efforts of D. Vindevogel in editing this text. We furthermore thank L. Avula for technical assistance with sample preparation.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Research Foundation - Flanders (FWO), grant No. G014906. B. Everaert is supported by a Ph.D. fellowship of the Reseach Foundation - Flanders (FWO). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Suzuki T, Higgins PJ, Crawford DR. Control selection for RNA quantitation. Biotechniques. 2000;29:332–337. doi: 10.2144/00292rv02. [DOI] [PubMed] [Google Scholar]
- 2.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Jonge HJ, Fehrmann RS, de Bont ES Hofstra RM, Gerbens F, et al. Evidence based selection of housekeeping genes. PLoS One. 2007;2:e898. doi: 10.1371/journal.pone.0000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmittgen TD, Zakrajsek BA. Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods. 2000;46:69–81. doi: 10.1016/s0165-022x(00)00129-9. [DOI] [PubMed] [Google Scholar]
- 5.Brattelid T, Tveit K, Birkeland JA, Sjaastad I, Qvigstad E, et al. Expression of mRNA encoding G protein-coupled receptors involved in congestive heart failure - a quantitative RT-PCR study and the question of normalisation. Basic Res Cardiol. 2007;102:198–208. doi: 10.1007/s00395-007-0648-1. [DOI] [PubMed] [Google Scholar]
- 6.Pilbrow AP, Ellmers LJ, Black MA, Moravec CS, Sweet WE, et al. Genomic selection of reference genes for real-time PCR in human myocardium. BMC Med Genomics. 2008;1:64. doi: 10.1186/1755-8794-1-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brattelid T, Winer LH, Levy FO, Liestol K, Sejersted OM, et al. Reference gene alternatives to Gapdh in rodent and human heart failure gene expression studies. BMC Mol Biol. 2008;11:22. doi: 10.1186/1471-2199-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perez S, Royo LJ, Astudillo A, Escudero D, Alvarez F, et al. Identifying the most suitable endogenous control for determining gene expression in hearts from organ donors. BMC Mol Biol. 2007;8:114. doi: 10.1186/1471-2199-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao S, Fernald RD. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol. 2005;12:1047–1064. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 11.Lenth R. Java Applets for Power and Sample Size [Computer software]. Retrieved May 25th 2011, from http://www.stat.uiowa.edu/~rlenth/Power.
- 12.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 13.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 14.Kragelund C, Gronning B, Kober L, Hildebrandt P, Steffensen R. N-terminal pro-B-type natriuretic peptide and long-term mortality in stable coronary heart disease. N Engl J Med. 2005;352:666–675. doi: 10.1056/NEJMoa042330. [DOI] [PubMed] [Google Scholar]
- 15.Nahrendorf M, Swirski FK, Aikawa E, Stangenberg L, Wurdinger T, el al. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J Exp Med. 2007;204:3037–3047. doi: 10.1084/jem.20070885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derveaux S, Vandesompele J, Hellemans J. How to do successful gene expression analysis using real-time PCR. Methods. 2010;50:227–230. doi: 10.1016/j.ymeth.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Tarze A, Deniaud A, Le Bras M, Maillier E, Molle D, et al. GAPDH, a novel regulator of the pro-apoptotic mitochondrial membrane permeabilization. Oncogene. 2007;26:2606–2620. doi: 10.1038/sj.onc.1210074. [DOI] [PubMed] [Google Scholar]
- 18.Gross B, Gaestel M, Bohm H, Bielka H. cDNA sequence coding for a translationally controlled human tumor protein. Nucleic Acids Res. 1989;17:8367. doi: 10.1093/nar/17.20.8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thiele H, Berger M, Skalweit A, Thiele BJ. Expression of the gene and processed pseudogenes encoding the human and rabbit translationally controlled tumour protein (TCTP). Eur J Biochem. 2000;267:5473–5481. doi: 10.1046/j.1432-1327.2000.01609.x. [DOI] [PubMed] [Google Scholar]
- 20.Bommer UA, Heng C, Perrin A, Dash P, Lobov S, et al. Roles of the translationally controlled tumour protein (TCTP) and the double-stranded RNA-dependent protein kinase, PKR, in cellular stress responses. Oncogene. 2010;29:763–773. doi: 10.1038/onc.2009.380. [DOI] [PubMed] [Google Scholar]
- 21.Kosir R, Acimovic J, Golicnik M, Perse M, Majdic G, et al. Determination of reference genes for circadian studies in different tissues and mouse strains. BMC Mol Biol. 2010;11:60. doi: 10.1186/1471-2199-11-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Group characteristics. The data are shown as mean ± SD. HW: heart weight; BW: body weight; TL: tibia length; LVEDD: left ventricular end-diastolic diameter; AWT: anterior wall thickness. * p<0.05 vs. sham; **p<0.01 vs sham (student's t-test). Transthoracic echocardiography (AplioXV®, 13MHz linear probe, Toshiba) was performed on anesthetized mice one week post-myocardial infarction. Left ventricular end-diastolic diameters (LVEDD) and anterior wall thickness (AWT) were measured at the mid-papillary muscle level.
(DOCX)
MIQE checklist.
(XLS)