Abstract
Detection of human papillomavirus in head and neck cancer has therapeutic implications. In-situ hybridization and immuno-histochemistry for p16 are used by surgical pathologists. We compared the sensitivity and specificity of three popular commercial tests for human papillomavirus detection in head and neck squamous cell carcinomas to a “gold standard” human papillomavirus PCR assay. One hundred-and-ten prospectively collected, formalin fixed tumor specimens were compiled onto tissue microarrays and tested for human papillomavirus DNA by in-situ hybridization with two probe sets: a biotinylated probe for high-risk human papillomavirus types 16/18 (Dako, CA), and a probe cocktail for 16/18 plus 10 additional high-risk types (Ventana, AZ). P16INK4 expression was also assessed using a Pharmingen immuno-histochemistry antibody (BD Biosciences, CA). Tissue microarrays were stained and scored at expert laboratories. Human papillomavirus DNA was detected by MY09/11-PCR using Gold AmpliTaq and dot-blot hybridization on matched fresh frozen specimens in a research laboratory. Human papillomavirus 16 E6 and E7-RNA expression was also measured using RT-PCR. Test performance was assessed by receiver operating characteristic analysis. High-risk human papillomavirus DNA types 16, 18 and 35 were detected by MY-PCR in 28% of tumors, with the majority (97%) testing positive for type 16. Compared to MY-PCR, the sensitivity and specificity for high-risk human papillomavirus DNA detection with Dako in-situ hybridization was 21% (95%CI:7–42) and 100% (95%CI:93–100), respectively. Corresponding test results by Ventana in-situ hybridization were 59% (95%CI:39–78) and 58% (95%CI:45–71), respectively. P16 immuno-histochemistry performed better overall than Dako (p=0.042) and Ventana (p=0.055), with a sensitivity of 52% (95%CI:32–71) and specificity of 93% (95%CI:84–98). Compared to a gold standard human papillomavirus PCR assays, HPV detection by in-situ hybridization was less accurate for head and neck squamous cell carcinoma on tissue microarrays than p16 immuno-histochemistry. Further testing is warranted before these assays should be recommended for clinical human papillomavirus detection.
Keywords: human papillomavirus, head and neck neoplasms, PCR, in-situ hybridization, immuno-histochemistry, p16, diagnostic methods
Introduction
Head and neck cancer remains an important problem in the United States with over 30,000 cases and 7,800 deaths occurring annually 1. Detection of human papillomavirus (HPV) in head and neck squamous cell carcinoma has therapeutic implications. HPV, particularly type 16, is detected in a third of head and neck squamous cell carcinoma and is associated with improved survival and response to therapy 2,3. As a result, the most recent American Joint Committee on Cancer (AJCC) staging criteria recommends reporting HPV status especially for oropharyngeal cancer, where detection rates are highest 4. A number of commercial assays, including DNA, RNA and protein-based tests, are available. However, detection rates vary considerably across studies, and there is, no consensus for which, if any, is adequate for diagnostic evaluation of HPV in head and neck squamous cell carcinoma.
In-situ hybridization has been used for HPV DNA detection in both primary and metastatic tumors by pathologists for a number of years, although its effectiveness as a clinical diagnostic tool in head and neck squamous cell carcinoma has not been tested extensively 5. Alternatively, p16INK4a expression measured by immuno-histochemistry has also been suggested as a surrogate for transcriptionally active HPV16 6–10. The current gold standard used in HPV research includes PCR-based assays designed to detect either HPV DNA or RNA. Among the commercial tests readily available to pathologists, both p16 immuno-histochemistry and HPV in-situ hybridization can be performed on formalin fixed and paraffin embedded tissue and allow for direct visualization of target protein or nucleic acid in tumor cells. However, independent comparison of these multiple clinically utilized HPV detection methods is limited for head and neck squamous cell carcinoma 11–13. Critical evaluation of HPV detection assays in head and neck squamous cell carcinoma is growing in importance as HPV-associated tumors may require different therapeutic approaches. We compared the sensitivity and specificity of three popular commercial tests for HPV detection (two direct and one indirect) in prospectively collected head and neck squamous cell carcinoma primary tumors against gold standard HPV PCR assays.
Materials and methods
Collection of clinical samples and patient data
Matched fresh frozen and formalin fixed and paraffin embedded primary head and neck squamous cell carcinoma tumor samples were collected prospectively from 109 patients. Subsequent first and second head and neck squamous cell carcinoma primaries were collected from one patient. All patients with primary head and neck squamous cell carcinoma treated at Montefiore Medical Center were approached for participation at time of diagnosis. Tumor samples were collected following histological confirmation and snap-frozen in liquid nitrogen in the same pathology laboratory within minutes of surgical resection or biopsy. Additional biopsies were collected prior to therapy for research purposes and processed in the same fashion. Patient demographics and tobacco smoking and alcohol consumption histories were also collected by medical interview prior to treatment. The study protocol was approved by the Institutional Review Boards at Montefiore Medical Center and Albert Einstein College of Medicine, and all patients provided written informed consent. Histological confirmation of tumors was performed by a single pathologist (MBG) assessing tumor grade and staging based on the AJCC classification.
Tissue microarrays
Tissue microarray blocks were constructed from formalin fixed and paraffin embedded tissue using a semi-automatic tissue arrayer (Chemicon) and 1.0 mm cores, which contained by three or more cores per sample from the areas with maximal numbers of tumor cells. Microarray sections were deparaffinized, rehydrated, and washed in TBS (SIGMA tris buffer; T6664) prior to staining.
HPV in-situ hybridization
Tissue microarray sections were stained for HPV DNA by in-situ hybridization with two different probe sets: INFORM® HPV-III Fam16(B) probe cocktail (Ventana, AZ) for 12 high-risk (HR) types (HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58 and 66), and an HPV 16/18 biotinylated DNA probe set (Y1412) with GenPoint™ tyramide single amplification system (Dako, CA). Tissue microarray sections were stained and scored independently at two expert laboratories blinded to tumor site and results by the other assays. Examples of HPV DNA positive head and neck squamous cell carcinoma detected by Dako and Ventana are shown in Figure 1.
Figure 1.
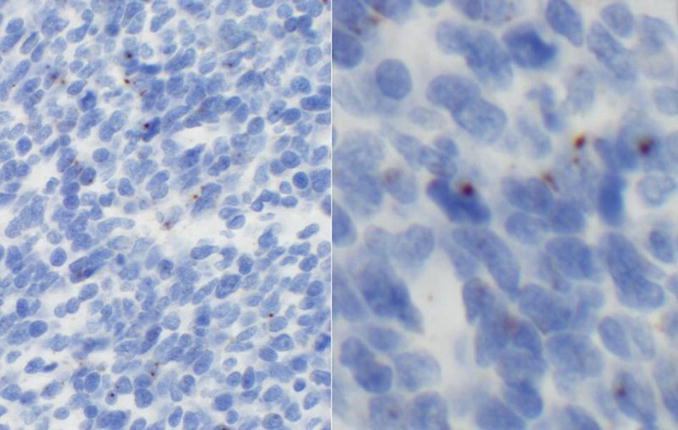
Top: Dako in-situ hybridization positive case showing brown intranuclear signals, this tumor was HPV16 positive by PCR (left panel: low power, right panel: high power). Bottom: Ventana in-situ hybridization positive case showing blue/purple intranuclear signals, this tumor was HPV16 positive by PCR (left panel: low power, right panel: high power).
p16 immuno-histochemistry
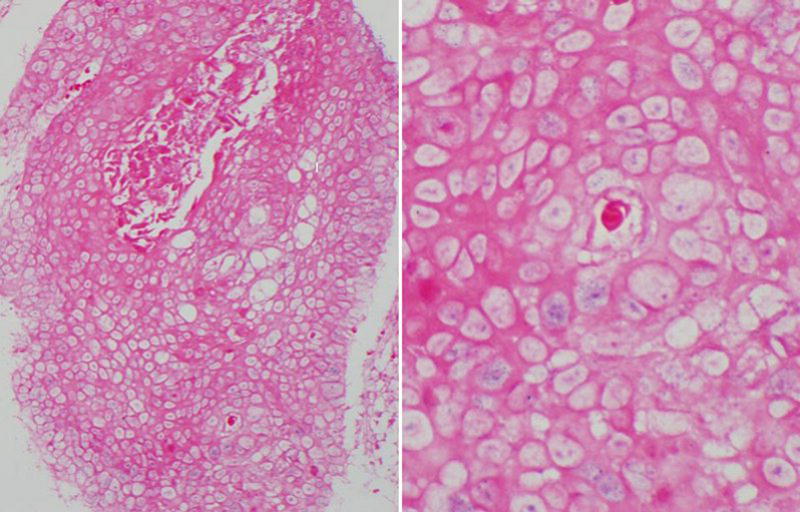
Deparaffinized tissue microarray sections were incubated with a Pharmingen immuno-histochemistry monoclonal mouse antibody (BD Biosciences, CA) at 1:50 dilution for 30 minutes at room temperature, followed by addition of secondary Dako EnVision™ biotinylated horseradish peroxidase-conjugated mouse antibody (K4001) for 30 minutes at room temperature without dilution, then stained with ultra-marque diaminobenzidine reagent 2–4 minutes (Dako, CA; K3468), and finally counterstained with hematoxylin (Harris formula; Surgipath, IL). An example of a p16 immuno-histochemistry positive staining head and neck squamous cell carcinoma is shown in Figure 2. All tissue microarray slides plus controls were stained in one assay. Positive and negative controls consisted of uterine cervix with severe dysplasia, with and without primary antibody, respectively. Immuno-histochemical expression for p16 was quantified according to cellular location (nuclear and cytoplasmic), staining intensity (on a scale of 0 to 3) according to the strongest signal that comprised at least 10% of each core, and distribution (0%, 25%, 50% or 75%) of antibody stains in tumor cells.
Figure 2.
P16 immuno-histochemistry positive case showing strong nuclear and cytoplasmic staining with a distribution of ≥75%, low power, this tumor was HPV16 positive by PCR.
HPV DNA detection and genotyping by PCR
Genomic DNA was isolated from 106 matched fresh frozen tissues using the DNEasyTissue Kit (Qiagen, CA). HPV-DNA was detected by a MY09/MY11/HMB01-PCR system using Gold AmpliTaq DNA polymerase that amplifies a 450bp segment in the L1-sequence of HPV as described 14. HPV genotyping was performed on all samples by dot-blot hybridization using biotinylated oligonucleotide probes for 40 HPV types, including those tested for by both in-situ hybridization assays. To verify the specificity of the hybridizations, a complete set of controls were included on all membranes. The integrity of specimen DNA was verified by amplification of a cellular gene (β-globin) 14. In addition, control samples ‘spiked’ with DNA from a cervical carcinoma cell-line with known quantity of HPV16 genomes were included in every assay. DNA viral load was also assessed by MY-PCR dot-blot intensity on a scale of 1–5 14.
HPV16 E6 and E7 mRNA detection
RNA samples, isolated from 86 matched fresh frozen tumors were tested for HPV16 E6 and E7 transcripts, found in over 80% of HPV positive head and neck squamous cell carcinomas. Total RNA was extracted and purified by a standardized protocol using TRIzol (Invitrogen, Carlsbad, CA). RNA was not available for 25 head and neck squamous cell carcinoma. Samples were subjected to a multiplex RT-PCR assay using a series of oligonucleotide primers that span the 204 to 525 nucleotide E6 and E7 regions of the HPV16 genome following a standardized protocol 15,16. The primers were designed to detect the full-length and spliced E6 mRNA transcripts. E7-encoding mRNA primers were designed based on HPV16 sequence NC_001526 (complete genome) using the web-based Primer3 design program (available at http://frodo.wi.mit.edu/primer3). The performance of the primers was confirmed using control RNA samples from HPV positive and negative head and neck squamous cell carcinoma and cervical carcinoma cell lines: FaDu, SCC-25, CAL27, SCC-15, UM-SCC-1, UM-SCC-47, UPCI-SCC-090, SiHa, and CaSki. In addition, GAPDH primers were included in each assay to verify the integrity of the specimen mRNA (based on sequence AY340484:Homo sapiens glyceraldehyde-3-phosphate dehydrogenase gene, complete cds).
Statistical Analyses
Stained tissue microarray sections were scored at independent expert laboratories: Ventana probes were used at Department of Pathology Laboratory, Ohio State University College of Medicine, OH (GN), and Dako probes at Genzyme, NY (ML). Positive nuclear staining was ascertained over multiple core replicates from the same primary tumor biopsy or resection specimen. A third pathologist (MBG), blinded to HPV PCR and in-situ hybridization results and tumor site, read the tissue microarrays for p16 expression in the cytoplasm and nucleus. Mean staining was then calculated across multiple core replicates from the same primary tumor biopsy or resection. P16 positivity was defined by a mean intensity cut-off of ≥2 and diffuse (≥75%) staining distribution in either the nuclei or cytoplasm derived by sensitivity analysis and corresponding to published criteria 12. Histological assessment of keratinization was also completed for 66 tumors, measured on a four-point scale from high (≥75%) to low (<25%).
Test performance was assessed by receiver operating characteristic analysis against the HPV PCR results as the gold standard. Areas under the receiver operating characteristic curves and 95% confidence intervals (CI) were estimated to assess differences in overall test performances 17. In-situ hybridization and immuno-histochemistry results were compared to HR-HPV DNA genotyping results by MY-PCR and HPV16 E6/E7 RNA expression by RT-PCR. Concordance between PCR and in-situ hybridization test results was assessed by Kappa statistic and Spearman correlation. Differences in keratinization across tumors were assessed using chi square tests.
Results
One hundred and ten prospectively collected primary head and neck squamous cell carcinoma tumor specimens were tested for HPV DNA by PCR, HPV in-situ hybridization and p16 immuno-histochemistry. Specimens were collected prior to initiation of chemotherapy or radiation by biopsy (N=36) or surgical resection (N=74, including 21 laser resection cases). Head and neck squamous cell carcinoma tumor sites represented include oropharynx (N=30), hypopharynx (N=8), larynx (N=32), lip/oral cavity (N=38) and nasopharynx (N=2). The majority of head and neck squamous cell carcinoma were stage IV (N=66, 60%), followed by stage III (N=19, 17%), II (N=14, 13%) and I (N=11, 10%). The majority of patients were male (N=76, 70%) with a median age at head and neck squamous cell carcinoma diagnosis between 62 and 63 years. Non-Hispanic White (N=43, 39%) followed by Hispanic White (N=28, 26%) and non-Hispanic African-American (N=26, 24%) patients represented the three largest race and ethnic groups. A history of tobacco smoking and alcohol drinking was also reported by the majority of patients (N=94, 86%), of which less than half continued to smoke (N=41, 37%) and drink (N=21, 19%) at time of diagnosis.
High-risk HPV DNA was detected by MY-PCR in 28% (N=30/106) of tumors. HPV16 was found in all but one of the HR-HPV positive cases. Table 1 shows the performance of each of the HPV testing methods (in-situ hybridization and immuno-histochemistry) relative to HPV MY-PCR and RT-PCR. Compared to MY-PCR, the sensitivity and specificity of HR-HPV DNA detection by in-situ hybridization using Ventana probes was 59% and 58%, respectively. Corresponding test results for HR-HPV DNA detection by Dako were 21% and 100%. The results for the Dako in-situ hybridization test were unchanged when compared to HPV16/18 detection only as the single case positive for a HR-HPV type other than 16/18 was not discernable by Dako in-situ hybridization due to insufficient tumor tissue or core drop-out on the corresponding tissue microarray section (see Supplemental Material)
Table 1.
Performance of HPV testing methods relative to PCR in head and neck squamous cell carcinoma
| No. HPV+ by test (%) | No. tested | Gold standard: High-risk HPV DNA by MY-PCR
|
||||
|---|---|---|---|---|---|---|
| Sensitivity (95%CI)1 | Specificity (95%CI) | Area under the curve2 (95%CI) | p-value3 | |||
| All sites combined | ||||||
| P16 immuno-histochemistry | 19 (20%) | 97 | 52% (32–71) | 93% (84–98) | 0.72 (0.62–0.82) | (reference) |
| In-situ hybridization Ventana | 41 (47%) | 87 | 59% (39–78) | 58% (45–71) | 0.59 (0.47–0.70) | 0.055 |
| In-situ hybridization Dako | 5 (6.6%) | 76 | 21% (7–42) | 100% (93–100) | 0.60 (0.52–0.69) | 0.042 |
|
| ||||||
| Oropharynx only | ||||||
| P16 immuno-histochemistry | 10 (37%) | 27 | 69% (39–91) | 93% (66–100) | 0.81 (0.66–0.96) | (reference) |
| In-situ hybridization Ventana | 16 (70%) | 23 | 67% (35–90) | 27% (6–61) | 0.47 (0.27–0.67) | 0.000 |
| In-situ hybridization Dako | 4 (21%) | 19 | 40% (12–74) | 100% (66–100) | 0.70 (0.54–0.86) | 0.046 |
|
| ||||||
| No. HPV+ by test (%) | No. tested |
Gold standard: HPV16 RNA by RT-PCR
|
||||
| Sensitivity (95%CI)1 | Specificity (95%CI) | Area under the curve2 (95%CI) | p-value3 | |||
|
| ||||||
| All sites combined | ||||||
| P16 immuno-histochemistry | 18 (23%) | 80 | 56% (35–76) | 93% (82–98) | 0.74 (0.64–0.85) | (reference) |
| In-situ hybridization Ventana | 35 (48%) | 73 | 67% (45–84) | 61% (46–75) | 0.64 (0.52–0.76) | 0.600 |
| In-situ hybridization Dako | 4 (6%) | 67 | 18% (5–40) | 100% (92–100) | 0.59 (0.51–0.67) | 0.045 |
|
| ||||||
| Oropharynx only | ||||||
| P16 immuno-histochemistry | 9 (43%) | 21 | 90 (56–100) | 100% (72–100) | 0.95 (0.85–1.00) | (reference) |
| In-situ hybridization Ventana | 13 (68%) | 19 | 67% (30–93) | 30% (7–65) | 0.48 (0.26–0.71) | 0.007 |
| In-situ hybridization Dako | 3 (19%) | 16 | 38% (9–76) | 100% (63–100) | 0.69 (0.51–0.87) | 0.034 |
95% confidence intervals (CI).
Receiver operating characteristic area under the curve.
P-value for comparison of receiver operating characteristic curves for in-situ hybridization assays to p16 immuno-histochemistry assuming independence.
Overall performance of each test in detecting HR-HPV DNA (reflecting combined sensitivity and specificity) was assessed by comparing the areas under the receiver operating characteristic curves. An area under the curve of 0.5 or less indicates a diagnostic test that cannot distinguish HPV positive from HPV negative cases. Compared to p16 immuno-histochemistry (area under the curve=0.72), test performance by Ventana in-situ hybridization (area under the curve=0.59) and Dako in-situ hybridization (area under the curve=0.60) were statistically worse (p=0.055 and p=0.042, respectively). Sensitivity and specificity of p16 immuno-histochemistry for detection of HR-HPV (with an intensity cut-off of ≥2 and diffuse (≥75%) staining) was 52% and 93%, respectively. However, test performance for p16 immuno-histochemistry decreased if a lower intensity (<2) or distribution (<75%) cut-point was used, due to a proportionally larger change (loss) in specificity relative to sensitivity (gain). Agreement between Ventana and Dako probes was also poor (Kappa=0.14, 95%CI: 0.02–0.26), although the difference in overall performance between the two in-situ hybridization tests for detecting HR-HPV DNA was not significant (p=0.415). Test performance improved only slightly when formalin fixed and paraffin embedded and fresh frozen specimens were both procured by surgical resection (p16 immuno-histochemistry area under the curve=0.75, 95%CI: 0.63–0.86; Ventana in-situ hybridization area under the curve=0.64, 95%CI: 0.52–0.76, and Dako area under the curve=0.62, 95%CI: 0.51–0.72), although the differences were not significant.
Looking at HPV DNA prevalence by anatomic site, HR-HPV DNA detection by MY-PCR was highest amongst oropharyngeal tumors (N=14, 47%), followed by hypopharyngeal (N=3, 43%), laryngeal (N=8, 26%) and oral cavity (N=4, 11%) sites. HPV positive oropharyngeal tumors also tended to have higher viral levels measured by dot blot intensity (mean=4.2±1.2 and median=5, on a scale of 1–5), compared to laryngeal (mean=2.8±0.7, median=3) or oral cavity tumors (mean=3±0.8, median=3). HR-HPV DNA detection sensitivity in oropharyngeal tumors only was higher for all tests, although overall test performance was only somewhat improved for p16 immuno-histochemistry (area under the curve=0.81, 95%CI: 0.66–0.96) and Dako in-situ hybridization (area under the curve=0.70, 95%CI: 0.54–0.86), but not for Ventana in-situ hybridization (area under the curve=0.47, 95%CI: 0.27–0.67) (Table 1).
Test performance was also assessed against HPV E6/E7 RNA expression as measured by RT-PCR. HPV E6/E7 oncogene expression in cancer is associated with integration of the HPV genome into the host genome and over-expression of p16 following E7-mediated inactivation of pRb 18. HPV16 E6/E7 RNA expression was detected by RT-PCR in 31% of tumors tested (N=27/86). As observed with HPV DNA detection by MY-PCR, p16 immuno-histochemistry (area under the curve=0.74) performed significantly better than Dako in-situ hybridization (area under the curve=0.59, p=0.045), and somewhat better than Ventana in-situ hybridization (area under the curve=0.64, p=0.600). Overlap in HPV16 E6 and E7 expression was high (97%, Kappa=0.92, 95%CI 0.83–1.0), and little to no difference in performance was observed when HPV16 E7 and E6 RNA expression were assessed separately. Whereas test performance compared to HPV16 E6/E7 RNA by RT-PCR was little changed for in-situ hybridization (using either probe set) when restricting to oropharyngeal SCC cases, p16 immuno-histochemistry was significantly higher (area under the curve=0.95, 95%CI: 0.85–1.00) (Table 1).
We were also able to assess whether p16 expression by immuno-histochemistry correlated with tumor histology within the head and neck. Degree of keratinization across head and neck tumors was low (median between 50% and <25%), and was significantly less in oropharyngeal and laryngeal tumors compared to oral cavity, with 74%, 52% and 21% of tumors showing <25% keratinization, respectively, reflecting a mostly basaloid morphology among oropharyngeal tumors. We observed an inverse correlation between p16 staining intensity and degree of keratinization (rho=−0.215, p=0.102) overall and among oropharyngeal tumors in particular (rho=−0.490, p=0.054). P16 immuno-histochemistry continued to perform better than Ventana (p=0.002) or Dako (p=0.121) in-situ hybridization compared to high-risk HPV DNA detection by MY-PCR when restricted to non-keratinizing tumors.
Contrasting HPV16 DNA and RNA results (N=82), there was good agreement between HPV16 E6/E7 RT-PCR and HPV16-specific MY-PCR results (84%, Kappa=0.62, 95%CI: 0.44–0.81). Whereas most cases were HPV16 positive by both MY-PCR (that amplifies a target in L1) and E6/E7 RT-PCR, five (6%) did not show HPV16 E6 and E7 oncogene expression and eight (10%) had undetectable levels of HPV16 DNA. Three additional cases negative by MY-PCR tested positive for either E6 or E7 RNA, but not both – these were considered RT-PCR negative.
We also assessed the potential for using p16 immuno-histochemistry in a triage scenario to detect head and neck squamous cell carcinoma tumors expressing HPV oncogenes (i.e., E6/E7 RNA by RT-PCR) where HR-HPV DNA testing by either in-situ hybridization or MY-PCR is used to further discriminate cases using p16 expression with a cut-off for p16 of 1+ intensity and ≥10% distribution as positive. Setting this low threshold for p16 resulted in a detection rate of 94% and sensitivity and specificity of 96% and 7%, respectively (area under the curve=0.52, 95%CI: 0.46–0.57). We saw little to no improvement in detection performance when combining tests with p16 immuno-histochemistry as described with MY-PCR (area under the curve=0.76, 95%CI: 0.66–0.87), in-situ hybridization by Ventana (area under the curve=0.66, 95%CI: 0.55–0.78) or in-situ hybridization by Dako (area under the curve=0.58, 95%CI: 0.51–0.65) (Figure 3). These results were little changed when restricting to oropharyngeal SCC cases only (not shown).
Figure 3.
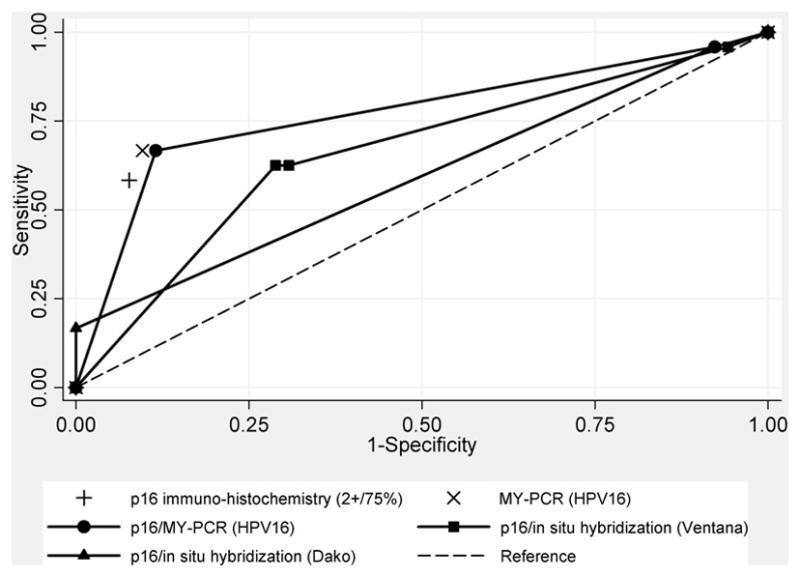
Receiver operating characteristic curves for head and neck squamous cell carcinomas by combined p16 immuno-histochemistry and HPV DNA testing by in-situ hybridization using the INFORM® HPV-III Fam16(B) (Ventana, CA) and HPV16/18 biotinylated GenPoint™ (Dako, CA) probes, or by MY-PCR, compared to HPV16 E6/E7 RT-PCR. Non-parametric receiver operating characteristic curves shown for combined test results with p16 immuno-histochemistry (assuming a 1+/≥10% cut-off) stratified by HPV Ventana in-situ hybridization (-■-), Dako in-situ hybridization (-▲-), or MY-PCR (-●-) results. Solitary markers shown reflect single test performance statistics for p16 IHC (assuming a 2+/≥75% cut-off) (+) and MY-PCR for HPV16 (X) alone. The dashed line indicates a reference test threshold with area under the receiver operating characteristic curve of 0.5.
There was little agreement between the in-situ hybridization and immuno-histochemistry test results among cases discordant by PCR. Among HPV16 RNA+/DNA− cases (N=8), half were positive by Ventana in-situ hybridization, two were positive by p16 immuno-histochemistry at the 2+/≥75% cut-off and none were positive by Dako in-situ hybridization, with no overlap between in-situ hybridization and immuno-histochemistry positives. Four of the remaining p16 cases stained with ≥2 intensity but with distributions of ≤50% (N=1) and ≤25% (N=3). Among HPV16 RNA−/DNA+ cases (N=5), only one each were found positive by Ventana in-situ hybridization and p16 immuno-histochemistry (at the 2+/≥75% cut-off), while the remaining four p16 cases also stained with ≥2 intensity but with distributions of ≤50% (N=2) and ≤25% (N=2). The majority of discordant cases (N=9/13) originated outside the oropharynx, including the anterior tongue, floor of mouth, retromolar trigone, alveolar ridge, posterior pharyngeal wall and supraglottis. Test performance increased only slightly when restricting cases to tumors concordant for HPV16 DNA and RNA detection by PCR (N=68): p16 immuno-histochemistry area under the curve=0.84 (95%CI: 0.73–0.96), Ventana in-situ hybridization area under the curve=0.66 (95%CI: 0.52–0.79), and Dako area under the curve=0.63 (95%CI: 0.52–0.75).
Discussion
Despite improvements over the last decade, five year survival rates for head and neck squamous cell carcinoma remain at 50%. Growing evidence indicates that, for a subset of head and neck squamous cell carcinoma, the presence of HPV in tumors constitutes a positive prognostic marker of disease 2,19. Detection of HPV in head and neck squamous cell carcinoma therefore has obvious therapeutic implications in treatment management, 4 or targeted intervention 20,21, and pathologists will play a central role in the identification and triaging of HPV positive head and neck squamous cell carcinoma cases.
We therefore asked, if detection of HPV in head and neck squamous cell carcinoma is important for treatment decisions, which test is most appropriate for accurate detection of HPV? To investigate this, we independently tested three commercial, clinically utilized HPV detection methods (two direct, one indirect) available to anatomic pathologists. This study represents one of the first systematic comparisons of these common HPV detection methods for head and neck squamous cell carcinoma 22.
High-risk HPV prevalence rates in our study varied considerably by detection method and anatomic site, with the highest estimates obtained for oropharyngeal tumors by in-situ hybridization using the Ventana Fam16 probe cocktail. Other studies, such as Agoston and colleagues13 have reported higher detection rates in oropharyngeal tumors using in-situ hybridization with the Ventana Fam16 probe cocktail, and PCR using L1 and E7 DNA primers, but did not provide analogous estimates of test performance as comparisons were made to combined test results assuming no false positives. Sensitivity and specificity estimates for p16 immuno-histochemistry using a Dako antibody (G175-405) relative to PCR were both higher (100%) and lower (35%), respectively, compared to our results with the Pharmingen antibody. Singhi and Westra12 used the Dako in-situ hybridization platform as the ‘gold standard’ for comparing p16 immuno-histochemistry in oropharyngeal cancers, which we show in this study to be least sensitive. Nonetheless, they reported higher (>80%) prevalence rates for HPV in a population of patients referred for suspicion of HPV; either to determine eligibility for clinical trials, guide clinical diagnosis, or requested by concerned patients and their partners.
The advantage of immuno-histochemistry for p16 is that it is readily available in surgical pathology laboratories, and relatively simple to evaluate. Typical p16 positive cases usually show diffuse, strong nuclear and cytoplasmic expression. Indeterminate staining patterns not reaching this cut-off value were relatively common in this study. This may in part be a sampling issue related to our use of tissue microarray cores; it is possible that the frequency of limited distribution of staining is less common when tissue sections are examined. This may also have been a factor for the in-situ hybridization staining, especially where positive cells are few and focal, as found with the Dako GenPoint probe.
Notably, there is a lack of published consensus as to what cut-off should be used for the determination of p16 positivity, with some groups setting low cut-off points 7,24–28. When restricting to the oropharynx, the head and neck site most likely to be associated with HPV, we detected no HPV16 RNA negative tumors with p16 staining with a distribution of ≥75%, whereas only one HPV16 RNA positive case had p16 staining with a distribution of <75%. Lewis and colleagues also reported p16 staining with a distribution of ≤75% in oropharyngeal SCC tumor cells in only 7% of p16 ‘positive’ cases 23.
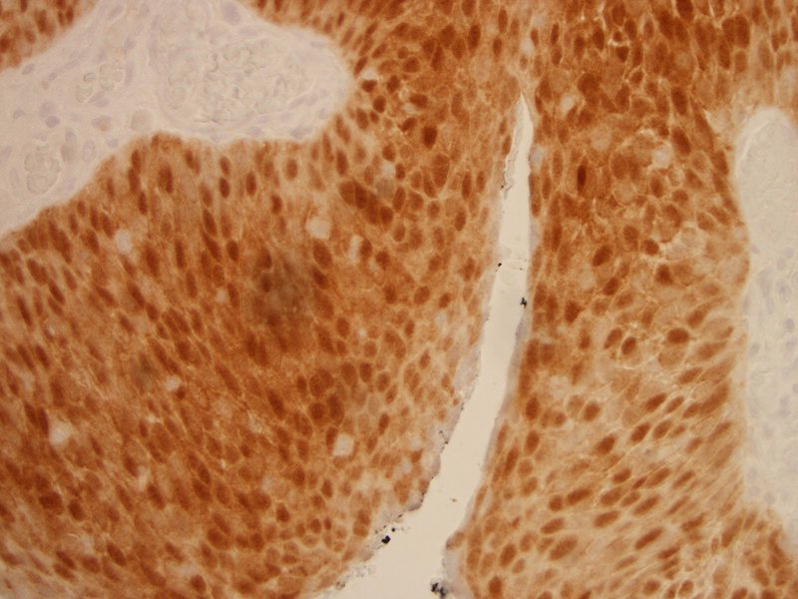
In contrast, in-situ hybridization probe signals can be more difficult to interpret than immuno-histochemistry staining and non-specific signals are a well recognized problem 29. Moreover, a distinction is made between intranuclear vs. paranuclear or cytoplasmic staining on in-situ hybridization. This specification allows for the identification of clinically relevant HPV infections in cancer cells. However, this can also lead to ambiguous staining results in situ and lower sensitivity compared to PCR. For example, whereas specific localization within the nucleus was needed for interpretation of Dako staining as positive, purple nuclear staining by Ventana probes was more frequently interpreted as positive in our experience. An example of an HPV PCR negative head and neck squamous cell carcinoma with discordant HPV in-situ hybridization results by Ventana (more sensitive test) and Dako (more specific test) is shown in Figure 4.
Figure 4.
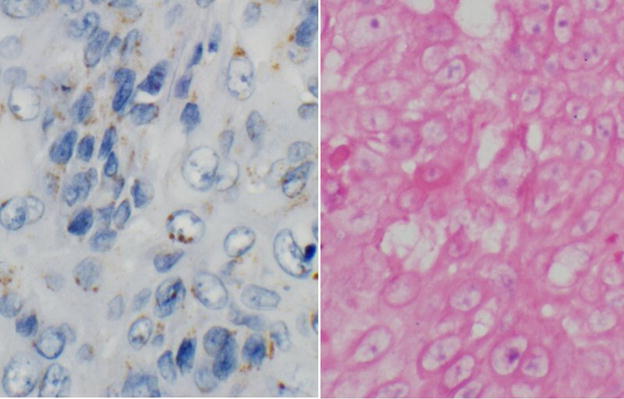
Dako in-situ hybridization negative tumor showing nonspecific cytoplasmic and paranuclear signals (left panel), this same case had purple intranuclear signals and was interpreted as positive by Ventana positive (right panel), but was HPV negative by PCR.
Results can also differ by pathologist. Comparison with blinded reading of the staining results by our laboratory pathologist (MBG) revealed higher sensitivity (33% vs. 21%) but lower specificity (94% vs. 100%) with Dako in-situ hybridization than the expert evaluation, and higher specificity (86% vs. 58%) and equal sensitivity with Ventana in-situ hybridization (59%) compared to MY-PCR. In response, automated imaging methods have recently been incorporated into commercial labs. However, even after removing individual subjectivity in interpretation, technical variability with hybridization can remain an obstacle. In contrast, PCR-based HPV tests have been in use in research for over 20 years and have the added advantage of providing type-specific information 30–32. PCR methods targeting the HPV L1 region are also used in clinical HPV testing of the cervix 14. However, this approach can be labor intensive and does not differentiate between integrated and episomal forms of the virus.
Molecular studies suggest that HPV-DNA in oropharyngeal tumors (overwhelmingly genotype 16) is present at high copy-numbers and is frequently integrated 33,34. Comparing HPV16 DNA and RNA detection by PCR, we observe a high concordance in DNA prevalence and E6/E7 expression, indicative of integration. However, the presence of E6/E7 RNA in DNA negative tumors, observed in two-thirds (N=8/13) of discordant cases by PCR, might suggest the presence of integrated virus with loss or disruption of the L1 open reading frame 18,35. Following integration, the intact portion of the HPV16 genome can produce multiple E6 and E7 oncogene transcripts 18,36. This is of interest as detection of HPV RNA in the tumor has recently been associated with better survival prediction than HPV DNA 8.
Alternatively, the up-regulation of p16 has been suggested as a marker of active HPV infection in head and neck squamous cell carcinoma 7, whereas inactivation of p16 by methylation is a common event in smoking associated cancers 37,38. P16 by immuno-histochemistry is a method commonly used by pathologists, and its use with in-situ hybridization for HPV DNA detection in a triage setting may help when HPV PCR analysis is not feasible 7,39–41. This combined testing may be of benefit where cross-linking and DNA degradation induced by formalin fixation is a concern, since primer systems that target larger HPV-DNA fragments tend to have poorer detection rates in formalin fixed and paraffin embedded samples compared with those targeting shorter DNA segments 42. For example, it has been suggested the specificity of p16 immuno-histochemistry in formalin fixed and paraffin embedded may be improved if followed by DNA analysis for p16-positive cases, when compared to E6 RT-PCR in fresh frozen tissue as the standard test 11. In our experience, p16 positive cases varied with respect to patterns of staining. However, relaxing the criteria for p16 positive immuno-histochemistry tests did not improve performance in HPV16 E6/E7 RNA detection when followed by in-situ hybridization, or by MY-PCR.
We should note that despite using expert staining and evaluations, tests were performed on tissue microarrays, which do not reproduce the clinical diagnostic setting. This approach was chosen because of the high-throughput capability for conducting in-situ hybridization and immuno-histochemistry assays simultaneously on multiple samples. Although, multiple tissue microarray cores were selected by a pathologist for each tumor to reflect the diagnostic histopathology specimen, results may differ for clinical formalin fixed and paraffin embedded tissue sections.
Typically, HPV positive oropharyngeal squamous cell carcinomas were non-keratinizing with basaloid type tumor cells. We found that p16 immuno-histochemistry continued to perform better than in-situ hybridization when compared to HPV detected by PCR in non-keratinizing squamous cell carcinomas and oropharyngeal tumors specifically. We also observed an inverse correlation between p16 expression and limited keratinization, which reflects non-keratinizing basaloid morphology.
In summary, while these results do not preclude the potential utility of assessing p16 expression as a biomarker for the clinical management of head and neck squamous cell carcinoma 23,43, particularly for oropharyngeal SCC, further analytical evaluation by blinded trials with multiple testing methods is warranted before these assays should be recommended for clinical HPV detection.
Supplementary Material
Acknowledgments
Sources of support: This project is supported in part by NIH grant CA115243 (to NFS) and the Department of Pathology, Albert Einstein College of Medicine/Montefiore Medical Center.
We thank the participants of this study; Catherine Sarta for her time and effort spent enrolling and following participants and with data entry; Gregory Rosenblatt for his assistance with data management; and Jaya Sunkara for her assistance with the immuno-histochemistry protocol.
List of commercial assays
Semi-automatic tissue arrayer: Chemicon (now part of Millipore). Millipore Inc., 290 Concord Road, Billerica, MA 01821
INFORM® HPV-III Fam16(B) probe set: Ventana Medical Systems, Inc., 1910 E. Innovation Park Drive Tucson, AZ 85755
Dako HPV16/18 biotinylated probe (Y1412) with GenPoint™ tyramide signal amplification system: Dako North America, Inc., 6392 Via Real Carpinteria, CA 93013
Pharmingen p16 immuno-histochemistry monoclonal mouse antibody: BD Biosciences, Pharmingen, 10975 Torreyana Road, San Diego, CA 92121
Footnotes
Disclaimer: The authors have no conflicts of interest to disclose. Genzyme Genetics and its logo are trademarks of Genzyme Corporation and used by Esoterix Genetic Laboratories, LLC, a wholly-owned subsidiary of LabCorp, under license. Esoterix Genetic Laboratories and LabCorp are operated independently from Genzyme Corporation.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, et al. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–30. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 2.Schlecht NF. Prognostic value of human papillomavirus in the survival of head and neck cancer patients: an overview of the evidence. Oncol Rep. 2005;14:1239–47. [PubMed] [Google Scholar]
- 3.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandwein-Gensler M, Smith RV. Prognostic indicators in head and neck oncology including the new 7th edition of the AJCC staging system. Head Neck Pathol. 2010;4:53–61. doi: 10.1007/s12105-010-0161-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fakhry C, Gillison ML. Clinical implications of human papillomavirus in head and neck cancers. J Clin Oncol. 2006;24:2606–11. doi: 10.1200/JCO.2006.06.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mulvany NJ, Allen DG, Wilson SM. Diagnostic utility of p16INK4a: a reappraisal of its use in cervical biopsies. Pathology. 2008;40:335–44. doi: 10.1080/00313020802035907. [DOI] [PubMed] [Google Scholar]
- 7.Smith EM, Wang D, Kim Y, Rubenstein LM, Lee JH, Haugen TH, et al. P16INK4a expression, human papillomavirus, and survival in head and neck cancer. Oral Oncol. 2008;44:133–42. doi: 10.1016/j.oraloncology.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Shi W, Kato H, Perez-Ordonez B, Pintilie M, Huang S, Hui A, et al. Comparative prognostic value of HPV16 E6 mRNA compared with in situ hybridization for human oropharyngeal squamous carcinoma. J Clin Oncol. 2009;27:6213–21. doi: 10.1200/JCO.2009.23.1670. [DOI] [PubMed] [Google Scholar]
- 9.Kong CS, Balzer BL, Troxell ML, Patterson BK, Longacre TA. p16INK4A immunohistochemistry is superior to HPV in situ hybridization for the detection of high-risk HPV in atypical squamous metaplasia. Am J Surg Pathol. 2007;31:33–43. doi: 10.1097/01.pas.0000213347.65014.ee. [DOI] [PubMed] [Google Scholar]
- 10.Holladay EB, Logan S, Arnold J, Knesel B, Smith GD. A comparison of the clinical utility of p16(INK4a) immunolocalization with the presence of human papillomavirus by hybrid capture 2 for the detection of cervical dysplasia/neoplasia. Cancer. 2006;108:451–61. doi: 10.1002/cncr.22284. [DOI] [PubMed] [Google Scholar]
- 11.Smeets SJ, Hesselink AT, Speel EJ, Haesevoets A, Snijders PJ, Pawlita M, et al. A novel algorithm for reliable detection of human papillomavirus in paraffin embedded head and neck cancer specimen. Int J Cancer. 2007;121:2465–72. doi: 10.1002/ijc.22980. [DOI] [PubMed] [Google Scholar]
- 12.Singhi AD, Westra WH. Comparison of human papillomavirus in situ hybridization and p16 immunohistochemistry in the detection of human papillomavirus-associated head and neck cancer based on a prospective clinical experience. Cancer. 2010;116:2166–73. doi: 10.1002/cncr.25033. [DOI] [PubMed] [Google Scholar]
- 13.Agoston ES, Robinson SJ, Mehra KK, Birch C, Semmel D, Mirkovic J, et al. Polymerase chain reaction detection of HPV in squamous carcinoma of the oropharynx. Am J Clin Pathol. 2010;134:36–41. doi: 10.1309/AJCP1AAWXE5JJCLZ. [DOI] [PubMed] [Google Scholar]
- 14.Castle PE, Schiffman M, Gravitt PE, Kendall H, Fishman S, Dong H, et al. Comparisons of HPV DNA detection by MY09/11 PCR methods. J Med Virol. 2002;68:417–23. doi: 10.1002/jmv.10220. [DOI] [PubMed] [Google Scholar]
- 15.Braakhuis BJ, Snijders PJ, Keune WJ, Meijer CJ, Ruijter-Schippers HJ, Leemans CR, et al. Genetic patterns in head and neck cancers that contain or lack transcriptionally active human papillomavirus. J Natl Cancer Inst. 2004;96:998–1006. doi: 10.1093/jnci/djh183. [DOI] [PubMed] [Google Scholar]
- 16.van Houten VM, Snijders PJ, van den Brekel MW, Kummer JA, Meijer CJ, van Leeuwen B, et al. Biological evidence that human papillomaviruses are etiologically involved in a subgroup of head and neck squamous cell carcinomas. Int J Cancer. 2001;93:232–5. doi: 10.1002/ijc.1313. [DOI] [PubMed] [Google Scholar]
- 17.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating curves: A nonparametric approach. Biometrics. 1988;44:837–45. [PubMed] [Google Scholar]
- 18.zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–50. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 19.Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, Pinto H, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100:261–9. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- 20.Hallez S, Simon P, Maudoux F, Doyen J, Noel JC, Beliard A, et al. Phase I/II trial of immunogenicity of a human papillomavirus (HPV) type 16 E7 protein-based vaccine in women with oncogenic HPV-positive cervical intraepithelial neoplasia. Cancer Immunol Immunother. 2004;53:642–50. doi: 10.1007/s00262-004-0501-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang XG, Revskaya E, Bryan RA, Strickler HD, Burk RD, Casadevall A, et al. Treating cancer as an infectious disease--viral antigens as novel targets for treatment and potential prevention of tumors of viral etiology. PLoS One. 2007;2:e1114. doi: 10.1371/journal.pone.0001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen CT, Lewis JS, Jr, El-Mofty SK, Haughey BH, Nussenbaum B. Human papillomavirus and oropharynx cancer: biology, detection and clinical implications. Laryngoscope. 2010;120:1756–72. doi: 10.1002/lary.20936. [DOI] [PubMed] [Google Scholar]
- 23.Lewis JS, Jr, Thorstad WL, Chernock RD, Haughey BH, Yip JH, Zhang Q, et al. p16 positive oropharyngeal squamous cell carcinoma:an entity with a favorable prognosis regardless of tumor HPV status. Am J Surg Pathol. 2010;34:1088–96. doi: 10.1097/PAS.0b013e3181e84652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wittekindt C, Gultekin E, Weissenborn SJ, Dienes HP, Pfister HJ, Klussmann JP. Expression of p16 protein is associated with human papillomavirus status in tonsillar carcinomas and has implications on survival. Adv Otorhinolaryngol. 2005;62:72–80. doi: 10.1159/000082474. [DOI] [PubMed] [Google Scholar]
- 25.El-Mofty SK, Patil S. Human papillomavirus (HPV)-related oropharyngeal nonkeratinizing squamous cell carcinoma: characterization of a distinct phenotype. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2006;101:339–45. doi: 10.1016/j.tripleo.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Kim SH, Koo BS, Kang S, Park K, Kim H, Lee KR, et al. HPV integration begins in the tonsillar crypt and leads to the alteration of p16, EGFR and c-myc during tumor formation. Int J Cancer. 2007;120:1418–25. doi: 10.1002/ijc.22464. [DOI] [PubMed] [Google Scholar]
- 27.Charfi L, Jouffroy T, de Cremoux P, Le Peltier N, Thioux M, Freneaux P, et al. Two types of squamous cell carcinoma of the palatine tonsil characterized by distinct etiology, molecular features and outcome. Cancer Lett. 2008;260:72–8. doi: 10.1016/j.canlet.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 28.Kuo KT, Hsiao CH, Lin CH, Kuo LT, Huang SH, Lin MC. The biomarkers of human papillomavirus infection in tonsillar squamous cell carcinoma-molecular basis and predicting favorable outcome. Mod Pathol. 2008;21:376–86. doi: 10.1038/modpathol.3800979. [DOI] [PubMed] [Google Scholar]
- 29.Unger ER. In situ diagnosis of human papillomaviruses. Clin Lab Med. 2000;20:289–301. vi. [PubMed] [Google Scholar]
- 30.Shibata DK, Arnheim N, Martin WJ. Detection of human papilloma virus in paraffin-embedded tissue using the polymerase chain reaction. J Exp Med. 1988;167:225–30. doi: 10.1084/jem.167.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manos MM, Ting Y, Wright DK, Lewis AJ, Broker TR, Wolinski SM. Use of polymerase chain reaction amplification for the detection of genital human papillomaviruses. Cancer Cells. 1989;7:209–14. [Google Scholar]
- 32.Gravitt PE, Manos MM. Polymerase chain reaction-based methods for the detection of human papillomavirus DNA. IARC Sci Publ. 1992;119:121–33. [PubMed] [Google Scholar]
- 33.Gillison ML, Koch WM, Shah KV. Human papillomavirus in head and neck squamous cell carcinoma: are some head and neck cancers a sexually transmitted disease? Curr Opin Oncol. 1999;11:191–9. doi: 10.1097/00001622-199905000-00010. [DOI] [PubMed] [Google Scholar]
- 34.Lindel K, Beer KT, Laissue J, Greiner RH, Aebersold DM. Human papillomavirus positive squamous cell carcinoma of the oropharynx: a radiosensitive subgroup of head and neck carcinoma. Cancer. 2001;92:805–13. doi: 10.1002/1097-0142(20010815)92:4<805::aid-cncr1386>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 35.Klaes R, Woerner SM, Ridder R, Wentzensen N, Duerst M, Schneider A, et al. Detection of high-risk cervical intraepithelial neoplasia and cervical cancer by amplification of transcripts derived from integrated papillomavirus oncogenes. Cancer Res. 1999;59:6132–6. [PubMed] [Google Scholar]
- 36.Chung CH, Gillison ML. Human papillomavirus in head and neck cancer: its role in pathogenesis and clinical implications. Clin Cancer Res. 2009;15:6758–62. doi: 10.1158/1078-0432.CCR-09-0784. [DOI] [PubMed] [Google Scholar]
- 37.Soria JC, Rodriguez M, Liu DD, Lee JJ, Hong WK, Mao L. Aberrant promoter methylation of multiple genes in bronchial brush samples from former cigarette smokers. Cancer Res. 2002;62:351–5. [PubMed] [Google Scholar]
- 38.Toyooka S, Maruyama R, Toyooka KO, McLerran D, Feng Z, Fukuyama Y, et al. Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int J Cancer. 2003;103:153–60. doi: 10.1002/ijc.10787. [DOI] [PubMed] [Google Scholar]
- 39.Klingenberg B, Hafkamp HC, Haesevoets A, Manni JJ, Slootweg PJ, Weissenborn SJ, et al. p16 INK4A overexpression is frequently detected in tumour-free tonsil tissue without association with HPV. Histopathology. 2010;56:957–67. doi: 10.1111/j.1365-2559.2010.03576.x. [DOI] [PubMed] [Google Scholar]
- 40.Licitra L, Perrone F, Bossi P, Suardi S, Mariani L, Artusi R, et al. High-risk human papillomavirus affects prognosis in patients with surgically treated oropharyngeal squamous cell carcinoma. J Clin Oncol. 2006;24:5630–6. doi: 10.1200/JCO.2005.04.6136. [DOI] [PubMed] [Google Scholar]
- 41.Kumar B, Cordell KG, Lee JS, Worden FP, Prince ME, Tran HH, et al. EGFR, p16, HPV Titer, Bcl-xL and p53, Sex, and Smoking As Indicators of Response to Therapy and Survival in Oropharyngeal Cancer. J Clin Oncol. 2008;26:3128–37. doi: 10.1200/JCO.2007.12.7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baay MF, Quint WG, Koudstaal J, Hollema H, Duk JM, Burger MP, et al. Comprehensive study of several general and type-specific primer pairs for detection of human papillomavirus DNA by PCR in paraffin-embedded cervical carcinomas. J Clin Microbiol. 1996;34:745–7. doi: 10.1128/jcm.34.3.745-747.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lassen P, Eriksen JG, Hamilton-Dutoit S, Tramm T, Alsner J, Overgaard J. HPV-associated p16-expression and response to hypoxic modification of radiotherapy in head and neck cancer. Radiother Oncol. 2010;94:30–5. doi: 10.1016/j.radonc.2009.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


