Abstract
Background
Rare mutations of the epithelial sodium channel (ENaC) lead to Mendelian forms of salt-sensitive hypertension or salt-wasting hypotension. We aimed to examine the association between common variants in the ENaC genes and salt-sensitivity of blood pressure (BP).
Methods and Results
A total of 1,906 Han Chinese participated in the Genetic Epidemiology Network of Salt Sensitivity (GenSalt) study, which includes a 7-day low-sodium intake (51.3 mmol sodium/day) followed by a 7-day high-sodium intake (307.8 mmol sodium/day). Nine BP measurements were obtained at baseline and each intervention period using a random-zero sphygmomanometer. Single nucleotide polymorphisms (SNPs), both tagging and functional, from the three ENaC subunits, α, β, and γ (SCNN1A, SCNN1B, and SCNN1G), were genotyped. Multiple common SNPs in SCNN1G were significantly associated with BP response to low-sodium intervention (rs4073930, p=1.7×10-5; rs4073291, p=1.1×10-5; rs7404408, p=1.9×10-5; rs5735, p=3.0×10-4; rs4299163, p=0.004; and rs4499238, p=0.002) even after correcting for multiple testing. For example, under an additive model, the minor allele G of SNP rs4073291 was associated with 1.33 mmHg lower systolic BP (SBP) reduction during low-sodium intervention.
Conclusions
This large dietary sodium intervention study indicates that common variants of ENaC subunits may contribute to the variation of BP response to dietary sodium intake. Future studies are warranted to confirm these findings in an independent population and to identify functional variants for salt-sensitivity.
Keywords: blood pressure, epithelial sodium channel, genetic variant, salt-sensitivity
Interactions among numerous genetic and environmental factors play an important role in blood pressure (BP) regulation. High dietary sodium intake is one of the most important environmental risk factors for elevated BP.1–3 However, BP response to dietary sodium intake varies considerably among individuals, a phenomenon known as salt-sensitivity.4, 5 Salt-sensitivity of BP has been associated with an increased risk of hypertension, cardiovascular disease, and premature death.6, 7 Previous studies have suggested that an individual's genetic profile may contribute to their BP responses to dietary sodium intake.8–13 Thus, the investigation of genetic determinants of salt-sensitivity will help to understand the potential interaction between genetic factors and dietary sodium intake on the regulation of BP.
The renal epithelial sodium channel (ENaC) mediates net renal reabsorption of sodium in the distal tubule and is critical for the control of sodium balance, blood volume, and thereby of BP.14, 15 ENaC is composed of three partly homologous subunits, α, β, and γ.16 It has been documented that gain-of-function mutations of ENaC genes lead to Mendelian forms of hypertension or hypotension. For example, mutations in the cytoplasmic C terminus of either the β- or γ-subunit of ENaC cause Liddle's syndrome, which is an autosomal dominant form of salt-sensitive hypertension and associated with increased ENaC activity and sodium reabsorption of the kidney.17–20 On the other hand, mutations in ENaC subunits may also cause pseudohypoaldosteronism type I, which is an inherited form of severe hypotension and salt wasting.21
Recently, researchers have tried to identify common variants of ENaC genes which may influence the risk of essential hypertension and reported inconsistent results.22–25 Iwai et al reported that SNP rs5718, also known as G(−173)A, in the promoter region of the ENaC γ-subunit gene was associated with low SBP in a Japanese population.22 However, this finding was not replicated in two independent studies of white Australians.23, 26 In addition, two studies conducted in Chinese Kazakhs reported inconsistent findings about the association of rs879605 in the α-subunit gene with hypertension.24, 25 These discrepancies might reflect differences in genetic architecture and linkage disequilibrium (LD) patterns among different ethnicities and heterogeneity of hypertension etiology among individuals even within the same ethnicity. Furthermore, none of these studies took into account the gene-sodium interaction on BP. Animal experiments suggested that variants of the ENaC genes and sodium intake might interactively determine the development of hypertension.27 We examined the association between common variants in ENaC genes and BP responses to dietary sodium intervention in the Genetic Epidemiology Network of Salt-Sensitivity (GenSalt) study.
Methods
Study Participants
The GenSalt study was a family-based dietary feeding study conducted in rural areas of northern China from 2003 to 2005. A community-based BP screening was conducted among persons aged 18-60 years in the study villages to identify potential probands and their families. The probands who had a mean SBP between 130-160 mmHg and/or a DBP between 85-100 mmHg and no use of antihypertensive medications and their spouses, siblings and offspring were recruited for the dietary feeding study. All participants were of Han Chinese ethnicity. Individuals who had stage-2 hypertension, secondary hypertension, a history of clinical cardiovascular disease or diabetes, used antihypertensive medications, or were pregnant, heavy alcohol drinkers, or currently on a low-sodium diet were excluded from the study. More information about study design and participants has been published elsewhere.28 Among 1,906 eligible participants for the dietary intervention, 1,871 (98.2%) and 1,860 (97.6%) completed the low-sodium and high-sodium interventions, respectively, and were included in the current analysis. Institutional Review Boards or ethics committees at all participating institutes approved the study protocol. Written informed consents for the baseline observation and for the intervention were obtained from each participant prior to data collection or intervention, respectively.
Dietary Intervention
After a 3-day baseline observation, the study participants received a 7-day low-sodium diet (3 grams of sodium chloride or 51.3 mmol of sodium or 1,179.9 mg of sodium per day) followed by a 7-day high-sodium diet (18 grams of sodium chloride or 307.8 mmol of sodium or 7,079.4 mg of sodium per day). All foods were cooked without salt, and prepackaged salt was added to the individual study participant's meal when it was served by the study staff. To ensure study participants' compliance to the intervention program, they were required to avoid consuming any foods and beverages that were not provided by the study. Dietary compliance by the participants was confirmed by measurements of 24-hour urinary excretion of sodium and potassium. One 24-hour and two overnight urine specimens were collected at the baseline and the last 3 days of each intervention phase. The overnight urinary sodium excretion was converted to 24-hour values using the formulas developed from a random subsample of 238 subjects who collected overnight and 24-hour urine samples on the same days. The mean (standard deviation) of 24-hour urinary excretions of sodium and potassium were 242.4 (66.7) mmol and 36.9 (9.6) mmol at baseline, 47.5 (16.0) mmol and 31.4 (7.7) mmol during the low-sodium intervention, and 244.3 (37.7) mmol and 35.7 (7.5) mmol during the high-sodium intervention, respectively.
BP Measurements
Three sitting BP measurements were obtained each morning of the 3-day baseline observation and on days 5, 6, and 7 of each intervention period by the trained and certified observers using a random-zero sphygmomanometer according to a standard protocol.29 BP levels at baseline and during intervention were calculated as the mean of nine measurements from each period.
Previous GenSalt analysis showed that BP responses to low-sodium and to high-sodium interventions shared some but not all genetic determinants (genetic correlation coefficients ρ= -0.43, p= 0.03 for SBP and ρ= -0.58, p=0.0003 for DBP).30 BP response to high-sodium intervention from low-sodium intervention may provide a more valid phenotype measure for salt-sensitivity because participants' sodium intake was controlled during both phases. On the other hand, identifying genetic determinants of BP response to low-sodium from usual diet should have more direct clinical and public health implications. Therefore, we used both BP responses, to low-sodium intervention from baseline and to high-sodium intervention from low-sodium intervention, in this study. In the following text and tables, the terms ‘BP response to low-sodium intervention from baseline’ and ‘BP response to high-sodium intervention from low-sodium intervention’ are simplified as ‘BP response to low-sodium’ and ‘BP response to high-sodium’, respectively.
Single Nucleotide Polymorphism (SNP) Selection and Genotyping
Both SCNN1B and SCNN1G genes are located on chromosome 16, whereas SCNN1A is located on chromosome 12. Tag SNPs from these genes were selected based on empirical patterns of linkage disequilibrium structure in the Chinese Han of Beijing (CHB) HapMap sample using Tagger software. The r2 threshold used for selecting each tagSNP was 0.8. We also included SNPs that were previously reported to be associated with BP or hypertension. SNPs were genotyped using SNPlex assays (Applied Biosystems, Foster City, California, USA) based on oligonucleotide ligation assay for capillary electrophoresis on ABI 3700 DNA Analyzers (Applied Biosystems). To provide better coverage of these three genes, we included additional SNPs genotyped on the Affymetrix 6.0 platform (Affymetrix, Santa Clara, California, USA), which had minor allele frequencies greater than 0.01. In total, 46 SNPs passed quality control and were included in this analysis. The detailed information for these SNPs concerning their genome and gene locations, allele frequencies, and p-values for Hardy-Weinberg equilibrium (HWE) test is presented in Table S1 (please see http://hyper.ahajournal.org).
Statistical Analysis
The Mendelian consistency of the SNP genotype data was assessed by PLINK (version 1.05; http://pngu.mgh.harvard.edu/∼purcell/plink/).31 We used Haploview software (version 4.2; http://www.broadinstitute.org/haploview) to test HWE for each SNP and estimate the extent of pairwise linkage disequilibrium (LD) between SNPs.32 The solid spine LD method, as implemented in Haploview, was used to define LD blocks.
Baseline characteristics and BP response variables of intervention participants were summarized as means (standard deviations) and medians (ranges) for continuous variables and as percentages for categorical variables. Analyses of the association between SNPs and BP response phenotypes were conducted using a linear mixed-effects model implemented in the Proc Mixed procedure of SAS (version 9.1; SAS Institute, Cary, NC). A ‘sandwich’ option was used to compute the estimated variance-covariance matrix of the fixed-effects (genetic variant effects) parameters by using the asymptotically consistent estimator. Because most of the studied families (random effects) only included sib pairs, we selected compound symmetry as the covariance structure which assumes the same degree of dependency among family members. Age, sex, BP measurement room temperature, and study site were adjusted in multivariable analyses. Additive genetic models were assumed. The false discovery rate (FDR) method was used to adjust for multiple testing.33 A FDR q value is calculated in this method to estimate the proportion of rejected null hypotheses which are erroneously rejected. We used the Proc Multtest procedure, along with the FDR option, in SAS to calculate q value for each SNP. A q value of 0.05 was used as the threshold for statistical significance in our study.
Results
Table 1 shows study participants' baseline characteristics, BP levels during low-sodium and high-sodium interventions, and BP responses to dietary sodium interventions. Overall, BP levels decreased from baseline to low-sodium intervention and increased from low-sodium to high-sodium intervention. BP levels were similar at baseline and during the high-sodium intervention. BP responses to dietary sodium intervention were significantly different from zero.
Table 1.
Characteristics of 1,906 participants.
| Variable | Mean ± SD | Median (Range)* |
|---|---|---|
| Age, yrs | 38.7 ± 9.6 | 39.0 (16.0, 62.0) |
| Male, No (%) | 1010 (53.0) | |
| BMI, kg/m2 | 23.3 ± 3.2 | 22.9 (14.5, 37.8) |
| Baseline BP, mm Hg | ||
| Systolic | 116.9 ± 14.2 | 115.8 (80.9, 176.9) |
| Diastolic | 73.7 ± 10.3 | 73.3 (38.9, 109.3) |
| BP during low-sodium intervention, mmHg | ||
| Systolic | 111.4 ± 12.2 | 110.0 (82.4, 159.3) |
| Diastolic | 71.0 ± 9.7 | 70.7 (41.1, 112.2) |
| BP during high-sodium intervention, mmHg | ||
| Systolic | 116.3 ± 13.6 | 114.4 (82.0, 181.1) |
| Diastolic | 72.9 ±10.3 | 72.4 (41.3, 116.0) |
| BP response to low-sodium, mmHg† | ||
| Systolic | -5.5 ± 7.0§ | -4.4 (-45.8, 24.2) |
| Diastolic | -2.8 ± 5.5§ | -2.6 (-23.1, 18.2) |
| BP response to high-sodium, mmHg‡ | ||
| Systolic | 4.9 ± 6.0§ | 4.7 (-11.3, 36.2) |
| Diastolic | 1.9 ± 5.4§ | 1.8 (-20.0, 27.1) |
Median and range is for continuous variables;
BP change from baseline to the low-sodium intervention;
BP change from the low-sodium to high-sodium intervention;
P value was less than 0.0001 when compared with no BP change during sodium interventions. SD, standard deviation; BMI, body mass index; BP, blood pressure.
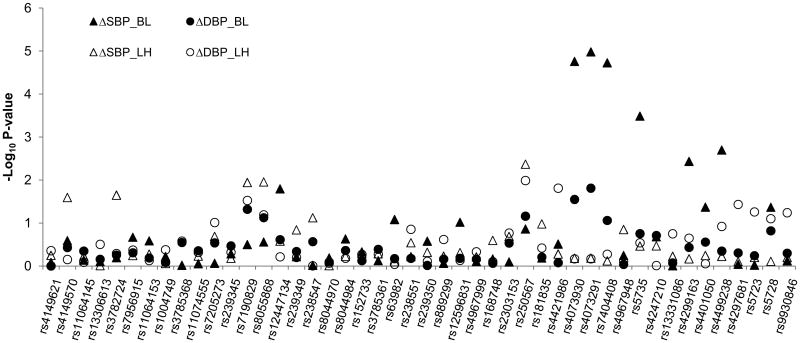
Figure 1 shows the association of each SNP with SBP and DBP responses to low-sodium and high-sodium interventions. After adjusting for age, sex, BP measurement room temperature, and study site, multiple SNPs in the SCNN1A, SCNN1B and SCNN1G genes were associated with BP responses to dietary sodium intervention with p values <0.05. The regression coefficient estimates and standard errors are shown in Table 2. Several SNPs in the SCNN1A and SCNN1B genes were associated with BP responses to high-sodium intervention although the statistical significance did not remain after correcting for multiple comparisons. For example, the minor A allele of SNP rs250567 of SCNN1B was associated with 0.95 mmHg (p=0.004, q=0.18) and 0.82 mmHg (p=0.01, q=0.36) decreased SBP and DBP responses to high-sodium intervention, respectively, compared to its major G allele. After adjusting for multiple testing, six SNPs from SCNN1G were significantly associated with SBP response to low-sodium intervention. The four SNPs with the smallest p values (rs4073930, rs4073291, rs7404408, and rs5735) were in the same LD block and highly correlated with each other (Figure S1, please see http://circgenetics.ahajournals.org). The minor alleles of these four SNPs were associated with a smaller SBP decrease in response to low-sodium intervention. For example, the minor allele G of SNP rs4073291 was associated with 1.33 mmHg lower SBP reduction during low-sodium intervention (p=1.1×10-5, q=3.0×10-4).
Figure 1.
The log P values for the association between 46 single nucleotide polymorphisms of SCNN1A (a), SCNN1B (b), and SCNN1G (c) genes and blood pressure responses to low- and high-sodium interventions. The horizontal dash line indicates P value of 0.05. ΔSBP_BL, systolic BP changes from baseline to low-sodium intervention; ΔDBP_BL, diastolic BP changes from baseline to low-sodium intervention; ΔSBP_LH, systolic BP changes from low- to high-sodium intervention; ΔDBP_LH, diastolic BP changes from low- to high-sodium intervention.
Table 2. Regression coefficients (standard errors) of blood pressure responses to low-sodium and high-sodium interventions associated with minor alleles from linear mixed-effects models.
| SNP | Alleles* | MAF | BP response to low sodium, mmHg† | BP response to high sodium, mmHg† | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Systolic | Diastolic | Systolic | Diastolic | |||||||
| Beta (s.e.) | P-value | Beta (s.e.) | P-value | Beta (s.e.) | P-value | Beta (s.e.) | P-value | |||
| SCNN1A | ||||||||||
| rs4149570 | C:A | 0.50 | 0.23 (0.21) | 0.26 | 0.16 (0.17) | 0.37 | 0.43 (0.19) | 0.03 | 0.07 (0.18) | 0.71 |
| rs3782724 | A:G | 0.24 | 0.12 (0.26) | 0.64 | -0.13 (0.22) | 0.56 | -0.48 (0.21) | 0.02 | -0.13 (0.20) | 0.51 |
| SCNN1B | ||||||||||
| rs7190829 | A:G | 0.18 | -0.27 (0.27) | 0.32 | -0.47 (0.24) | 0.05 | 0.59 (0.23) | 0.01 | 0.50 (0.23) | 0.03 |
| rs8055868 | G:A | 0.20 | -0.29 (0.27) | 0.28 | -0.41 (0.23) | 0.08 | 0.61 (0.24) | 0.01 | 0.41 (0.22) | 0.06 |
| rs12447134 | A:C | 0.08 | 0.96 (0.40) | 0.02 | 0.39 (0.34) | 0.24 | -0.38 (0.34) | 0.26 | 0.16 (0.31) | 0.61 |
| rs250567 | G:A | 0.10 | 0.58 (0.39) | 0.14 | 0.58 (0.32) | 0.07 | -0.95 (0.33) | 0.004 | -0.82 (0.32) | 0.01 |
| SCNN1G | ||||||||||
| rs4421986 | C:T | 0.06 | -0.51 (0.50) | 0.31 | -0.08 (0.39) | 0.84 | 0.29 (0.45) | 0.53 | -0.88 (0.36) | 0.02 |
| rs4073930 | T:C | 0.16 | 1.28 (0.30) | 1.7E-5 /3.0E-4‡ | 0.56 (0.26) | 0.03 | 0.11 (0.24) | 0.66 | 0.11 (0.26) | 0.68 |
| rs4073291 | T:G | 0.16 | 1.33 (0.30) | 1.1E-5 /3.0E-4‡ | 0.63 (0.26) | 0.02 | 0.11 (0.24) | 0.65 | 0.11 (0.27) | 0.68 |
| rs7404408 | C:T | 0.16 | 1.29 (0.30) | 1.9E-5 /3.0E-4‡ | 0.44 (0.26) | 0.09 | 0.07 (0.24) | 0.76 | 0.16 (0.26) | 0.53 |
| rs5735 | T:C | 0.17 | 1.18 (0.33) | 3.0E-4 /0.004‡ | 0.36 (0.27) | 0.18 | 0.24 (0.26) | 0.34 | 0.29 (0.27) | 0.29 |
| rs4299163 | G:C | 0.10 | 1.09 (0.38) | 0.004 /0.02‡ | 0.29 (0.32) | 0.37 | 0.14 (0.33) | 0.68 | 0.38 (0.31) | 0.23 |
| rs4401050 | C:T | 0.09 | 0.86 (0.42) | 0.04 | 0.36 (0.33) | 0.28 | 0.20 (0.33) | 0.57 | 0.05 (0.35) | 0.89 |
| rs4499238 | G:A | 0.10 | 1.11 (0.36) | 0.002 /0.03‡ | 0.23 (0.31) | 0.45 | 0.17 (0.31) | 0.59 | 0.47 (0.30) | 0.12 |
| rs4297681 | C:A | 0.06 | -0.05 (0.47) | 0.91 | 0.25 (0.36) | 0.50 | 0.11 (0.41) | 0.78 | -0.72 (0.34) | 0.04 |
| rs5728 | A:G | 0.19 | 0.60 (0.30) | 0.04 | 0.36 (0.25) | 0.15 | 0.07 (0.26) | 0.78 | -0.43 (0.25) | 0.08 |
Major allele: minor allele.
BP response to low-sodium = mean BP of last 3 days during low-sodium intervention − mean BP during baseline; BP response to high sodium = mean BP of last 3 days during high-sodium intervention − mean BP of last 3 days during low sodium intervention.
FDR q values less than 0.05 are listed in bold. Beta is the effect size on BP response in mmHg, per minor allele based on the additive genetic model. Age, sex, BP measurement room temperature, and study site were adjusted in the mixed-effects models. SNP, single nucleotide polymorphism; MAF, minor allele frequency; SBP, systolic blood pressure; DBP, diastolic blood pressure.
Discussion
Our study identified several common variants in the ENaC genes which may contribute to the BP response to dietary sodium intake. To our knowledge, this is the first report of a comprehensive analysis of association between common variants in the ENaC genes and BP response to dietary sodium intervention.
A previous study in an animal model indicated that ENaC genes might play a critical role in salt-sensitive hypertension.20 Pradervand et al. generated a mouse model of Liddle's syndrome with deletion of the C terminus of ENaC β-subunit by Cre/loxP-mediated recombination.27 Under normal sodium diet, mice heterozygous (L/+) and homozygous (L/L) for Liddle mutation (L) develop normally during the first three months of life and have BP levels similar to wild-type. Under high sodium intake, the mice with Liddle's syndrome mutation develop high BP, metabolic alkalosis, and hypokalemia accompanied by cardiac and renal hypertrophy.
Multiple mutations in the ENaC β and γ subunits have been related to Liddle's syndrome in humans. Most of them cause the deletion of a conserved proline-rich PY motif (codon 611 to 623) in the cytoplasmic C-terminus region of the β or γ subunits, resulting in increased ENaC activity.34 A mutation Asn530Ser of the γ subunit, outside of the PY motif, was also reported to be associated with Liddle's syndrome.35 In the single SNP analysis of our study, six SNPs of the SCNN1G gene, one in an exon and five in introns, showed significant association with SBP response to low-sodium intervention. The significant exonic SNP rs5735 is synonymous and does not change the amino acid encoded. A study of a Japanese population and two studies of Caucasian populations did not find an association between this synonymous mutation and BP.22, 26, 36 To speculate on the functional implication of the five significant intronic SNPs, we used the web tool FastSNP which could analyze potential SNP function using a variety of biological databases and analytic tools.37 Two of the five SNPs, rs4073930 and rs4299163, are possible intronic enhancers for the regulation of gene expression. On the other hand, it is a possibility that none of these SNPs are causal and they are simply in high LD with the true causal variant which was not tested in the current study. In order to fully interpret these associations, further dissection of the regions surrounding these SNPs and functional studies are warranted in the future.
Investigators have expended much effort to sequence the exons of SCNN1B gene in order to find mutations associated with BP in the general population. T594M is the most studied mutation and has been implicated as a gain-of-function mutation causing impaired renal sodium excretion and salt-sensitive hypertension.38, 39 However, this mutation seems unique for people of African descent and was absent in selected European American or Japanese populations.38, 40 T594M was not genotyped in our study. In addition, some other mutations of the SCNN1B gene were detected in a Japanese population but none of them was associated with BP levels.41
Recently, several common variants of SCNN1B were reported to be associated with BP or hypertension in white Europeans (rs239345), Chileans (a GT short tandem repeat in intron 8), and Koreans (rs7205273 and rs8044970).42–44 In our study, these SNPs were not significantly associated with salt-sensitivity of BP. However, SNP rs250567 of the SCNN1B gene showed a trend toward association with BP response to high-sodium intervention. This SNP is located at the 3′ flanking region and has no inferred functional implication based on the analysis by FastSNP. Further resequencing to discover new genetic variants around this SNP may help to detect stronger signals and identify functional variants.
Mutations in the SCNN1A gene could lead to pseudohypoaldosteronism type I.21 However, we did not observe any associations between common variants of SCNN1A and salt-sensitivity of BP after adjusting for multiple testing. Inconsistent results on the association of BP to a common promoter SNP rs879605, also known as G2139A have been reported across studies. The G2139A allele was associated with higher BP level in a Japanese population.45 A haplotype containing this allele was found to be associated with essential hypertension in a sample of Kazakhs, a Chinese minority group.25 However, neither single marker nor haplotype analysis of this SNP was associated with hypertension in another sample of Chinese Kazakhs.24 In our study, the association between one of its adjacent SNPs, rs11064153, and salt-sensitivity of BP was not significant.
We reported here for the first time significant associations between common variants of the ENaC genes and BP response to dietary sodium intervention. These findings suggest that the variants in ENaC genes play an important role in the development of salt-sensitive hypertension. Furthermore, low sodium intervention might be particularly effective in lowering BP among individuals with specific variants of ENaC genes. Replication of these results in other populations and further functional studies are critically important for identifying true causal variants.
Supplementary Material
Acknowledgments
Funding Sources: The Genetic Epidemiology Network of Salt Sensitivity (GenSalt) is supported by research grants (U01HL072507, R01HL087263, and R01HL090682) from the National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, Maryland.
Footnotes
Conflict of Interest Disclosures: None.
References
- 1.Elliott P, Stamler J, Nichols R, Dyer AR, Stamler R, Kesteloot H, Marmot M. Intersalt revisited: Further analyses of 24 hour sodium excretion and blood pressure within and across populations. Intersalt Cooperative Research Group. BMJ. 1996;312:1249–1253. doi: 10.1136/bmj.312.7041.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chobanian AV, Hill M. National heart, Lung, and Blood Institute Workshop on Sodium and Blood Pressure: A critical review of current scientific evidence. Hypertension. 2000;35:858–863. doi: 10.1161/01.hyp.35.4.858. [DOI] [PubMed] [Google Scholar]
- 3.He FJ, MacGregor GA. Salt, blood pressure and cardiovascular disease. Curr Opin Cardiol. 2007;22:298–305. doi: 10.1097/HCO.0b013e32814f1d8c. [DOI] [PubMed] [Google Scholar]
- 4.Weinberger MH. Salt sensitive human hypertension. Endocr Res. 1991;17:43–51. doi: 10.1080/07435809109027188. [DOI] [PubMed] [Google Scholar]
- 5.Luft FC, Weinberger MH. Heterogeneous responses to changes in dietary salt intake: the salt-sensitivity paradigm. Am J Clin Nutr. 1997;65:612S–617S. doi: 10.1093/ajcn/65.2.612S. [DOI] [PubMed] [Google Scholar]
- 6.Morimoto A, Uzu T, Fujii T, Nishimura M, Kuroda S, Nakamura S, Inenaga T, Kimura G. Sodium sensitivity and cardiovascular events in patients with essential hypertension. Lancet. 1997;350:1734–1737. doi: 10.1016/S0140-6736(97)05189-1. [DOI] [PubMed] [Google Scholar]
- 7.Weinberger MH, Fineberg NS, Fineberg SE, Weinberger M. Salt sensitivity, pulse pressure, and death in normal and hypertensive humans. Hypertension. 2001;37:429–432. doi: 10.1161/01.hyp.37.2.429. [DOI] [PubMed] [Google Scholar]
- 8.Miller JZ, Weinberger MH, Christian JC, Daugherty SA. Familial resemblance in the blood pressure response to sodium restriction. Am J Epidemiol. 1987;126:822–830. doi: 10.1093/oxfordjournals.aje.a114719. [DOI] [PubMed] [Google Scholar]
- 9.Svetkey LP, McKeown SP, Wilson AF. Heritability of salt sensitivity in black Americans. Hypertension. 1996;28:854–858. doi: 10.1161/01.hyp.28.5.854. [DOI] [PubMed] [Google Scholar]
- 10.Beeks E, Kessels AG, Kroon AA, van der Klauw MM, de Leeuw PW. Genetic predisposition to salt-sensitivity: A systematic review. J Hypertens. 2004;22:1243–1249. doi: 10.1097/01.hjh.0000125443.28861.0d. [DOI] [PubMed] [Google Scholar]
- 11.Gu D, Rice T, Wang S, Yang W, Gu C, Chen CS, Hixson JE, Jaquish CE, Yao ZJ, Liu DP, Rao DC, He J. Heritability of blood pressure responses to dietary sodium and potassium intake in a Chinese population. Hypertension. 2007;50:116–122. doi: 10.1161/HYPERTENSIONAHA.107.088310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu D, Kelly TN, Hixson JE, Chen J, Liu D, Chen JC, Rao DC, Mu J, Ma J, Jaquish CE, Rice TK, Gu C, Hamm LL, Whelton PK, He J. Genetic variants in the renin-angiotensin-aldosterone system and salt sensitivity of blood pressure. J Hypertens. 2010;28:1210–1220. [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Q, Hixson JE, Rao DC, Gu D, Jaquish CE, Rice T, Shimmin LC, Chen J, Cao J, Kelly TN, Hamm LL, He J. Genetic variants in the apelin system and blood pressure responses to dietary sodium interventions: A family-based association study. J Hypertens. 2010;28:756–763. doi: 10.1097/HJH.0b013e3283370d32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pratt JH. Central role for ENaC in development of hypertension. J Am Soc Nephrol. 2005;16:3154–3159. doi: 10.1681/ASN.2005050460. [DOI] [PubMed] [Google Scholar]
- 15.Shehata MF. Regulation of the epithelial sodium channel ]ENaC[ in kidneys of salt-sensitive Dahl rats: insights on alternative splicing. Int Arch Med. 2009;2:28. doi: 10.1186/1755-7682-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Canessa CM, Schild L, Buell G, Thorens B, Gautschi I, Horisberger JD, Rossier BC. Amiloride-sensitive epithelial Na+ channel is made of three homologous subunits. Nature. 1994;367:463–467. doi: 10.1038/367463a0. [DOI] [PubMed] [Google Scholar]
- 17.Shimkets RA, Warnock DG, Bositis CM, Nelson-Williams C, Hansson JH, Schambelan M, Gill JR, Jr, Ulick S, Milora RV, Findling JW, Canessa CM, Rossier BC, Lifton RP. Liddle's syndrome: heritable human hypertension caused by mutations in the beta subunit of the epithelial sodium channel. Cell. 1994;79:407–414. doi: 10.1016/0092-8674(94)90250-x. [DOI] [PubMed] [Google Scholar]
- 18.Hansson JH, Nelson-Williams C, Suzuki H, Schild L, Shimkets R, Lu Y, Canessa C, Iwasaki T, Rossier B, Lifton RP. Hypertension caused by a truncated epithelial sodium channel gamma subunit: genetic heterogeneity of Liddle syndrome. Nat Genet. 1995;11:76–82. doi: 10.1038/ng0995-76. [DOI] [PubMed] [Google Scholar]
- 19.Hansson JH, Schild L, Lu Y, Wilson TA, Gautschi I, Shimkets R, Nelson-Williams C, Rossier BC, Lifton RP. A de novo missense mutation of the beta subunit of the epithelial sodium channel causes hypertension and Liddle syndrome, identifying a proline-rich segment critical for regulation of channel activity. Proc Natl Acad Sci U S A. 1995;92:11495–11499. doi: 10.1073/pnas.92.25.11495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furuhashi M, Kitamura K, Adachi M, Miyoshi T, Wakida N, Ura N, Shikano Y, Shinshi Y, Sakamoto K, Hayashi M, Satoh N, Nishitani T, Tomita K, Shimamoto K. Liddle's syndrome caused by a novel mutation in the proline-rich PY motif of the epithelial sodium channel beta-subunit. J Clin Endocrinol Metab. 2005;90:340–344. doi: 10.1210/jc.2004-1027. [DOI] [PubMed] [Google Scholar]
- 21.Chang SS, Grunder S, Hanukoglu A, Rosler A, Mathew PM, Hanukoglu I, Schild L, Lu Y, Shimkets RA, Nelson-Williams C, Rossier BC, Lifton RP. Mutations in subunits of the epithelial sodium channel cause salt wasting with hyperkalaemic acidosis, pseudohypoaldosteronism type 1. Nat Genet. 1996;12:248–253. doi: 10.1038/ng0396-248. [DOI] [PubMed] [Google Scholar]
- 22.Iwai N, Baba S, Mannami T, Katsuya T, Higaki J, Ogihara T, Ogata J. Association of sodium channel gamma-subunit promoter variant with blood pressure. Hypertension. 2001;38:86–89. doi: 10.1161/01.hyp.38.1.86. [DOI] [PubMed] [Google Scholar]
- 23.Morris B, Benjafield A, Ishikawa K, Iawi N. Polymorphism (-173G>A) in promoter of human epithelial sodium channel gamma subunit gene (SCNN1G) and association analysis in essential hypertension. Hum Mutat. 2001;17:157. doi: 10.1002/1098-1004(200102)17:2<157::AID-HUMU23>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 24.Wang XF, Lu XM, Lin RY, Wang SZ, Zhang LP, Qian J, Lu DR, Wen H, Jin L. Lack of association of functional variants in alpha-ENaC gene and essential hypertension in two ethnic groups in China. Kidney Blood Press Res. 2008;31:268–273. doi: 10.1159/000151286. [DOI] [PubMed] [Google Scholar]
- 25.Xu H, Li NF, Hong J, Zhang L, Zhou L, Li T, Ou Yang WJ, Cheng QY. Relationship between four single nucleotide polymorphisms of epithelial sodium channel alpha subunit gene and essential hypertension of Kazakhs in Xinjiang. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2009;31:740–745. doi: 10.3881/j.issn.1000-503X.2009.06.018. [DOI] [PubMed] [Google Scholar]
- 26.Busst CJ, Scurrah KJ, Ellis JA, Harrap SB. Selective genotyping reveals association between the epithelial sodium channel gamma-subunit and systolic blood pressure. Hypertension. 2007;50:672–678. doi: 10.1161/HYPERTENSIONAHA.107.089128. [DOI] [PubMed] [Google Scholar]
- 27.Pradervand S, Wang Q, Burnier M, Beermann F, Horisberger JD, Hummler E, Rossier BC. A mouse model for Liddle's syndrome. J Am Soc Nephrol. 1999;10:2527–2533. doi: 10.1681/ASN.V10122527. [DOI] [PubMed] [Google Scholar]
- 28.Gensalt: rationale, design methods and baseline characteristics of study participants. GenSalt Collaborative Research Group. J Hum Hypertens. 2007;21:639–646. doi: 10.1038/sj.jhh.1002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perloff D, Grim C, Flack J, Frohlich ED, Hill M, McDonald M, Morgenstern BZ. Human blood pressure determination by sphygmomanometry. Circulation. 1993;88:2460–2470. doi: 10.1161/01.cir.88.5.2460. [DOI] [PubMed] [Google Scholar]
- 30.Mei H, Rice TK, Gu D, Hixson JE, Jaquish CE, Zhao Q, Chen JC, Cao J, Li J, Kelly TN, Rao DC, He J. Genetic correlation of blood pressure responses to dietary sodium and potassium intervention and cold pressor test in Chinese population. J Hum Hypertens. 2010 doi: 10.1038/jhh.2010.88. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 33.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Roy Statist Soc Ser B. 1995;57:298–300. [Google Scholar]
- 34.Fajac I, Viel M, Sublemontier S, Hubert D, Bienvenu T. Could a defective epithelial sodium channel lead to bronchiectasis. Respir Res. 2008;9:46. doi: 10.1186/1465-9921-9-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rossi E, Farnetti E, Debonneville A, Nicoli D, Grasselli C, Regolisti G, Negro A, Perazzoli F, Casali B, Mantero F, Staub O. Liddle's syndrome caused by a novel missense mutation (P617L) of the epithelial sodium channel beta subunit. J Hypertens. 2008;26:921–927. doi: 10.1097/HJH.0b013e3282f85dfe. [DOI] [PubMed] [Google Scholar]
- 36.Tobin MD, Tomaszewski M, Braund PS, Hajat C, Raleigh SM, Palmer TM, Caulfield M, Burton PR, Samani NJ. Common variants in genes underlying monogenic hypertension and hypotension and blood pressure in the general population. Hypertension. 2008;51:1658–1664. doi: 10.1161/HYPERTENSIONAHA.108.112664. [DOI] [PubMed] [Google Scholar]
- 37.Yuan HY, Chiou JJ, Tseng WH, Liu CH, Liu CK, Lin YJ, Wang HH, Yao A, Chen YT, Hsu CN. FASTSNP: An always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34:W635–641. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su YR, Rutkowski MP, Klanke CA, Wu X, Cui Y, Pun RY, Carter V, Reif M, Menon AG. A novel variant of the beta-subunit of the amiloride-sensitive sodium channel in African Americans. J Am Soc Nephrol. 1996;7:2543–2549. doi: 10.1681/ASN.V7122543. [DOI] [PubMed] [Google Scholar]
- 39.Baker EH, Dong YB, Sagnella GA, Rothwell M, Onipinla AK, Markandu ND, Cappuccio FP, Cook DG, Persu A, Corvol P, Jeunemaitre X, Carter ND, MacGregor GA. Association of hypertension with T594M mutation in beta subunit of epithelial sodium channels in black people resident in London. Lancet. 1998;351:1388–1392. doi: 10.1016/s0140-6736(97)07306-6. [DOI] [PubMed] [Google Scholar]
- 40.Matsubara M, Ohkubo T, Michimata M, Hozawa A, Ishikawa K, Katsuya T, Nagai K, Tsuji I, Higaki J, Araki T, Satoh H, Hisamichi S, Ito S, Ogihara T, Imai Y. Japanese individuals do not harbor the T594M mutation but do have the P592S mutation in the C-terminus of the beta-subunit of the epithelial sodium channel: The Ohasama study. J Hypertens. 2000;18:861–866. doi: 10.1097/00004872-200018070-00007. [DOI] [PubMed] [Google Scholar]
- 41.Matsubara M, Metoki H, Suzuki M, Fujiwara T, Kikuya M, Michimata M, Ohkubo T, Hozawa A, Tsuji I, Hisamichi S, Araki T, Imai Y. Genotypes of the betaENaC gene have little influence on blood pressure level in the Japanese population. Am J Hypertens. 2002;15:189–192. doi: 10.1016/s0895-7061(01)02266-x. [DOI] [PubMed] [Google Scholar]
- 42.Hannila-Handelberg T, Kontula K, Tikkanen I, Tikkanen T, Fyhrquist F, Helin K, Fodstad H, Piippo K, Miettinen HE, Virtamo J, Krusius T, Sarna S, Gautschi I, Schild L, Hiltunen TP. Common variants of the beta and gamma subunits of the epithelial sodium channel and their relation to plasma renin and aldosterone levels in essential hypertension. BMC Med Genet. 2005;6:4. doi: 10.1186/1471-2350-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gonzalez AA, Carvajal CA, Riquelme E, Krall PM, Munoz CR, Mosso LM, Kalergis AM, Fardella CE. A polymorphic GT short tandem repeat affecting beta-ENaC mRNA expression is associated with low renin essential hypertension. Am J Hypertens. 2007;20:800–806. doi: 10.1016/j.amjhyper.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 44.Jin HS, Hong KW, Lim JE, Hwang SY, Lee SH, Shin C, Park HK, Oh B. Genetic variations in the sodium balance-regulating genes enac, nedd4l, ndfip2 and usp2 influence blood pressure and hypertension. Kidney Blood Press Res. 33:15–23. doi: 10.1159/000275706. [DOI] [PubMed] [Google Scholar]
- 45.Iwai N, Baba S, Mannami T, Ogihara T, Ogata J. Association of a sodium channel alpha subunit promoter variant with blood pressure. J Am Soc Nephrol. 2002;13:80–85. doi: 10.1681/ASN.V13180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.