Abstract
Precise regulation of DNA damage response is crucial for cellular survival after DNA damage, and its abrogation often results in genomic instability in cancer. Phosphorylated histone H2AX (γH2AX) forms nuclear foci at sites of DNA damage and facilitates DNA damage response and repair. MicroRNAs are short, non-protein-encoding RNA molecules, which post-transcriptionally regulate gene expression by repressing translation of and/or degrading mRNA. How microRNAs modulate DNA damage response is largely unknown. In this study, we developed a cell-based screening assay utilizing ionizing radiation-induced γH2AX foci formation in a human osteosarcoma cell line, U2OS, as the readout. By screening a library of human microRNA mimics, we identified several microRNAs that inhibited γH2AX foci formation. Among them, miR-138 directly targeted the histone H2AX 3′-UTR, reduced histone H2AX expression and induced chromosomal instability after DNA damage. Overexpression of miR-138 inhibited homologous recombination and enhanced cellular sensitivity to multiple DNA damaging agents (cisplatin, camptothecin, and ionizing radiation). Reintroduction of histone H2AX in miR-138 overexpressing cells attenuated miR-138-mediated sensitization to cisplatin and camptothecin. Our study suggests that miR-138 is an important regulator of genomic stability and a potential therapeutic agent to improve the efficacy of radiotherapy and chemotherapy with DNA damaging agents.
Keywords: microRNA, DNA damage response, γH2AX, drug resistance, glioma
Introduction
DNA damage response pathways are composed of sensors that sense damaged DNA, mediators or transducers that amplify signals, and effectors that determine the fate of cells by modulating gene expression or inducing cell cycle arrest, DNA repair, apoptosis or senescence (1). DNA double strand breaks (DSBs) can be generated by various endogenous and exogenous stresses, such as replication fork collapse and an exposure to ionizing radiation (IR) (2). MRE11-RAD50-NBS1 (MRN) complex detects DSBs, recruits ATM kinase and triggers phosphorylation of histone H2AX at Serine 139 (γH2AX) by ATM kinase (3, 4). Subsequently, numerous signaling and repair proteins, such as MDC1 and 53BP1, accumulate at DNA breaks to form discrete foci. Among these proteins, MDC1 interacts directly with γH2AX and recruits more MRN-bound ATM kinase to phosphorylate histone H2AX molecules surrounding the DNA breaks (5–7). This positive feedback leads to augmentation of the γH2AX signaling, which can be spread to 1–2 Mega bases away from DNA lesions (8). Therefore, by recruiting DNA damage signaling and repair proteins, γH2AX plays an important role in the repair of DNA lesions. H2AX-deficient cells are sensitive to IR and exhibit genomic instability, DSB repair defects and mild DNA damage checkpoint dysfunction (9, 10).
MicroRNAs (miRNA) are small non-protein-encoding RNAs that post-transcriptionally regulate gene expression. They originate from longer primary miRNA transcripts that are processed through two steps of cleavage mediated by the endonucleases Drosha and Dicer (11). The mature miRNA is incorporated into an RNA-induced silencing complex (RISC) and directs RISC to the 3′-UTR of its targets through complementarity to the seed region of the miRNA (i.e., nucleotides 2–8), leading to translational inhibition and/or degradation of mRNA targets (11). Aberrant expression of miRNAs is often seen in cancer (12), where specific miRNAs function as tumor suppressors or oncogenes and modulate many aspects of carcinogenesis (13). These miRNAs may have diagnostic or prognostic value and are also potential therapeutic targets in the treatment of specific types of cancer.
Several recent studies support the notion that miRNAs play important roles in DNA damage response and DNA repair (14–17). These miRNA-mediated regulations of DNA damage response have the potential to improve the efficacy of cancer therapies such as chemotherapy and radiotherapy which rely on the induction of DNA damage. However, understanding of the role of miRNAs in DNA damage response is incomplete. To further understand the role of miRNAs in the regulation of DNA damage response, we performed a cell-based miRNA library screen utilizing IR-induced γH2AX foci formation as the readout. Among the hits from the screen, we focused on miR-138, which we found to directly downregulate histone H2AX expression.
Materials and methods
Cell lines
U2OS, HeLa, U87, LN-229, MDA-MB-231 and M059J were purchased from the American Type Culture Collections. SiHa and normal human foreskin fibroblasts were provided by the Galloway lab (Fred Hutchison Cancer Research Center). All cell lines were grown in DMEM with 10% FBS, 2 mM L-glutamine and 100 units/ml penicilin and 100 μg/ml streptomycin in a humidified 5% CO2-containing atmosphere at 37°C.
Glioblastoma (GBM) tissue samples
GBM samples, kindly provided by Dr. Ryo Nishikawa (Department of Neuro-Oncology, International Medical Center, Saitama Medical University, Japan) and Dr. David James (Department of Neurological Surgery, University of California, San Francisco) were obtained from patients undergoing surgical treatment after written informed consents. RNA was isolated from frozen tumor samples using Trizol (Invitrogen) following the manufacturer’s instructions. The use of glioma clinical samples in this study was approved by the institutional review board of the University of California at San Diego.
Plasmids, siRNAs, miRNA mimics/inhibitors and virus production
3′-UTR of H2AX (position -19 to 1046) was amplifed by PCR and cloned into pGL3-control (Promega) to obtain pGL3-H2AX 3′UTR WT plasmid. A putative binding site of miR-138 in H2AX 3′-UTR was mutated using the QuikChange site-direct mutagenesis kit (Stratagene). H2AX coding sequence without 3′-UTR was PCR amplified and cloned into pMMPpuro vector and the FLAG signal was added to the N-terminus of H2AX to obtain pMMP-Flag-H2AX plasmid. To obtain constructs expressing miR-138 precursors, miR-138-1 and miR-138-2 and the spanning sequences (150 bp on each end) were amplified by PCR and inserted into either pSM30-GFP (a gift of Dr. Guangwei Du) or pLemiR (Open Biosystems) vectors. Plasmids were delivered into cells using TransIT-LT-1 reagent (Mirus). Specific siRNAs were used to knock down H2AX (5′-AACAACAAGAAGACGCGAATC-3′) and BRCA2 (5′-AACAACAATTACGAACCAAAC-3′) (18) (Qiagen). Transfection of siRNAs or miRNA mimics (Dharmacon) was done at 20 nM using HiPerFect (Qiagen). MiRNA mimic negative control (miR-neg, Dharmacon) or nontargeting siRNA (siCtrl, 5′-AATTCTCCGAACGTGTCACGT-3′) (Qiagen) were used as negative controls. Locked nucleic acid (LNA)-based negative or miR-138 inhibitor (Exiqon) were transfected with Lipofectamine 2000 (Invitrogen) at 100 nM. To produce retrovirus generating H2AX, pMMP-Flag-H2AX was co-transfected with the packaging plasmid Ampo-V into HEK293T cells. Supernatants were collected at 48 h and 72 h after transfection for retrovirus preparation. To produce lentivirus expressing miR-138, pLemiR-138-1 was cotransfected with packaging plasmids psPAX2 and pMD2.G into HEK293T cells. Supernatants were collected at 72 h for lentivirus preparation.
Immunofluorescence microscopy
Cells were grown on coverslips and treated with IR using a JL Shepherd Mark I Cesium Irradiator (JL Shepherd & associates, San Fernando, CA). Cells were simultaneously fixed and permeabilized with 1% paraformaldehyde containing 0.5% Triton X-100 in PBS for 30 min, blocked with 3% bovine serum albumin containing 0.1% Tween-20 for 30 min, and incubated with antibodies against γH2AX (#05-636, 1:2000; Millipore) and 53BP1 (NB100-134, 1:2000; Novus Biologicals) for 2 hours at room temperature or overnight at 4°C. Then, cells were incubated with species-specific Alexa488 or Alexa594-conjugated secondary antibodies (1:2000; Invitrogen) for 1 h at room temperature. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI, 1 μg/mL). Coverslips were mounted on slides in Vectashield (Vector Laboratories). Two-dimensional acquisitions were made with a microscope (TE2000, Nikon) equipped with a 40× immersion objective (1.3 numerical aperture) and a CCD camera (CoolSNAP ES, Photometrics). Images were acquired with MetaVue (Universal Imaging) and analyzed using ImageJ (National Institute of Health). At least 200 cells per experimental point were scored for presence of foci, and each experiment was repeated at least three times independently.
MiRNA mimic library screening
Human miRIDIAN miRNA mimic library (v10.1) was purchased from Dharmacon. U2OS cells were reversely transfected with 20 nM miRNA mimics in glass-bottom 96-well plates (#655892, Greiner Bio-one) using HiPerFect transfection reagent (Qiagen). After incubation for two days, cells were subjected to ionizing irradiation at 2 Gy, and processed as described above for immunofluorescent staining with mouse anti-γH2AX 1 h following irradiation. Images were captured with the automated Cellomics Array Scan microscope (Thermo Scientific), and processed for thresholding, background correction, and segmentation. DAPI staining was used to define the nucleus. Average number of γH2AX foci per nucleus was counted using the automated counter. The z-score value was calculated based on the formula Z= (X-μnc)/σ, where X was the score of individual sample, μnc was the mean of negative controls in each plate and σ was the standard deviation of the whole population. The screening was done three times. Average Z-scores from three independent screens were calculated. Z-scores below -2 or above 2 were considered significantly decreasing or increasingγH2AX foci, respectively.
Western blot analysis
Whole-cell extracts were obtained by direct lysis of cells in lysis buffer (0.05 mol/L Tris-HCl (pH 6.8), 2% SDS, 6% β-mercaptoethanol) boiled for 5 min. SDS-PAGE electrophoresis was done using NuPAGE 3% to 8% Tris-acetate or NuPAGE 4% to 12% Tris-glycine gels (Invitrogen), and proteins were transferred on nitrocellulose membranes. Mouse antibodies against γH2AX (1:2000), pATM1981 (#4526S, 1:2000, Cell Signaling), BRCA2 (Ab-1, 1:500, Calbiochem), Vinculin (V9131, 1:20000; Sigma), rabbit antibodies against histone H2AX (#07–627, 1:2000; Millipore), histone H3 (ab12079, 1:20000; Abcam), DNA-PK (ab32566, 1:2000; Abcam), ATM (#17995, 1:4000, GeneTex), pChk2 (#2661, 1:1000; Cell Signaling), Chk2 (#07–057, 1:500; Upstate), Actin (Sc-1616-R, 1:10000; Santa Cruz), and horseradish peroxidase-conjugated anti-mouse and anti-rabbit IgG (1:5,000; Amersham) were used. Chemiluminescence was used for detection (Perkin-Elmer Life Sciences). Films were digitalized using a standard scanner and images were processed using Photoshop CS (Adobe Systems, Inc.).
Real-time RT-PCR
Total RNAs were extracted using Trizol reagent (Invitrogen), chloroform extraction and isopropanol precipitation. Ten nanograms of RNA were reverse-transcribed using the Taqman miRNA Reverse Transcription Kit (Applied Biosystems) with a miRNA-specific primer. For H2AX gene expression, 100 ng of total RNA were reverse-transcribed with random hexamer. The Taqman MiRNA Assay Kit or Gene Expression Kit (Applied Biosystems) was used for quantitative PCR reaction on the AB-Prism 7900 (Applied Biosystems) in a 96-well plate according to the manufacturer’s instruction. The comparative Ct value was employed for quantification of transcripts. MiR-138 expression was normalized to the values of RNU24. H2AX expression was normalized to the values of 18S rRNA.
Luciferase assay
U2OS cells in 24-well plates were co-transfected with miR-neg or miR-138 and pGL3-control firefly luciferase vectors containing empty, wild-type or mutant H2AX 3′-UTR sequence. pRL-TK Renilla plasmid was also co-transfected as an internal control for transfection efficiency. Two days post-transfection, Firefly and Renilla luciferase activities were measured using the Dual-Luciferase Assay kit (Promega) according to the manufacturer’s instructions. Relative luciferase activity was calculated by normalizing the ratio of Firefly/Renilla luciferase to that of negative control-transfected cells.
Chromosomal breakage assay
Cells, at density of 60–80%, were irradiated with 2 Gy IR and then cultured for 15 h. After further incubation with 0.1 μg/ml colcemid (Sigma) for 3 h, cells were trypsinized and collected for metaphase chromosome preparation as described (19). 30 μl of the chromosome preparation was dropped on a pre-chilled microslide and let spread by gravity. Slides were stained by 1 μg/mL DAPI and mounted. Metaphase chromosome images were acquired using MetaVue software under a microscope (TE2000, Nikon) with a 100× oil immersion lens and a CCD camera (CoolSNAP ES, Photometrics). Seventy-five metaphase spreads from three independent experiments were analyzed for chromosome breaks, dicentric chromosomes, radial chromosomes and other abnormalities.
Homologous recombination (HR) assay
A U2OS cell clone stably expressing HR reporter direct repeat of GFP (DR-GFP) was a gift from Drs. Maria Jasin and Koji Nakanishi (20). This reporter consists of two differentially mutated GFP genes oriented as direct repeats. Expression of I-SceI endonuclease will generate a site-specific DSB within one of the mutated GFP genes, which, when repaired by gene conversion, will result in a functional GFP gene. Briefly, U2OS DR-GFP cells were sequentially transfected with siRNAs or miRNA mimics and pCBASce (a HA-tagged I-SceI expression vector). Two days later, cells were harvested, fixed in 2% paraformadehyde, and subjected to sequential immunostaining with mouse anti-HA (Covance) and Allophycocyanin donkey anti-mouse antibodies (Jackson ImmunoResearch). Flow cytometry analysis was performed in a Benchtop FACS Calibur Analyzer to determine the percentages of GFP-positive cells in HA-positive population, which resulted from HR repair induced by DSBs.
Cell cycle analysis
Cells were pulse labeled with 30 μmol/L 5-bromo-2′-deoxyuridine (BrdU, Sigma) for 15 min, washed twice with PBS, and fixed with 70% ice-cold ethanol. Cells were then stained for DNA content (propidium iodide) and BrdU incorporation with anti-BrdU rat monoclonal antibody (MAS250, Harlan Sera-Lab) followed by FITC-conjugated goat anti-rat antibody (Jackson ImmunoResearch). Flow cytometry analysis was then performed to determine the distribution of cell cycle.
Survival assay
Radiosensitivity and chemosensitivity were determined by crystal violet assay (21). Cells were seeded onto 12-well plates at 6 × 103 cells/well (radiosensitivity) or 2× 104/well (chemosensitivity) and irradiated with IR or treated with cisplatin, camptothecin (Sigma), paclitaxel (Sigma) or AZD2281 (Axon Medchem) at indicated doses. After incubation for 6 days (radiosensitivity) or 3 days (chemosensitivity), monolayers were fixed in 10% methanol containing 10% acetic acid. Adherent cells were stained with 0.5% crystal violet in methanol. The absorbed dye was resolubilized with methanol containing 0.1% SDS, which was transferred into 96-well plates and measured photometrically (595 nm) in a microplate reader. Cell survival was calculated by normalizing the absorbance to that of non-treated controls.
Statistical analysis
The student’s t-test was used to evaluate the significant difference between two groups of data in all in vitro experiments. The Pearson correlation test was used to analyze the correlation between H2AX and miR-138 expression in GBM samples. A P-value < 0.05 was considered significant.
Results
MiR-138 downregulates IR-induced histone H2AX phosphorylation and foci formation
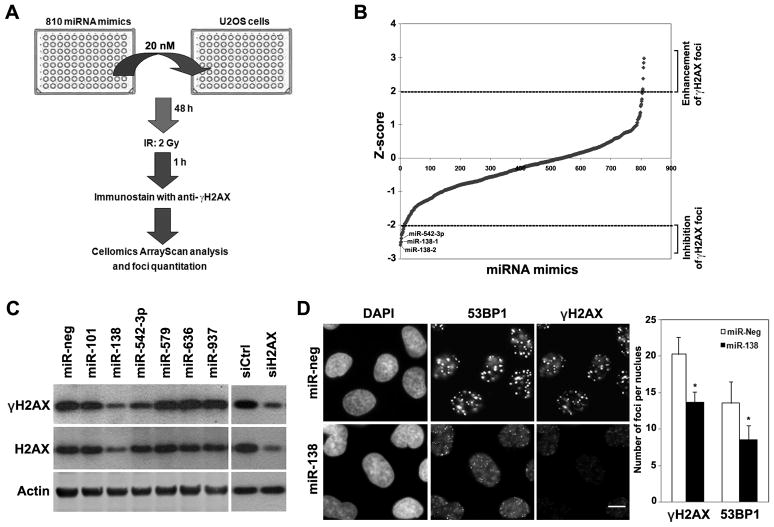
Initially, we performed cell-based screening using IR-induced γH2AX foci formation as readout. A library of 810 human miRNA mimics was used to individually transfect each miRNA into U2OS cells in 96-well format, followed by IR treatment (2 Gy). One hour after IR treatment, γH2AX foci were quantitated with an automated high-throughput fluorescence microscope equipped with an automatic foci counter (Fig. 1A), based on our observation of rapid induction of γH2AX foci after IR (Supplemental Fig. S1A). Several miRNA mimics either reduced (Z-score < −2) or increased (Z-score > 2) IR-induced γH2AX foci (Fig. 1B and Supplemental Table 1). Validation of the miRNA mimics that inhibited γH2AX foci formation showed that only miR-138 and miR-542-3p reproducibly reduced the γH2AX foci formation (Fig. 1D and Fig. S1B). In addition, both miR-138 and miR-542-3p substantially reduced γH2AX protein abundance (Fig. 1C). We focused on miR-138 in the following studies, because miR-138 had the most robust effect on γH2AX level and foci formation. The expression levels of miR-138 in U2OS cells transfected with miR-138 mimics were validated by real-time RT-PCR (Supplemental Fig. S2A). Consistent with the role of γH2AX in regulating 53BP1 foci formation, overexpression of miR-138 also inhibited IR-induced 53BP1 foci formation (Fig. 1D).
Figure 1. Identification of miRNAs that regulate γH2AX foci formation.
(A) Scheme of miRNA gain-of-function screening for IR-induced γH2AX foci formation. (B) Z-score plot of miRNA mimics in the regulation of IR-induced γH2AX foci formation. Several miRNA mimics were identified to positively or negatively affect γH2AX foci formation (Z-score >2 or <−2). (C) U2OS cells were transfected with indicated miRNA mimics at 20 nM. Forty-eight hours post transfection, cells were irradiated with 2 Gy IR and collected for western blot analysis of γH2AX and H2AX 1 hour after IR. Immunoblots for actin served as loading controls. (D) U2OS cells were transfected with negative control (miR-neg) or miR-138 mimics, irradiated with 2 Gy IR and then fixed for immunofluorescent staining of γH2AX and 53BP1 1 hour after IR. The number of foci per nucleus was quantitated (mean ± SD, n=3). Scale bar, 20 μm.
MiR-138 negatively regulates the expression of histone H2AX
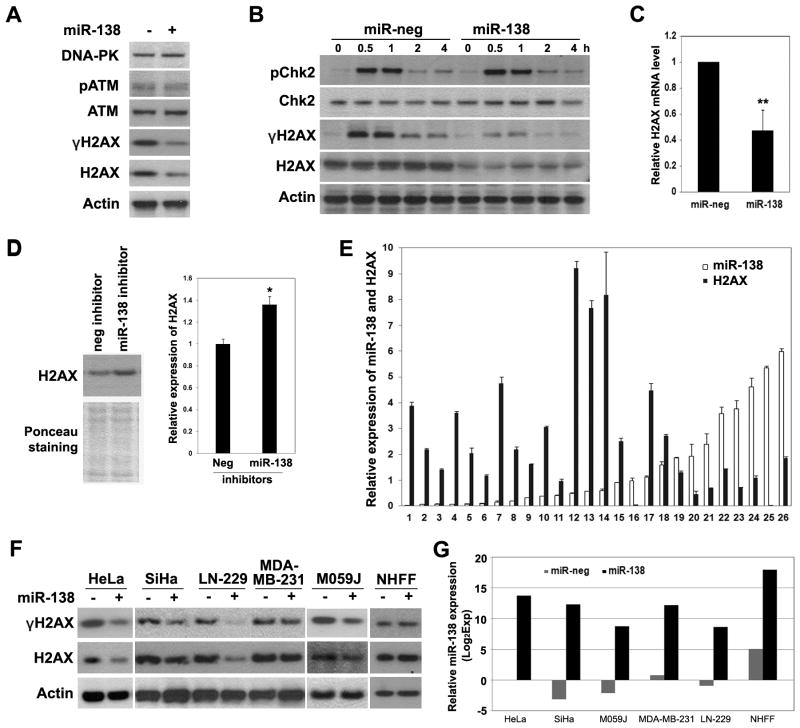
ATM and DNA-PK are the major kinases that phosphorylate H2AX after IR (22), but overexpression of miR-138 had no effect on the expression of ATM and DNA-PK in U2OS cells (Fig. 2A). In addition, the phosphorylation of ATM and CHK2, another important substrate of ATM, was unaffected in miR-138-overexpressing cells (Fig. 2A and 2B), suggesting an intact ATM activation. In contrast, miR-138 overexpression significantly downregulated H2AX protein (Fig. 1C, 2A and 2B) and mRNA levels (Fig. 2C), indicating that miR-138 decreases γH2AX mainly through downregulation of the expression of H2AX itself. We also transfected U2OS cells with plasmids containing miR-138 genomic sequence from either of the two genomic loci from which it is encoded, miR-138-1 (3p21) and miR-138-2 (16q13). Both constructs produced mature miR-138 in U2OS cells (Supplemental Fig. S2B), and reduced H2AX phosphorylation and expression (Supplemental Fig. S2C).
Figure 2. MiR-138 negatively regulates the expression of histone H2AX expression.
(A, B) U2OS cells transfected with miR-neg or miR-138 mimics (20 nM) were irradiated with 2 Gy IR, and harvested at 1 h (A) or indicated time points (B) for western blot analysis using indicated antibodies. (C) Expression levels of H2AX mRNA in miR-neg or miR-138-transfected U2OS cells were determined by qRT-PCR (mean ± SD, n=3) and normalized to the values of 18S rRNA. (D) U87 cells were transfected with miR-138 inhibitors for 72 hours and harvested for western blot analysis. Quantitation of H2AX level was done with Image J software. (E) The expression levels of miR-138 and H2AX in RNA samples from glioblastoma tissues were quantitated with qRT-PCR and normalized to the levels of RNU24 and 18S rRNA. (Pearson correlation test, r=−0.38, p = 0.053 (two-tailed)). (F, G) Multiple cancer cell lines and normal human foreskin fibroblasts (NHFF) were transfected with miR-neg or miR-138 mimics (20 nM), irradiated with 2Gy IR and harvested for western blotting and qRT-PCR analyses 1 hour after IR. MiR-138 expression was assayed with qRT-PCR and normalized to that of negative control (miR-neg) transfected HeLa cells. * P < 0.05, ** P < 0.01
Since miR-138 is highly expressed in brain tissues (37), we then examined the expression of miR-138 in 10 glioma cell lines. Normal brain tissue and several glioma cell lines expressed higher level of miR-138 compared to U2OS cells (Supplemental Fig. S3). U87 cells with the highest expression of miR-138 had very low level of H2AX protein expression (Supplemental Fig. S3). Inhibition of miR-138 with antisense inhibitor mildly but reproducibly increased H2AX protein level in U87 cells (Fig. 2D), suggesting that miR-138 is one of the negative regulators of H2AX expression at physiological level. Consistently, a marginally significant inverse correlation between miR-138 and H2AX mRNA levels was observed in 26 glioblastoma tissue samples (Pearson correlation test, r=−0.38, p = 0.053 (two-tailed)) (Fig. 2E). In addition, overexpression of miR-138 also significantly reduced γH2AX and H2AX in two glioma cell lines M059J and LN229 with low miR-138 expression and two cervical cancer cell lines HeLa and SiHa (Fig. 2F and 2G), but had little effect in normal human foreskin fibroblasts and a breast cancer cell line, MDA-MB-231 (Fig. 2F). Thus, the regulatory effect of miR-138 on H2AX is not limited to U2OS cells.
Histone H2AX is a direct target of miR-138
The miRNA target prediction algorithm TargetScan 5.0 predicted that the 3′-UTR of H2AX mRNA contains one putative miR-138 binding site (Fig. 3A). This potential binding site and the flanking sequences are highly conserved across mammals including human, monkey, mouse and rat (Supplemental Fig. S4). To determine whether miR-138 regulates H2AX through binding to its 3′-UTR, we inserted the H2AX 3′-UTR behind the 3′ end of the luciferase gene of pGL3 vector (H2AX 3′-UTR WT) and transfected either this construct or the parent luciferase expression vector, together with miR-138 mimic or negative control mimic, into U2OS cells. MiR-138 mimic significantly downregulated the luciferase activity of the construct in which the luciferase gene was fused with the H2AX 3′-UTR, while it had no significant effect on the parent luciferase expression vector (Fig. 3B). Mutation of the predicted binding site of miR-138 within H2AX 3′-UTR (H2AX 3′-UTR mut) abolished the inhibitory effect of miR-138 on luciferase activity (Fig. 3B). In addition, overexpression of miR-138 by transfection of constructs expressing miR-138-1 or miR-138-2, the two genomic sequences from which miR-138 is transcribed, had similar effects on the luciferase activity of H2AX 3′-UTR-containing vector (Supplemental Fig. S5). Furthermore, in U2OS cells expressing Flag-tagged H2AX cDNA lacking its native 3′-UTR, overexpression of miR-138 could not reduce the expression of exogenous Flag-H2AX (Fig. 3C, lane 1 vs. lane 2, upper band), while it downregulated endogenous H2AX expression (Fig. 3C, lane 1 vs. lane2 and lane 3 vs. lane 4, lower band). Taken together, these data suggest that H2AX 3′-UTR is a direct target of miR-138.
Figure 3. H2AX is a direct target of miR-138.
(A) Sequences of miR-138 and the predicted miR-138 binding site at H2AX 3′-UTR (wild-type (WT) and mutant (mut), 921–944 nt of the H2AX 3′-UTR). (B) Wild-type or mutant H2AX 3′-UTR (position 19 to 1046) were cloned into the pGL3 vector, as 3′ fusions to the luciferase gene. U2OS were co-transfected with the indicated miRNA mimics and luciferase vectors. Luciferase activity was assayed 48 h later and normalized to that of negative control-transfected cells (mean ± SD, n=3). (C) U2OS cells stably transfected with empty vector or Flag-H2AX construct were transfected with miR-neg or miR-138. Expression levels of H2AX were then determined by western blotting. The immunoblot for histone H3 served as a loading control. * P < 0.05
MiR-138 reduces genomic stability and HR repair
Since miR-138 downregulated γH2AX, we asked whether miR-138 modulates genomic stability. U2OS cells were exposed to IR (2Gy) and then allowed to recover and arrest at the first metaphase following IR. MiR-138-overexpressing or H2AX-knockdown cells had a significantly higher amount of chromosome breaks compared to negative control transfected cells after IR (Fig. 4A and 4B). Thus, overexpression of miR-138 resulted in increased genomic instability after DNA damage.
Figure 4. MiR-138 affects genomic stability and homologous recombination repair.
(A–B) U2OS cells were transfected with miRNA mimics or H2AX siRNA, irradiated with 2 Gy IR, allowed to recover for 15 hours and then arrested at metaphase. Cells were harvested for metaphase spreading and analysis of chromosome abnormalities. (A) A representative metaphase spread image of U2OS cells overexpressing miR-138. Arrows point out the chromosome abnormalities as indicated. (B) Statistical analysis of chromosome abnormalities (mean ± SD, n=3). Other abnormalities included radial-, dicentric and rejoining chromosomes. (C–F) U2OS DR-GFP cells were sequentially transfected with miRNA mimics or siRNAs and the pBASceI plasmid. Cells were incubated for 48 h, fixed and stained for HA-I-SceI expression and analyzed by FACS of GFP-positive cells in the HA-positive population (C, D) (mean ± SD, n=4), for western blot analysis of BRCA2 and H2AX levels (E), and for cell cycle analysis (F). * P < 0.05, *** P < 0.001
H2AX knockout cells are deficient in HR (23) and sensitive to IR (9). Phosphorylation of H2AX on S139 is essential for the HR activity of H2AX (23). Therefore, we examined the effect of miR-138 on HR repair activity by monitoring the percentage of GFP-positive cells in U2OS DR-GFP cells, where the acquisition of GFP expression reflects HR activity (24). While depletion of BRCA2, a crucial protein for HR (25), substantially reduced the HR efficiency (Fig. 4C, 16% vs 4%), overexpression of miR-138 more moderately but significantly (p<0.05) reduced the HR efficiency (16% vs. 10%), producing a reduction similar to that observed with knockdown of H2AX (16% vs. 9.5%) (Fig. 4C, 4D and 4E). The data are consistent with previous reports showing that depletion of H2AX by siRNA causes a mild HR defect (26). In addition, overexpression of miR-138 had no strong effect on cell cycle distribution in both U2OS DR-GFP cells (Fig. 4F) and the parental U2OS cells (Supplemental Fig. S6A), although it modestly inhibited cell growth (Supplemental Fig. S6B). Thus, the HR defect in miR-138-transfected cells was not due to an alteration of the cell cycle and may contribute to the observed genomic instability.
MiR-138 enhances cellular sensitivity to DNA damaging agents
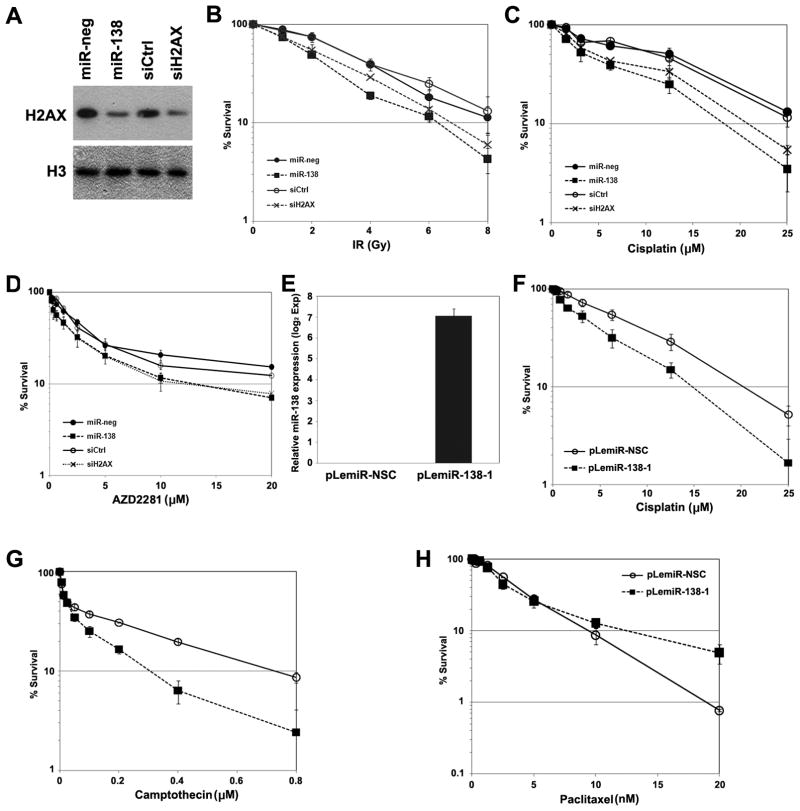
Next we examined the effect of modulating miR-138 on sensitivity of U2OS cells to DNA damaging agents. H2AX siRNA-transfected and miR-138 mimic-transfected U2OS cells were both mildly sensitive to IR compared to control cells (Fig. 5A and 5B), consistent with the role of H2AX in DSB repair and radiosensitivity (9). HR-deficient cells are more sensitive than HR-proficient cells to cisplatin and poly(ADP-ribose) polymerase (PARP) inhibitors (27–29). Consistent with this, H2AX siRNA-transfected U2OS cells and miR-138 mimic-transfected U2OS cells were sensitized to cisplatin (Fig. 5C) and a PARP inhibitor, AZD2281 (Fig. 5D).
Figure 5. MiR-138 sensitizes cells to DNA damaging agents.
(A–C) U2OS cells were transfected with miRNA mimics or siRNAs as indicated. Cells were reseeded for western blot analysis of H2AX level (A), IR (B), cisplatin (C) and AZD2281 (D) sensitivity assays (mean ± SEM, n=4). (E–H) U2OS cells were infected with lentivirus producing non-target small RNA control (pLemiR-NSC) or miR-138 (pLemiR-138-1). Cells were harvested for quantitation of miR-138 level using real-time PCR (E) or seeded for chemosensitivity assays as indicated (F–H) (mean ± SEM, n=3).
Delivery of miR-138 using lentivirus expressing miR-138-1 from genomic sequence (pLemiR-138-1) efficiently produced miR-138 in U2OS cells (Fig. 5E) and also rendered cells sensitive to both cisplatin and another DNA damaging agent, camptothecin, compared to cells infected with control lentivirus (pLemiR-NSC) (Fig. 5F and 5G). Furthermore, pLemiR-138-1 also sensitized two glioblastoma cell lines, LN229 and M059J, to cisplatin and camptothecin (Supplemental Fig. S7). In contrast, pLemiR-138-1 did not sensitize U2OS cells to paclitaxel (Fig. 5H), an agent that inhibits mitosis by stabilizing microtubules (30), suggesting that miR-138-mediated sensitization to DNA damage agents was not a result of general cellular toxicity.
Importantly, reintroduction of H2AX lacking 3′-UTR (Fig. 6A) significantly reduced the extent of miR-138-mediated sensitization to both cisplatin and camptothecin in miR-138 overexpressing cells (Fig. 6B and 6C), while it had a minimal effect on drug resistance in the control cells. These findings suggest that miR-138 sensitizes cells to cisplatin and camptothecin mostly by downregulating H2AX.
Figure 6. Reintroduction of histone H2AX attenuates sensitivity of miR-138 overexpressing cells to cisplatin and camptothecin.

(A–C) U2OS cells stably expressing empty vector or Flag-H2AX were infected with lentivirus expressing non-targeting miRNA or miR-138 and seeded for western blot analysis of H2AX level (A), cisplatin (B) and camptothecin (C) sensitivity (mean ± SEM, n=3). The immunoblot for histone H3 served as a loading control.
Discussion
We identified miR-138 as a potent negative regulator of γH2AX foci formation. MiR-138 overexpression directly targeted the H2AX 3′-UTR, reduced H2AX expression, induced genomic instability after DNA damage and sensitized cells to DNA damaging agents. Other groups have also reported that miRNAs regulate DNA damage response. For example, miR-421 induces an S-phase checkpoint defect by suppressing ATM expression (15). MiR-16 controls Cdc25A and Wip1 phosphatases after DNA damage, thereby regulating the ATM/ATR pathway (16, 17). MiR-24 is upregulated during post-mitotic differentiation of hematopoietic cells and directly downregulates H2AX expression to inhibit DNA damage response and enhance chemosensitivity (14). However, miR-24 was not a strong hit in our miRNA library screening using U2OS cells (Z score, −0.515 and −1.216) (Supplemental Table 1 and Fig. S8). This discrepancy might be due to different cell lines used in these studies. Function and targets of miRNAs can be context dependent (31), and accordingly, miR-138 had little effect on H2AX level in normal human fibroblasts and MDA-MB-231 cancer cells. The mechanism of this context dependency is an important topic for future studies. Nevertheless, all these studies support the notion that DNA damage response is controlled by miRNAs, which may ultimately modulate genome stability and tumorigenesis.
Expression levels of many miRNAs are altered in response to DNA damage (16, 32–35). MiR-138 level is not significantly altered upon IR treatment for up to 24 h in IM9 B-cell lyphoblastoma cells (34, 35), or A549 non-small cell lung cancer cells (34, 35). MiR-138 level was only slightly reduced in U2OS cells after IR (Supplemental Fig. S9). In contrast, Weidhaas et al found that miR-138 level was rapidly induced in response to IR in A549 cells and a HPV E6/E7-transformed lung epithelial cell line, CRL-2741 (33). This discrepancy might be due to the dosage of IR and/or cell lines used by different groups. Interestingly, miR-138 is downregulated in a multidrug-resistant leukemia cell line HL-60/VCR compared with parental HL-60 cells, which is essential for the resistance to vincristine and cisplatin (36). In line with our findings, this study highlights the important role of miR-138 in the development of drug resistance although whether H2AX is involved in the drug resistance of HL-60/VCR is not known.
MiR-138 is broadly conserved among vertebrates (37). It is highly expressed in brain tissues and is involved in the development of several organs (38–40). Two genomic loci encoding miR-138 (miR-138-1 and miR-138-2) have been identified. The miR-138-1 gene is located at chromosome 3p21, where allelic loss is frequently detected in nasopharyngeal cancer (41, 42). MiR-138 is frequently downregulated in several cancer types. MiR-138 is downregulated in anaplastic thyroid carcinoma, and downregulation of miR-138 is correlated with upregulation of hTERT (43). MiR-138 is also downregulated in metastatic head and neck squamous cell carcinoma (44, 45) and in hepatocellular carcinoma (46). Ectopic expression of miR-138 in head and neck squamous cell carcinoma cells leads to cell cycle arrest, apoptosis, and suppresses cell invasion (44, 45). However, given that in our work miR-138 had no significant effect on the cell cycle distribution of U2OS cells, although it mildly inhibited cell proliferation, the effect of miR-138 overexpression on cell cycle appears context-dependent. MiR-138-mediated cell growth inhibition may be a result of defective DNA repair due to downregulation of H2AX. Indeed, H2AX deficient mouse embryonic fibroblasts exhibit impaired growth (10). On the other hand, loss of one H2AX allele enhances tumor susceptibility in mice in the absence of p53 (47). Therefore, miR-138 overexpression may promote malignant transformation due to genomic instability caused by H2AX downregulation, or may inhibit tumor progression by slowing down proliferation. The fact that miR-138 is frequently downregulated in several types of cancer suggests that downregulation of miR-138, which can lead to increased cell proliferation and invasion, may be advantageous for cancer cell survival and growth.
Considering the fact that one individual miRNA can target multiple oncogenic pathways, restoration of miRNAs downregulated in cancer has attracted a lot of attention as a potential therapeutic option (48, 49). Our study suggests that overexpression of miR-138 will sensitize tumor cells to DNA damaging agents and that miR-138 is a potential therapeutic agent for cancer treatment. Future study will address the possibility that miR-138 mimic may be useful as a chemosensitizer/radiosensitizer in therapy for certain types of cancer.
Supplementary Material
Acknowledgments
Financial support: Howard Hughes Medical Institute, the National Institutes of Health/National Heart, Lung and Blood Institute (R21 HL092978 to T.T.), Fanconi Anemia Research Fund (to T.T.), Fred Hutchinson Cancer Research Center and Listwin Family Foundation funding (New Development funds to M.T.). Y.W. is a research fellow supported by Canadian Institute of Health Research. J.W. is supported by PHS NRSA 2T32 GM007270 from NIGMS.
We thank Drs. Maria Jasin, Koji Nakanishi, Guangwei Du, Matthew Fero and Denise Galloway for reagents, Drs. Ryo Nishikawa and David James for clinical samples, the Imaging Facility of Fred Hutchinson Cancer Research Center for screening, and Dr. German Gomez and members of Taniguchi Lab and Tewari Lab for technical support and discussions.
Footnotes
Potential conflicts of interest: none.
References
- 1.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–9. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 2.Lobrich M, Shibata A, Beucher A, et al. gammaH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle. 2010;9:662–9. doi: 10.4161/cc.9.4.10764. [DOI] [PubMed] [Google Scholar]
- 3.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–68. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 4.Dickey JS, Redon CE, Nakamura AJ, Baird BJ, Sedelnikova OA, Bonner WM. H2AX: functional roles and potential applications. Chromosoma. 2009;118:683–92. doi: 10.1007/s00412-009-0234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 Directly Binds Phosphorylated Histone H2AX to Regulate Cellular Responses to DNA Double-Strand Breaks. Cell. 2005;123:1213–26. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 6.Lukas C, Melander F, Stucki M, et al. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. Embo J. 2004;23:2674–83. doi: 10.1038/sj.emboj.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lou Z, Minter-Dykhouse K, Franco S, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 8.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–16. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bassing CH, Chua KF, Sekiguchi J, et al. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc Natl Acad Sci U S A. 2002;99:8173–8. doi: 10.1073/pnas.122228699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Celeste A, Petersen S, Romanienko PJ, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–7. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.Trang P, Weidhaas JB, Slack FJ. MicroRNAs as potential cancer therapeutics. Oncogene. 2008;27 (Suppl 2):S52–7. doi: 10.1038/onc.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28:369–78. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 14.Lal A, Pan Y, Navarro F, et al. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat Struct Mol Biol. 2009;16:492–8. doi: 10.1038/nsmb.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu H, Du L, Nagabayashi G, Seeger RC, Gatti RA. ATM is down-regulated by N-Myc-regulated microRNA-421. Proc Natl Acad Sci U S A. 2010;107:1506–11. doi: 10.1073/pnas.0907763107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pothof J, Verkaik NS, van IW, et al. MicroRNA-mediated gene silencing modulates the UV-induced DNA-damage response. EMBO J. 2009;28:2090–9. doi: 10.1038/emboj.2009.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Wan G, Mlotshwa S, et al. Oncogenic Wip1 phosphatase is inhibited by miR-16 in the DNA damage signaling pathway. Cancer Res. 2010;70:7176–86. doi: 10.1158/0008-5472.CAN-10-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakai W, Swisher EM, Karlan BY, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–20. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S, Kuhne WW, Kulharya A, et al. Involvement of p54(nrb), a PSF partner protein, in DNA double-strand break repair and radioresistance. Nucleic Acids Res. 2009;37:6746–53. doi: 10.1093/nar/gkp741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weinstock DM, Nakanishi K, Helgadottir HR, Jasin M. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 2006;409:524–40. doi: 10.1016/S0076-6879(05)09031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taniguchi T, Garcia-Higuera I, Xu B, et al. Convergence of the Fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;109:459–72. doi: 10.1016/s0092-8674(02)00747-x. [DOI] [PubMed] [Google Scholar]
- 22.Stiff T, O’Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–6. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 23.Xie A, Puget N, Shim I, et al. Control of sister chromatid recombination by histone H2AX. Mol Cell. 2004;16:1017–25. doi: 10.1016/j.molcel.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang F, Romanienko PJ, Weaver DT, Jeggo PA, Jasin M. Chromosomal double-strand break repair in Ku80-deficient cells. Proc Natl Acad Sci U S A. 1996;93:8929–33. doi: 10.1073/pnas.93.17.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moynahan ME. The cancer connection: BRCA1 and BRCA2 tumor suppression in mice and humans. Oncogene. 2002;21:8994–9007. doi: 10.1038/sj.onc.1206177. [DOI] [PubMed] [Google Scholar]
- 26.Yuan J, Chen J. MRE11-RAD50-NBS1 complex dictates DNA repair independent of H2AX. J Biol Chem. 2010;285:1097–104. doi: 10.1074/jbc.M109.078436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raschle M, Knipscheer P, Enoiu M, et al. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–80. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bryant HE, Helleday T. Inhibition of poly (ADP-ribose) polymerase activates ATM which is required for subsequent homologous recombination repair. Nucleic Acids Res. 2006;34:1685–91. doi: 10.1093/nar/gkl108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lord CJ, McDonald S, Swift S, Turner NC, Ashworth A. A high-throughput RNA interference screen for DNA repair determinants of PARP inhibitor sensitivity. DNA Repair (Amst) 2008;7:2010–9. doi: 10.1016/j.dnarep.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 30.Horwitz SB. Mechanism of action of taxol. Trends Pharmacol Sci. 1992;13:134–6. doi: 10.1016/0165-6147(92)90048-b. [DOI] [PubMed] [Google Scholar]
- 31.Liu H, Kohane IS. Tissue and process specific microRNA-mRNA co-expression in mammalian development and malignancy. PLoS One. 2009;4:e5436. doi: 10.1371/journal.pone.0005436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009;69:1221–9. doi: 10.1158/0008-5472.CAN-08-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weidhaas JB, Babar I, Nallur SM, et al. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007;67:11111–6. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cha HJ, Shin S, Yoo H, et al. Identification of ionizing radiation-responsive microRNAs in the IM9 human B lymphoblastic cell line. Int J Oncol. 2009;34:1661–8. doi: 10.3892/ijo_00000297. [DOI] [PubMed] [Google Scholar]
- 35.Shin S, Cha HJ, Lee EM, et al. Alteration of miRNA profiles by ionizing radiation in A549 human non-small cell lung cancer cells. Int J Oncol. 2009;35:81–6. [PubMed] [Google Scholar]
- 36.Zhao X, Yang L, Hu J, Ruan J. miR-138 might reverse multidrug resistance of leukemia cells. Leuk Res. 34:1078–82. doi: 10.1016/j.leukres.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. Rna. 2006;12:1161–7. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morton SU, Scherz PJ, Cordes KR, Ivey KN, Stainier DY, Srivastava D. microRNA-138 modulates cardiac patterning during embryonic development. Proc Natl Acad Sci U S A. 2008;105:17830–5. doi: 10.1073/pnas.0804673105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siegel G, Obernosterer G, Fiore R, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–16. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Z, Bian C, Zhou H, et al. MicroRNA hsa-miR-138 inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells through EID-1. Stem Cells Dev. 2010 doi: 10.1089/scd.2010.0072. on line. [DOI] [PubMed] [Google Scholar]
- 41.Ko JY, Lee TC, Hsiao CF, et al. Definition of three minimal deleted regions by comprehensive allelotyping and mutational screening of FHIT, p16(INK4A), and p19(ARF) genes in nasopharyngeal carcinoma. Cancer. 2002;94:1987–96. doi: 10.1002/cncr.10406. [DOI] [PubMed] [Google Scholar]
- 42.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitomo S, Maesawa C, Ogasawara S, et al. Downregulation of miR-138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Sci. 2008;99:280–6. doi: 10.1111/j.1349-7006.2007.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X, Jiang L, Wang A, Yu J, Shi F, Zhou X. MicroRNA-138 suppresses invasion and promotes apoptosis in head and neck squamous cell carcinoma cell lines. Cancer Lett. 2009;286:217–22. doi: 10.1016/j.canlet.2009.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang L, Liu X, Kolokythas A, et al. Downregulation of the Rho GTPase signaling pathway is involved in the microRNA-138-mediated inhibition of cell migration and invasion in tongue squamous cell carcinoma. Int J Cancer. 2010;127:505–12. doi: 10.1002/ijc.25320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding J, Huang S, Wu S, et al. Gain of miR-151 on chromosome 8q24. 3 facilitates tumour cell migration and spreading through downregulating RhoGDIA. Nat Cell Biol. 2010;12:390–9. doi: 10.1038/ncb2039. [DOI] [PubMed] [Google Scholar]
- 47.Celeste A, Difilippantonio S, Difilippantonio MJ, et al. H2AX Haploinsufficiency Modifies Genomic Stability and Tumor Susceptibility. Cell. 2003;114:371–83. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70:7027–30. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petrocca F, Lieberman J. Promise and Challenge of RNA Interference-Based Therapy for Cancer. J Clin Oncol. 2011;29:747–54. doi: 10.1200/JCO.2009.27.6287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.