Abstract
Background
Dilated cardiomyopathy (DCM) and hypertrophic cardiomyopathy (HCM) are classic forms of systolic and diastolic heart failure, respectively. Mutations in genes encoding sarcomere and cytoskeletal proteins are major causes of HCM and DCM. MURC, encoding muscle-restricted coiled-coil, a Z line protein, regulates cardiac function in mice. We investigated potential causal role of MURC in human cardiomyopathies.
Methods and Results
We sequenced MURC in 1,199 individuals including 383 probands with DCM, 307 with HCM and 509 healthy controls. We found six heterozygous DCM-specific missense variants (p.N128K, p.R140W, p.L153P, p.S307T, p.P324L and p.S364L) in eight unrelated probands. Variants p.N128K and p.S307T segregated with inheritance of DCM in small families (χ2=8.5, p=0.003). Variants p.N128K, p.R140W, p.L153P and p.S364L were considered probably or possibly damaging. Variant p.P324L recurred in three independent probands, including one proband with a TPM1 mutation (p.M245T). A deletion variant (p.L232-R238del) was present in three unrelated HCM probands but it did not segregate with HCM in a family who also had a MYH7 mutation (p.L970V). The phenotype in mutation carriers was notable for progressive heart failure leading to heart transplantation in four patients, conduction defects and atrial arrhythmias. Expression of mutant MURC proteins in neonatal rat cardiac myocytes transduced with recombinant adenoviruses was associated with reduced RhoA activity, lower mRNA levels of hypertrophic markers and smaller myocyte size as compared to wild type MURC.
Conclusions
MURC mutations impart loss-of-function effects on MURC functions and are likely causal variants in human DCM. The causal role of a deletion mutation in HCM is uncertain.
Keywords: heart failure, genetics, cardiomyopathy, mutation, RhoA
Heart failure is a major cause of mortality and morbidity1. It is associated with over 270,000 deaths in the US alone1. Primary dilated cardiomyopathy (DCM) and hypertrophic cardiomyopathy (HCM) are prototypic genetic forms of systolic and diastolic heart failure, respectively. Molecular genetic studies have led to partial elucidation of causal genes and identification of several hundred mutations in families and cases with cardiomyopathies2–5. Accordingly, mutations in genes coding for sarcomere and cytoskeletal proteins have emerged as important causes of primary cardiomyopathies2–5. Genetic studies also have highlighted the genetic heterogeneity of cardiomyopathies, particularly for DCM3,4. Accordingly, the most common known gene for DCM account for ~5% of all primary HCM cases4. Collectively, the known causal genes are responsible for approximately 2/3rd of HCM families and a much smaller fraction of DCM families4,5. Thus, the causal genes for a significant number of HCM and DCM cases and families remain to be identified.
The conventional approach for identification of the causal genes for single gene disorders is genetic linkage analysis. However, the approach does not provide sufficient resolution to identify the causal mutations in small families or sporadic cases. Direct DNA sequencing of the candidate genes, particularly in view of the recent advances in DNA sequencing technologies, is an alternative approach that is being utilized increasingly to identify the causative mutations/genes. Among the candidate genes for cardiomyopathies are genes encoding the Z disc proteins6,7. We recently identified and characterized Muscle-Restricted Coiled-Coil (MURC), which encodes a Z-line component protein8,9. We showed that MURC activates RhoA/ROCK pathway, expression of atrial natriuretic peptide (ANP) and regulates myofiber organization8. MURC is also a member of the cavin complex associated with sarcolemmal caveaolae of muscle cells10. Sub-cellular distribution of MURC is altered in myopathic fibers10. Collectively, the experimental data implicate MURC as a biologically plausible gene for human cardiomyopathies10. To delineate the causal role of MURC in cardiomyopathies, we sequenced the coding regions and the splice junctions of MURC in 690 cases with cardiomyopathies and 509 controls and whenever available, we extended the genetic analysis to family members. We complemented the genetic studies with in vitro functional studies in cardiac myocytes. The findings, collectively, implicate MURC as a novel gene for DCM.
Methods
Study population
The study protocol was approved by the Institutional Review Board and was in accord with the Human Subjects' Committee guidelines. The participating individuals signed informed consents. The main study population was comprised of 383 cases with DCM, 307 cases with HCM and 277 normal individuals7. We obtained phenotypic data including a twelve-lead electrocardiograms and echocardiograms in all participants. Cardiomyopathies were diagnosed according to the conventional criteria11. Given the relatively small number of the African Americans in the main control group, we included a second group of 232 African-Americans who had normal electrocardiograms and echocardiograms12. Upon identification of a putative mutation, defined as insertion/deletion or frame-shift, non-synonymous, or splice sequence variants, we recruited and phenotyped additional family members, whenever available.
DNA sequencing
We sequenced all exons and exon-intron boundaries of MURC in 1,199 participants by Sanger method in sense and antisense directions (Primer sequence is provided in Online Table 1) using the Big Dye Terminator Reactions in an Applied Biosystems 3730×l Genetic Analyzer (Applied Biosystems, Inc. Foster City, CA). We analyzed the output using Variant Reporter software (Applied Biosystems, Inc.). In addition, two investigators analyzed every sequence printout to detect the variants. We compared the sequence with the published GenBank sequence for MURC (GRCh37 reference genome assembly, chromosome 9, region 103340336..103350180). To reduce the possibility of sequencing errors, we repeated the sequencing reactions in all samples that contained a sequence variant. Only variants that were detected in sense and anti-sense directions and confirmed in independent sequencing reactions were considered as real variants.
Mutation calling
To ascertain a causal role, we analyzed segregation of the variants with the phenotype in the families, whenever available and analyzed evolutionary conservation of the involved amino acid using PRRN (http://align.genome.jp/prrn/), charge change and hydropathy index. We used PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/index.shtml) to predict functional effects of the variants13. Non-synonymous variants that were present only in the cases with cardiomyopathies but absent in the normal controls were considered likely disease-causing mutations and selected for in vitro functional studies.
Sequencing of known common causal genes for cardiomyopathies
We sequenced the coding regions and exon-intron boundaries of LMNA, encoding Lamin A/C, a known gene for DCM in 103 proband with a compound phenotype of DCM and conduction defect and/or atrial fibrillation4. Likewise, we had previously sequenced all exons and exon-intron boundaries of MYH7, MYBPC3, ACTC1, TNNT2, TNNI3, TPM1 and MYOZ2 in 81 probands with familial HCM. Finally, to detect compound mutations in those who carried a mutation in MURC, we sequenced MYH7, MYBPC3, TNNT2, TNNI3, TPM1, ACTC1 and LMNA, known relatively common genes for HCM and DCM in all MURC mutation carriers5, 14, 15.
Genotyping for short tandem repeat (STR) markers
To determine whether the recurring mutations occurred de novo or shared a common ancestral allele, we genotyped the individuals for three STR markers (D9S180, D9S910 and D9S176) that spanned an approximately 2.5 Mbp genomic region on 9q31.1 locus. Genotyping was performed by PCR using fluorescent-labeled primers and capillary electrophoresis on an ABI 3730×l Genetic Analyzer and analyzed using the GeneMapper v4.1 (Applied Biosystems).
Plasmid constructs
We cloned human FLAG-tagged MURC cDNA into a pcDNA3.1 vector (pcDNA3.1-hMURC). We introduced the p.N128K, p.R140W, p.L153P, p.S307T, p.P324L and p.S364L mutations by site directed mutagenesis8. Sequences of mutant-specific oligonucleotide primers are shown in Online Table 1.
Isolation and culture of neonatal rat cardiac myocytes (NRCM)
We isolated and prepared NRCM from 1-day-old Sprague Dawley rats as described8. Briefly, we digested ventricular tissues enzymatically and separated cardiac myocytes over a Percoll gradient. We changed the culture medium to serum-free medium after 24 hours and maintained the cells under serum-free conditions before experiments.
Replication-deficient recombinant adenoviruses and gene transfer
We generated recombinant adenoviruses expressing FLAG-tagged human wild type (Ad-hMURC-WT), each of the mutant MURC protein or β-galactosidase (Ad-LacZ), as described previously8. The Ad-LacZ and Ad-hMURC-WT were served as controls. We infected the NRCM with the recombinant adenoviruses at a multiplicity of infection (MOI) of 10. After incubation at 37°C for 1 hour, we removed the viral suspension and cultured the cells with serum-depleted culture media for 48 hours.
Immunoblotting
To detect expression of the WT or mutant MURC, we electrophoresed the cell lysates in 10% SDS-polyacrylamide gels and transferred the proteins to polyvinylidene difluoride membranes (Millipore, Billerica, MA). We incubated the membranes with primary antibodies against FLAG (SIGMA, St Louis, MO) and Gapdh to detect FLAG-tagged MURC and Gapdh proteins, respectively. The secondary antibody was Horseradish Peroxidase (HRP)-conjugated anti-mouse IgG (GE Healthcare, Waukesha, WI).
RhoA activation assay
We determined RhoA activity in the protein extracts from NRCM transduced with the recombinant adenoviruses using an absorbance-based G-LISA RhoA activation assay biochemistry kit (Cytoskeleton, Denver, CO), as described8. We isolated total protein two days after transduction of NRCM with the recombinant adenoviruses per instruction of the manufacturer. We equalized the total protein concentrations among the samples at 1 mg/ml for the assay and measured the signals at an absorbance of 490 nm using a microplate spectrometer.
RNA extraction and quantitative reverse transcriptase-PCR (qPCR)
We isolated total cellular RNA from NRCM using an RNeasy Mini Kit (QIAGEN, Inc.). We performed qPCR to quantify mRNA expression levels of rat ANP (Nppa), B-type natriuretic peptide (BNP or Nppb) and rat skeletal α-actin (SkA or Acta1) in NRCM transduced with WT or mutant MURC constructs, as described8. Sequences of oligonucleotide primers are shown in Online Table 1.
Cardiac myocyte size
Cardiac myocytes were infected with the recombinant adenoviruses and cultured under serum-free conditions for 48 hr. Cells were then fixed with 4% paraformaldehyde and stained with fluorescein isothiocyanate-conjugated phalloidin (SIGMA) for the detection of actin filaments. DNA was stained with 4',6-diamidino-2-phenylindole (DAPI). We measured myocyte surface area using Image software (National Institutes of Health, Bethesda, MD) in at least 100 cells per each group.
Statistics
Statistical analyses were similar to those published recently7, 16. We expressed the continuous variables that followed normality distribution as mean ± SD. We compared differences among the continuous variables that satisfied the normality distribution by one-way analysis of variance and applied the Bonferroni correction to multiple-comparison tests. Variables that were not normally distributed were compared by Kruskal-Wallis test. We analyzed differences in the categorical variables by Chi Square test.
All in vitro experiments were performed at least 3 times and the mean values of the three measurements were used. Data is expressed as mean ± standard error and analyzed by one-way ANOVA with Sheffe's post hoc analysis. A value of P<0.05 was considered significant. All statistical analyses were performed using STATA v. 10.1.
Results
Characteristics of the study population
Phenotypic characteristics of the study population are shown in Table 1 and are partly published7, 12.
Table 1.
Baseline Characteristics
| Control (*) | DCM | HCM | P | |
|---|---|---|---|---|
| N=967 (1199*) | 277 (509*) | 383 | 307 | NA |
| Demographics | ||||
| Male/Female | 129(46) / 148 (54) | 214(56) / 169(44) | 167(54) / 140(46) | 0.046 |
| Caucasian / African-American | 193(70) / 84 (30) | 204(53) / 179(47) | 258(84) / 49(16) | <0.001 |
| Age (Years) | 44.24±17.50 | 51.43±13.92 | 52.40±16.23 | <0.0001 |
| BMI (Kg/m2) | 28.04±7.22 | 30.15±7.48 | 30.19±7.21 | 0.0003 |
| BSA (m2) | 1.96±0.32 | 2.07±0.33 | 2.05±0.29 | <0.0001 |
| DM (%) | 26 (10) | 90 (23) | 35 (12) | <0.001 |
| Smoker (%) | 26 (10) | 57 (15) | 49 (17) | <0.001 |
| Heart Rate (bpm) | 71.3±12.4 | 80.1±16.2 | 70.4±12.0 | <0.0001 |
| Systolic BP (mmHg) | 123.5±15.7 | 116.8±16.8 | 128.5±17.5 | <0.0001 |
| Diastolic BP (mmHg) | 73.3±9.9 | 73.4±12.4 | 74.3±13.4 | 0.5315 |
| Echocardiographic data | ||||
| ST (cm) | 0.96±0.18 | 1.01±0.22 | 1.89±0.48 | <0.0001 |
| PWT (cm) | 0.97±0.2 | 1.08±0.23 | 1.34±0.32 | <0.0001 |
| LVEDD (cm) | 4.51±0.53 | 6.28±0.97 | 4.15±0.67 | <0.0001 |
| LVESD (cm) | 2.90±0.53 | 5.39±1.12 | 2.45±0.70 | <0.0001 |
| LVM (g) | 184.26±65.97 | 350.83±142.24 | 355.89±128.24 | <0.0001 |
| LVMI (g/m2) | 94.02±25.12 | 169.50±63.22 | 173.30±63.22 | <0.0001 |
| LA size (cm) | 3.46±0.76 | 4,43±1.00 | 4.25±0.79 | <0.0001 |
| LVEF | 61.47±5.37 | 24.42±8.04 | 67.03±5.15 | <0.0001 |
Denotes the total number of the control group with data that are included in the table. We added a second control group of African-American individuals (N=232) who had normal electrocardiograms and echocardiograms but did not have the detailed data.
Abbreviations: HCM: Hypertrophic cardiomyopathy; DCM: Dilated cardiomyopathy; NA: not applicable; BMI; Body mass index; BSA: Body surface area; DM: Diabetes mellitus; BP: Blood pressure; ST: Septal thickness; PWT: Posterior wall thickness; LVEDD: Left ventricular end diastolic diameter; LVESD: Left ventricular systolic diameter; LV Mass; Left ventricular mass; LVMI: Left ventricular mass indexed to body surface area; LVEF: Left ventricular ejection fraction; LA: left atrium.
MURC sequence variants
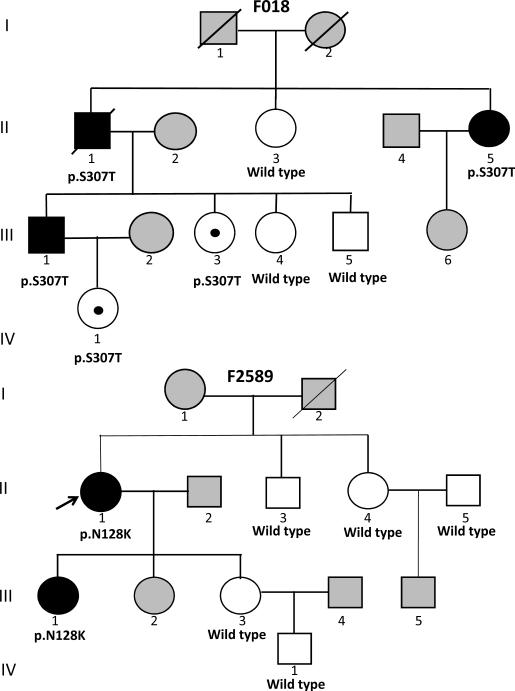
We found six DCM-specific heterozygous missense variants (p.N128K, p.R140W, p.L153P, p.S307T, p.P324L and p.S364L) in 8 probands (Table 2). These variants were absent in 509 normal individuals. The p.N128K variant co-segregated with DCM in 2 affected family members and was absent in 5 unaffected family members (Figure 1). The p.S307T was present in 4 adult family members of whom 3 had severe DCM (Figure 1). One young adult 31 years old and one child also had the p.S307T variant, while three phenotypically normal adults had the wild type genotype. Segregation analysis that includes healthy individuals in cardiomyopathies is subject to age-dependent and incomplete penetrance. The absence of a clinical phenotype in young mutation carriers in this family may reflect partial and age-dependent penetrance of the variant. Collectively, the p.N128K and p.S307T variants were present in 5 affected family members and absent in 8 out of 9 clinically normal members (X2=8.5, p=0.003). The small size of these two families and the incomplete penetrance of the p.S307T variant impeded the power to calculate the LOD score, as a measure of linkage to strengthen the causal role of these variants. The p.P324L was a recurring variant, as it was detected in 3 probands. To determine whether the probands shared a common ancestral allele at the MURC locus, we genotyped the three families for 3 locus-specific STR markers. The three probands did not share the genotypes for the MURC locus, indicating independent origins of the mutation in these individuals (Online Figure 1).
Table 2.
Non-Synonymous Variants Detected in Probands With Dilated Cardiomyopathy
| Amino acid | Nucleotide | Controls | DCM | HCM | |||
|---|---|---|---|---|---|---|---|
| N=509 | N=383 | N=307 | |||||
| A | C | A | C | A | C | ||
| 316 | 193 | 179 | 204 | 49 | 258 | ||
| p.N128K | g.474C>G | 0 | 0 | 1 | 0 | 0 | 0 |
| p.R140W | g.7721C>T | 0 | 0 | 0 | 1 | 0 | 0 |
| p.L153P | g.7761T>C | 0 | 0 | 1 | 0 | 0 | 0 |
| p.S307T | g.8223G>C | 0 | 0 | 0 | 1 | 0 | 0 |
| p.P324L | g.8274C>T | 0 | 0 | 3 | 0 | 0 | 0 |
| p.S364L | g.8394C>T | 0 | 0 | 1 | 0 | 0 | 0 |
| Total number of non-synonymous variants per group and ethnic background | 0 | 0 | 6 | 2 | 0 | 0 | |
| Total number of non-synonymous variants per group | 0 (0 %) | 8 (2.1%) | 0 (0%) | ||||
Abbreviations: As in Table 1., A: African Americans; C: Caucasians
Figure 1.
Pedigree of families with MURC mutations: Squares and circles represent male and female members. Full and open circles and squares indicated affected and unaffected (normal) individuals, respectively. Gray squares and circles indicate individuals that were not studied. The mutation and wild type codons are listed under those members that were studied. Individual III-3, who is 31 years old is a mutation carrier but is phenotypically normal (non-penetrance). Individual IV-1 in Pedigree F018 is only 5 years old and non-contributory to co-segregation analysis.
We also detected a deletion variant (p.L232-R238del), which excluded seven-amino acid long motif (ERLRQSG, Swiss-Prot ID: Q5BKX8.2) in three probands with HCM, one African American and two Caucasians. The deletion was absent in over 500 control individuals. Two affected members of a HCM family had the deletion variant. However, it was absent in an affected member of another family with the deletion variant (Online Figure 2). The two affected family members in the latter family had a p.L907V mutation in MHY7, a known gene for cardiomyopathies5. The p.L907V mutation was absent in over 500 normal individuals and affects a highly conserved domain in MYH7 protein. It is predicted to be probably damaging by PolyPhen-2 analysis (score 0.954, sensitivity: 0.74, specificity: 0.93).
In contrast to 7 phenotype-specific variants, we detected only one non-synonymous variant (p.R272H) that was present only in the control group (Table 2). The p.R272H was predicted to be a benign variant by PolyPhen-2 analysis. To test whether MURC was a disease-causing gene, we performed a gene-centric analysis by coalescing the non-synonymous variants in each group and comparing the prevalence of MURC variants in cases with DCM and controls. In support of a causal role of MURC variants in DCM, non-synonymous variants were over-represented by 25-fold in the cases (Pearson χ2 = 7.84, p=0.005).
As would be expected we identified several polymorphisms that were common to the cases and the controls including six non-synonymous variants, 14 synonymous and 8 non-coding variants (Online Tables 2 and 3). The frequencies of the non-synonymous or synonymous polymorphisms were not significantly different between the cases and controls. Likewise, the age distributions of the control individuals and the DCM cases with the non-synonymous variants were similar. Overall, the minor allele frequencies (MAFs) of the non-synonymous polymorphisms were <1% in the study population. The most common variant was the p.S78L, which had a MAF of 0.009 in the entire population and 0.033 in the African American sub-population. The p.N81K was detected in three African Americans, one with HCM, one with DCM and one normal individual. In each case, it co-segregated with the p.S78L variant.
Mutations in known genes for cardiomyopathies
We sequenced all coding regions and exon-intron boundaries of MYH7, MYBPC3, TNNT2, TNNI3, TPM1, ACTC1 and LMNA, known relatively common genes for cardiomyopathies4, 5, in all MURC mutation carriers. An African American patient with DCM had a compound mutation; the p.P324L variant in MURC and a novel non-synonymous variant p.M245T in the TPM1 gene. Likewise, one family member who had the p.L232-R238del variant in MURC also had a p.L907V mutation in the MYH7 gene (likely compound mutation).
We identified three non-synonymous variants in LMNA upon screening of 103 probands with DCM associated with conduction defect and/or atrial fibrillation4. None had a MURC mutation. Likewise, we identified thirty-two different mutations in MYH7 (14 mutations), MYBPC3 (11 mutations), TNNT2 (2 mutations), TNNI3 (1 mutation), TPM1 (2 mutations) and MYOZ2 (2 mutations) in screening of 81 probands with HCM. None had a mutation in MURC gene.
Phenotypic expression of MURC mutation carriers
The phenotype in the mutation carriers was notable for heart failure in the middle age (mean age 45.0 ± 6.5), leading to severely depressed left ventricular systolic function, which led to cardiac transplantation in 4 affected members. Cardiac tissues were not available to analyze histological correlates of MURC variants. Several affected members exhibited supra-ventricular arrhythmias and progressive conduction defects, requiring implantation of a permanent pacemaker and internal cardioverter/defibrillator. Detailed phenotypic data are shown in Table 3.
Table 3.
Demographic, Echocardiographic and Electrocardiographic Phenotypes in DCM Patients with MURC Mutations
| AA change | Ethnicity | Age at diagnosis | Sex | BMI (Kg/m2) | Clinical Phenotype | ST (cm) | PWT (cm) | LVEDD (cm) | LVEF % | ECG | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1* | N128K | A | 54 | F | 31.0 | Heart failure, recurrent non-sustained V. Tach, ICD implantation, Heart transplant | 0.9 | 0.8 | 6.8 | <10 | SR, 1st degree AV block, LAE, LVH with ST/T changes |
| 2* | A | 41 | F | 26.8 | Heart failure, Hypotension, non-sustained VT, SVT | 0.8 | 0.9 | 6.3 | 28 | SR, LVH with ST/T changes | |
| 3 | R140W | C | 41 | M | 25.9 | SCA, ICD implantation, Recurrent V Fib, Heart transplantation | 1.2 | 1 | 6.6 | 26.5 | A Fib, Atrial paced rhythm, LAE, LBBB, ST/T changes |
| 4 | L153P | A | 48 | M | 23.0 | Heart failure, NYHA functional class, cardiac thrombus | 0.5 | 0.5 | 9.1 | <10 | SR, 1st degree AV block, LAE, IVCD, V. Tach. |
| 5* | S307T | C | 48 | M | 34 | Heart failure, Syncope, PPI, Heart transplantation | 0.8 | 0.9 | 6.9 | <10 | Ventricular paced rhythm |
| 6* | C | 36 | M | 47.3 | Heart Failure, Palpitations, Class II NYHA functional class on therapy | 1.2 | 1.3 | 6.7 | 32 | SR, IVCD | |
| 7* | C | 33 | F | 24.8 | Heart failure, syncope and V. Tach, ICD implantation, Heart transplantation | 1.3 | 1.3 | 7.4 | <20 | SR, LAD, LBBB | |
| 8 | P324L (TPM1: M245T) | A | 52 | F | 25.9 | Heart failure, Hypotension and low cardiac output requiring inotropic support and LV assist device. Embolic stroke, PPI and ICD | 1.3 (0.9) | 1.4 (1) | 5 (5.5) | 22 | Dual-chamber paced rhythm, intermittent atypical AF, LVH with ST/T changes |
| 9 | P324L | A | 49 | F | 47.2 | Heart failure, Class II NYHA functional class on medical therapy | 0.8 | 1 | 5.4 | 32 | SR, LVH voltage |
| 10 | P324L | A | 46 | M | 27.4 | Heart failure and palpitations, Class IV NYHA functional class, evaluation for heart transplantation. | 0.8 | 1 | 5.4 | 32 | SB, LVH with ST/T changes |
| 11 | S364L | A | 47 | M | 49.7 | Heart failure, palpitations, Bi-ventricular pacing (cardiac resynchronization therapy), Ventricular support system, ICD | 0.8 | 1 | 6.4 | 22 | SR, LVH with ST/T, V. Tach. |
Individuals 1 and 2 as well as individuals 5, 6 and 7 are family members. Echocardiographic data are findings on presentation. Data in parenthesis indicate findings on a follow up echocardiogram. Individual # 8 had increased left ventricular wall thickness with decreased LVEF at the time of first evaluation. She subsequently developed a DCM phenotype.
Abbreviations: As in Tables 1 and 2. AA: amino acid; SCA: Sudden cardiac arrest; ECG: Electrocardiogram; M: Male; F: female; SR: Sinus rhythm; LAE: Left atrial enlargement; LVH: Left ventricular hypertrophy; IVCD; Intra-ventricular conduction delay; A Fib; Atrial fibrillation; LBBB: Left bundle branch block; V. Tach: Ventricular tachycardia; SB: Sinus bradycardia; AF: atrial flutter. NYHA: New York Heart Association, ICD: Internal cardioverter/defibrillator, SVT: supraventricular tachycardia
Predicted pathogenicity of the variants
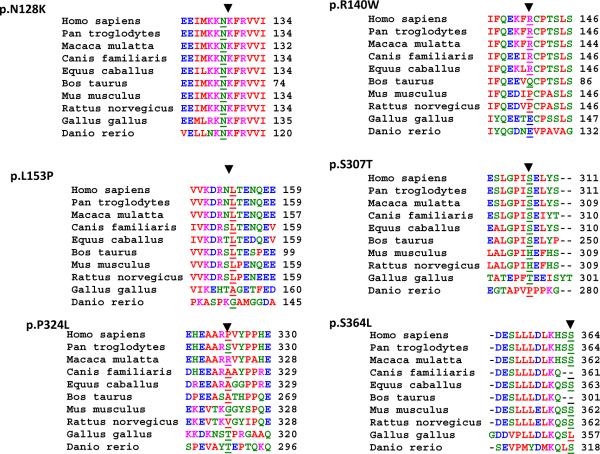
Conservation of the involved codon and domain across paralogous proteins from zebrafish to humans, a potential indicator of the biological significance of the codon, is shown in Figure 2. Likewise, the predicted effects of the non-synonymous variants on MURC protein are shown in Table 4. Codon N128K affected a totally conserved amino acid from zebra fish to man and had a very high PolyPhen-2 prediction score (0.980, sensitivity: 0.69; specificity: 0.94), placing it in the probably damaging category (highest category)13. The L153P also affected a highly conserved amino acid from rat (rattus norvegicus) to man and was categorized as probably damaging (score: 0.975, sensitivity: 0.70, specificity: 0.94). The p.R140W and p.S364L involved conserved amino acids and were predicted to be possibly damaging (score for p.R140W: 0.840, sensitivity: 0.80; specificity: 0.90 and score for p.S364L: 0.438, sensitivity: 0.438; specificity: 0.86). In contrast, p.S307T and p.P324L involved less or non-conserved amino acids and were predicted to be benign.
Figure 2.
Multiple sequence alignment (MSA) of MURC variants across species: MSA is performed using software package PRRN ((http://align.genome.jp/prrn/) to infer evolutionary conservation of the amino acids affected by the MURC non-synonymous variants identified in probands with DCM. All available sequences from different species are included.
Table 4.
Evolutionary conservation and Biophysical Effects of the Putative Causal Variants
| Amino Acid Change | Evolutionary conservation | Charge change | Change in hydropathy | PolyPhen-2 prediction |
|---|---|---|---|---|
| p.N128K | Conserved in all known proteins | Neutral to positive | −3.5 to −3.9 | Probably damaging |
| p.R140W | Partially conserved | Positive to Neutral | −4.5 to −0.9 | Possibly damaging |
| p.L153P | Highly conserved | Neutral | 3.8 to −1.6 | Probably damaging |
| p.S307T | Partially conserved | Neutral | −0.8 to −0.7 | Benign |
| p.P324L | Non-conserved | Neutral | −1.6 to 3.8 | Benign |
| p.S364L | Highly conserved | Neutral | −0.8 to 3.8 | Possibly damaging |
Functional characterization of MURC mutations
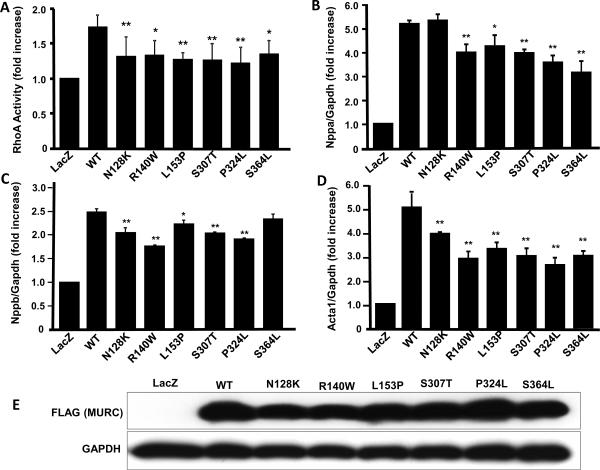
We have previously shown that MURC activates RhoA/ROCK pathway and its downstream target ANP8. Therefore, we transduced NRCM with recombinant viruses expressing either a WT or a mutant MURC protein and measured RhoA activity. Expression of the WT MURC increased RhoA activity by ~ 80%, a finding that is in accord with the previous data8. In contrast, RhoA activity was significantly less in myocytes expressing the mutant MURC proteins as compared to WT MURC group (Figure 3).
Figure 3.
Effects of MURC mutations on RhoA activation and expression of molecular markers of cardiac hypertrophy: Panel A. shows a bar graph representing RhoA activity in neonatal rat cardiac myocytes (NRCM) infected with recombinant adenoviruses at a multiplicity of infection (MOI) of 10. The groups included Ad-LacZ (expressing β-galactosidase, as a control), wild type human MURC (Ad-hMURC-WT) and each of the specific mutant MURCs (Ad-N128K, Ad-R140W, Ad-L153P, Ad-S307T, Ad-P324L, or Ad-S364L) that were identified in patients with DCM. (N=3 independent experiments per group. Panels B, C and D represent mRNA expression levels of atrial natriuretic peptide (ANP, gene Nppa), B-type or Brain natriuretic peptide (BNP, gene name Nppb) and skeletal α-actin (SkA, gene name Acta1) in cardiac myocytes transduced at an MOI of 10 with control recombinant adenoviruses or viruses expressing WT or each of the six mutant MURC proteins. Cells were harvested 48 hr after the infection for extraction of RNA and qPCR. Panel E. shows an immunoblot of myocytes proteins extracted 48 h after transduction with the recombinant viruses and probed with an anti-FLAG antibody to detect expression of the WT or mutant MURC proteins (upper blot). Immunoblotting was performed to assess equal expression level of the WT and mutant MURC proteins among the groups. The lower blot shows expression of Gapdh, which was used as a control for loading conditions.
* indicates p<0.05 and ** indicates p<0.01 as compared to WT MURC.
Likewise, expression of the WT MURC significantly increased mRNA levels of ANP (Nppa) and SkA (Acat1) by more than 5-fold and that of BNP (Nppb) by ~ 2.6-fold (Figure 3). In contrast, mRNA levels of the Nppa, Nppb and Acta1 in NRCM transduced with the mutant MURC recombinant viruses were attenuated significantly, as compared to WT MURC. The findings indicate that the mutations impart a loss-of-function effect on the activation of RhoA and expression of the hypertrophic markers by MURC.
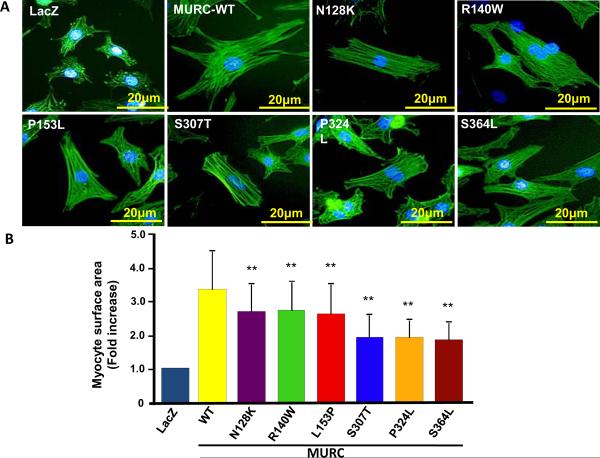
To further characterize the phenotypic consequence of the MURC mutations in vitro and complement the data on RhoA activity and expression of the molecular markers of hypertrophy, we measured the effects of expression of WT and mutant MURC proteins on cardiac myocyte size. Expression of the WT MURC increased cardiac myocyte surface area by about 3.5 fold. However, the mutant MURC had attenuated hypertrophic effects, as myocytes surface area was significantly less in the mutant groups as compared to the WT MURC group (Figure 4).
Figure 4.
Induction of cardiac myocyte hypertrophy: Panel A shows representative immunofluorescence images of NRCM transduced with a control virus or recombinant viruses, at an MOI of 10, expressing WT mutant MURC (Ad-LacZ, Ad-hMURC-WT, Ad-N128K, Ad-R140W, Ad-L153P, Ad-S307T, Ad-P324L, or Ad-S364L). The yellow bar indicates 20 μm scale. Panel B shows a bar graph depicting the mean and standard error of myocyte surface area in each experimental group. NRCM were infected with the recombinant viruses and stained with fluorescein isothiocyanate-conjugated phalloidin 48 hours after the transduction. Myocyte surface area was measured in at least 100 cells per each group.
* indicates p<0.05 and ** indicates p<0.01 as compared to WT MURC).
Discussion
We have identified six DCM-specific heterozygous missense variants including four that were predicted to be probably or possibly pathogenic. The mutations were associated with progressive heart failure, conduction defects and cardiac arrhythmias. In vitro functional studies indicate loss-of-function effects of the mutant MURC proteins on activation of RhoA, expression of hypertrophic markers and myocyte hypertrophy. Collectively, human molecular genetic and in vitro functional data implicate MURC as a causal gene for human DCM.
Several lines of evidence support the causal role of MURC mutations in DCM. All six non-synonymous variants were absent in over 500 control individuals who had no clinical, electrocardiographic or echocardiographic evidence of cardiomyopathies. The variants were also absent in the dbSNP (build 132). A gene-centric analysis showed that the missense variants were over-represented in patients with DCM by 25 folds. Likewise, two mutations co-segregated with the phenotype, albeit in small families. While the de novo recurring mutations in cases only imply a causal role in DCM, the p.P324L recurring variant affected a non-conserved amino acid and was predicted to be benign by PolyPhen-2 analysis. Functional data, assessed at multiple levels, showed concordant results and indicated loss-of-function effects of the MURC mutations. Furthermore, four variants either involved largely conserved paralogous codons and/or were predicted to affect structure and biological function of the encoded protein. The findings are also in accord with the in vitro and in vivo biological functions of MURC protein and its potential involvement in myopathic conditions8–10.
MURC is a biologically plausible candidate gene for cardiomyopathies. MURC is a Z-line component protein that is predominantly expressed in the heart8–10. Mutations in genes encoding sarcomere and cytoskeletal proteins are major causes of cardiomyopathies4, 5. MURC is involved in muscle protein homeostasis through regulating RhoA/ROCK and association with a multi-protein complex at the caveolae8–10. Murc interacts with extracellular signal-related kinases, RhoA/ROCK and serum response factor pathways, which are also implicated in heart failure8, 9, 17. RhoA appear to play a delicate role in regulating cardiac function, as its over-expression or its inhibition results in conduction defects and cardiac dysfunction18, 19. Likewise, cardiac-restricted over-expression of Murc in mice leads to heart failure, conduction defect and atrial arrhythmias8. Moreover, MURC is a component of a multi-protein complex in the caveolae that regulates cardiac function10. Its sub-cellular distribution is perturbed in human muscle disease associated with Caveolin-3 dysfunction10. Notably mutations in CAV3, encoding caveolin 3, are also known to cause cardiac and skeletal myopathy20, 21.
The causal role of the MURC deletion mutation in HCM is less clear. Evidence in support of a potential causal role include recurrence in two different ethnic populations and hence, occurring independently, its absence in > 500 normal individuals and the increasing recognition of prevalence of compound mutations in cardiomyopathies22–25. However, the deletion variant was not present in an affected family member with the phenotype and hence, conventionally can not be considered a causal variant in this family. The two affected members in this small family also had a missense mutation (p.L907V) in MYH7, which is predicted to be probably damaging. It is possible that the deletion variant co-dependently contributes to phenotypic expression of HCM in this family. These findings highlight the challenge of establishing the causal role of the DNA sequence variants in sporadic cases and small families, as opposed to large families, wherein co-segregation of the phenotype with inheritance of the variants (genetic linkage) could support the causality. While the actual LOD score is determined by the family structure and information content of the locus markers, typically more than seven affected family members are necessary to achieve a significant LOD score in a linkage analysis of a familial disease with an autosomal dominant mode of inheritance, assuming 100% penetrance. Unfortunately, neither the number of the affected individuals in our families is adequate nor the penetrance is complete to perform a meaningful linkage analysis. The challenge is expected to have considerable clinical implications, as whole-exome or whole-genome sequencing gains clinical applications. The task is daunting, as each diploid human genome contains over 10,000 non-synonymous variants, the majority of which are not associated with any discernible clinically phenotype (discussed in 26). Accordingly, several lines of genetic and functional data have to be incorporated in discerning clinical significance of the DNA sequence variants in sporadic cases or small families. Although our data strongly support the causal role of the MURC variants in DCM, one has to consider the possibility that the MURC variants may not be true disease-causing variants but are susceptibility alleles that require a second mutation or injury to cause the clinical phenotype. There is also a possibility that these variants are functional variants that do not play significant roles in susceptibility to cardiomyopathies. We submit that the strengths of the genetic and functional data favor the causal role of MURC variants in the pathogenesis of human cardiomyopathies.
Supplementary Material
Heart failure is a major cause of mortality and morbidity in the world. Genetic mutations are important causes of susceptibility to heart failure. Familial dilated cardiomyopathy (DCM) is a classic form of systolic heart failure caused by genetic mutations. The known causal genes for DCM typically code for sarcomere and cytoskeletal proteins. Prompted by the experimental data, which implicate Muscle-Restricted Coiled-Coil (MURC) in regulating cardiac function, we investigated the causal role of MURC gene in DCM. We sequenced the protein coding regions of MURC in 383 probands with DCM, 307 with hypertrophic cardiomyopathy (HCM) and 509 healthy controls. We found six missense mutations that were present only in DCM patients and were absent in controls. Two of these mutations segregated with inheritance of DCM in small families. One mutation recurred in three independent probands, including one proband who also had another mutation in the α-tropomyosin gene. We also found one deletion mutation in three unrelated HCM probands that might be a causative or susceptibility variant. The clinical features associated with MURC mutations included progressive heart failure that often led to heart transplantation, conduction defects and atrial arrhythmias. Studies in cultured cardiac myocytes showed that the mutations reduced RhoA activity, an important signal transducer in the heart and impaired myocyte hypertrophic response. We conclude that MURC is a likely causal gene for DCM. We suggest elucidation of the molecular genetic and pathogenesis of heart failure is an essential step in the ultimate treatment of this potentially deadly disease.
Acknowledgements
We thank T. Taniguchi, D. Naito, N. Nakanishi, and K. Miyagawa for their technical assistance.
Funding Sources: NHLBI (R01-088498); NIA (R21 AG038597-01), Burroughs Wellcome Award in Translational Research (#1005907) and TexGen Fund from Greater Houston Community Foundation. Additional support was provided by the Ministry of Education, Culture, Sports, Science, and Technology of Japan (T.U., T.O., and H.M.), Takeda Science Foundation (T.O.), Mitsubishi Pharma Research Foundation (T.U.), and Japan Heart Foundation/Novartis Grant for Research Award on Molecular and Cellular Cardiology (T.O.).
Footnotes
Conflict of Interest Disclosure: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roger VL, Go AS, Lloyd-Jones DM, Adams RJ, Berry JD, Brown TM, Carnethon MR, Dai S, de Simone G, Ford ES, Fox CS, Fullerton HJ, Gillespie C, Greenlund KJ, Hailpern SM, Heit JA, Ho PM, Howard VJ, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Makuc DM, Marcus GM, Marelli A, Matchar DB, McDermott MM, Meigs JB, Moy CS, Mozaffarian D, Mussolino ME, Nichol G, Paynter NP, Rosamond WD, Sorlie PD, Stafford RS, Turan TN, Turner MB, Wong ND, Wylie-Rosett J. Heart disease and stroke statistics--2011 update: A report from the american heart association. Circulation. 2011;123:459–463. doi: 10.1161/CIR.0b013e3182009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L, Seidman JG, Seidman CE. Harnessing molecular genetics for the diagnosis and management of hypertrophic cardiomyopathy. Ann Intern Med. 2010;152:513–520. W181. doi: 10.1059/0003-4819-152-8-201004200-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fatkin D, Graham RM. Molecular mechanisms of inherited cardiomyopathies. Physiol Rev. 2002;82:945–980. doi: 10.1152/physrev.00012.2002. [DOI] [PubMed] [Google Scholar]
- 4.Dellefave L, McNally EM. The genetics of dilated cardiomyopathy. Curr Opin Cardiol. 2010;25:198–204. doi: 10.1097/HCO.0b013e328337ba52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marian AJ. Hypertrophic cardiomyopathy: From genetics to treatment. Eur J Clin Invest. 2010;40:360–369. doi: 10.1111/j.1365-2362.2010.02268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knoll R, Hoshijima M, Hoffman HM, Person V, Lorenzen-Schmidt I, Bang ML, Hayashi T, Shiga N, Yasukawa H, Schaper W, McKenna W, Yokoyama M, Schork NJ, Omens JH, McCulloch AD, Kimura A, Gregorio CC, Poller W, Schaper J, Schultheiss HP, Chien KR. The cardiac mechanical stretch sensor machinery involves a z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002;111:943–955. doi: 10.1016/s0092-8674(02)01226-6. [DOI] [PubMed] [Google Scholar]
- 7.Osio A, Tan L, Chen SN, Lombardi R, Nagueh SF, Shete S, Roberts R, Willerson JT, Marian AJ. Myozenin 2 is a novel gene for human hypertrophic cardiomyopathy. Circ Res. 2007;100:766–768. doi: 10.1161/01.RES.0000263008.66799.aa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogata T, Ueyama T, Isodono K, Tagawa M, Takehara N, Kawashima T, Harada K, Takahashi T, Shioi T, Matsubara H, Oh H. Murc, a muscle-restricted coiled-coil protein that modulates the rho/rock pathway, induces cardiac dysfunction and conduction disturbance. Mol Cell Biol. 2008;28:3424–3436. doi: 10.1128/MCB.02186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tagawa M, Ueyama T, Ogata T, Takehara N, Nakajima N, Isodono K, Asada S, Takahashi T, Matsubara H, Oh H. Murc, a muscle-restricted coiled-coil protein, is involved in the regulation of skeletal myogenesis. Am J Physiol Cell Physiol. 2008;295:C490–498. doi: 10.1152/ajpcell.00188.2008. [DOI] [PubMed] [Google Scholar]
- 10.Bastiani M, Liu L, Hill MM, Jedrychowski MP, Nixon SJ, Lo HP, Abankwa D, Luetterforst R, Fernandez-Rojo M, Breen MR, Gygi SP, Vinten J, Walser PJ, North KN, Hancock JF, Pilch PF, Parton RG. Murc/cavin-4 and cavin family members form tissue-specific caveolar complexes. J Cell Biol. 2009;185:1259–1273. doi: 10.1083/jcb.200903053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maron BJ, Towbin JA, Thiene G, Antzelevitch C, Corrado D, Arnett D, Moss AJ, Seidman CE, Young JB. Contemporary definitions and classification of the cardiomyopathies: An american heart association scientific statement from the council on clinical cardiology, heart failure and transplantation committee; quality of care and outcomes research and functional genomics and translational biology interdisciplinary working groups; and council on epidemiology and prevention. Circulation. 2006;113:1807–1816. doi: 10.1161/CIRCULATIONAHA.106.174287. [DOI] [PubMed] [Google Scholar]
- 12.Cresci S, Kelly RJ, Cappola TP, Diwan A, Dries D, Kardia SL, Dorn GW., 2nd Clinical and genetic modifiers of long-term survival in heart failure. J Am Coll Cardiol. 2009;54:432–444. doi: 10.1016/j.jacc.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sunyaev S, Ramensky V, Koch I, Lathe W, 3rd, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 14.Alcalai R, Seidman JG, Seidman CE. Genetic basis of hypertrophic cardiomyopathy: From bench to the clinics. J Cardiovasc Electrophysiol. 2008;19:104–110. doi: 10.1111/j.1540-8167.2007.00965.x. [DOI] [PubMed] [Google Scholar]
- 15.Ashrafian H, Watkins H. Reviews of translational medicine and genomics in cardiovascular disease: New disease taxonomy and therapeutic implications cardiomyopathies: Therapeutics based on molecular phenotype. J Am Coll Cardiol. 2007;49:1251–1264. doi: 10.1016/j.jacc.2006.10.073. [DOI] [PubMed] [Google Scholar]
- 16.Lombardi R, Dong J, Rodriguez G, Bell A, Leung TK, Schwartz RJ, Willerson JT, Brugada R, Marian AJ. Genetic fate mapping identifies second heart field progenitor cells as a source of adipocytes in arrhythmogenic right ventricular cardiomyopathy. Circ Res. 2009;104:1076–1084. doi: 10.1161/CIRCRESAHA.109.196899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang J, Wei L, Otani T, Youker KA, Entman ML, Schwartz RJ. Inhibitory cardiac transcription factor, srf-n, is generated by caspase 3 cleavage in human heart failure and attenuated by ventricular unloading. Circulation. 2003;108:407–413. doi: 10.1161/01.CIR.0000084502.02147.83. [DOI] [PubMed] [Google Scholar]
- 18.Sah VP, Minamisawa S, Tam SP, Wu TH, Dorn GW, Ross J, Chien KR, Brown JH. Cardiac-specific overexpression of rhoa results in sinus and atrioventricular nodal dysfunction and contractile failure. The Journal Of Clinical Investigation. 1999;103:1627–1634. doi: 10.1172/JCI6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei L, Taffet GE, Khoury DS, Bo J, Li Y, Yatani A, Delaughter MC, Klevitsky R, Hewett TE, Robbins J, Michael LH, Schneider MD, Entman ML, Schwartz RJ. Disruption of rho signaling results in progressive atrioventricular conduction defects while ventricular function remains preserved. FASEB J. 2004;18:857–859. doi: 10.1096/fj.03-0664fje. [DOI] [PubMed] [Google Scholar]
- 20.Minetti C, Sotgia F, Bruno C, Scartezzini P, Broda P, Bado M, Masetti E, Mazzocco M, Egeo A, Donati MA, Volonte D, Galbiati F, Cordone G, Bricarelli FD, Lisanti MP, Zara F. Mutations in the caveolin-3 gene cause autosomal dominant limb-girdle muscular dystrophy. Nat Genet. 1998;18:365–368. doi: 10.1038/ng0498-365. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi T, Arimura T, Ueda K, Shibata H, Hohda S, Takahashi M, Hori H, Koga Y, Oka N, Imaizumi T. Identification and functional analysis of a caveolin-3 mutation associated with familial hypertrophic cardiomyopathy. Biochemical and Biophysical Research Communications. 2004;313:178–184. doi: 10.1016/j.bbrc.2003.11.101. [DOI] [PubMed] [Google Scholar]
- 22.Blair E, Price SJ, Baty CJ, Ostman-Smith I, Watkins H. Mutations in cis can confound genotype-phenotype correlations in hypertrophic cardiomyopathy. J Med Genet. 2001;38:385–388. doi: 10.1136/jmg.38.6.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Driest SL, Vasile VC, Ommen SR, Will ML, Tajik AJ, Gersh BJ, Ackerman MJ. Myosin binding protein c mutations and compound heterozygosity in hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 2004;44:1903–1910. doi: 10.1016/j.jacc.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 24.Bezzina CR, Rook MB, Groenewegen WA, Herfst LJ, van der Wal AC, Lam J, Jongsma HJ, Wilde AA, Mannens MM. Compound heterozygosity for mutations (w156x and r225w) in scn5a associated with severe cardiac conduction disturbances and degenerative changes in the conduction system. Circ. Res. 2003;92:159–168. doi: 10.1161/01.res.0000052672.97759.36. [DOI] [PubMed] [Google Scholar]
- 25.Jeschke B, Uhl K, Weist B, Schroder D, Meitinger T, Dohlemann C, Vosberg HP. A high risk phenotype of hypertrophic cardiomyopathy associated with a compound genotype of two mutated beta-myosin heavy chain genes. Hum. Genet. 1998;102:299–304. doi: 10.1007/s004390050695. [DOI] [PubMed] [Google Scholar]
- 26.Marian AJ. Nature's genetic gradients and the clinical phenotype. Circ Cardiovasc Genet. 2009;2:537–539. doi: 10.1161/CIRCGENETICS.109.921940. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.