Abstract
Ovarian cancer is the most lethal gynecologic malignancy with a five-year survival rate below 25% for patients with stage III–IV disease. Identifying key mediators of ovarian cancer invasion and metastasis is critical to the development of more effective therapeutic interventions. We previously identified Filamin A interacting protein 1-like (FILIP1L) as an important mediator of cell proliferation and migration. In addition, targeted expression of FILIP1L in tumors inhibited tumor growth in vivo. In our present study, we confirmed that both mRNA and protein expression of FILIP1L were down-regulated in ovarian cancer cells compared to normal ovarian epithelial cells. FILIP1L expression was inversely correlated with the invasive potential of ovarian cancer cell lines and clinical ovarian cancer specimens. We also provide evidence that DNA methylation is a mechanism by which FILIP1L is down-regulated in ovarian cancer. The CpG island in the FILIP1L promoter was heavily methylated in ovarian cancer cells. Methylation status of the FILIP1L promoter was inversely correlated with FILIP1L expression in ovarian cell lines and clinical ovarian specimens. Reduced methylation in the FILIP1L promoter following treatment with a DNA demethylating agent was associated with restoration of FILIP1L expression in ovarian cancer cells. A transcription activator, CREB was shown to bind to the CREB/ATF site in the CpG island of the FILIP1L promoter. Overall, these findings suggest that down-regulation of FILIP1L associated with DNA methylation is related with the invasive phenotype in ovarian cancer and that modulation of FILIP1L expression has the potential to be a target for ovarian cancer therapy.
Keywords: Ovarian cancer, FILIP1L, DNA methylation, Invasion, Cancer therapy
Introduction
Ovarian cancer is the second most common gynecologic malignancy but the most common cause of gynecologic cancer-related death. Most patients are diagnosed with advanced stage disease and the five-year survival rate is below 25% for stage III–IV disease (1). The standard cytotoxic systemic therapies following surgical cytoreduction are carboplatin and paclitaxel (www.nccn.org). Cure rates for this approach are less than 40% with overall survival averaging 50 months. Recurrences are common and therefore, gynecologic oncologists often enroll patients in clinical trials for platinum resistant disease recurrence. Researchers are making modest advances in the development of targeted therapy to improve the overall survival in ovarian cancer (2). A better understanding of the pathways involved in ovarian cancer invasion and spread are critical to the development of more effective therapies, especially for patients with advanced or refractory disease.
We identified Filamin A interacting protein 1-like (FILIP1L; previously known as down-regulated in ovarian cancer 1) as a mediator of cellular proliferation and migration in both normal and tumor cells (3, 4). We demonstrated that overexpression of FILIP1L resulted in inhibition of cell proliferation and migration and increased apoptosis (5). In addition, targeted expression of FILIP1L in tumors inhibited tumor growth in vivo (5). Overall, these findings suggested that the novel protein FILIP1L may be an important mediator of cell growth and migration and may play a role in defining the invasive potential of tumor cells.
FILIP1L mRNA was originally characterized by its presence in human ovarian surface epithelial (HOSE) cells and its absence in ovarian carcinoma cells (6). Using cDNA microarray analysis, FILIP1L was shown to be preferentially expressed in HOSE cells as opposed to ovarian carcinoma cells from patients with Stage IIIC and IV disease (7). Additionally, differential gene expression analysis revealed that the FILIP1L gene in ovarian cancer has several tagging single nucleotide polymorphisms (tSNPs) (8). FILIP1L was shown to be one of nine genes associated with functional suppression of tumorigenicity in ovarian cancer cell lines (9).
Based on these observations, we asked whether FILIP1L expression was inversely correlated with the degree of invasive potential or aggressive histologic morphology and behavior of ovarian cancer. Further, since epigenetic aberrations are intimately associated with ovarian tumorigenesis (10, 11), we examined whether or not the control of FILIP1L expression was mediated through epigenetic mechanisms.
In the present study, we demonstrate that cellular invasion and aggressive histology and behavior are inversely correlated with FILIP1L gene and protein expression in ovarian cell lines and clinical ovarian samples. We observed that overexpression of FILIP1L inhibited the invasive potential of an aggressive ovarian cancer cell line. We conclude that DNA methylation is a mechanism by which FILIP1L is down-regulated in ovarian cancer and that the DNA methylation status of the FILIP1L promoter is inversely correlated with FILIP1L expression in ovarian cell lines as well as clinical ovarian specimens. Taken together, these data suggest that the degree of FILIP1L expression may be a predictor of ovarian cancer behavior and further, the modulation of FILIP1L expression in ovarian cancer may be a useful target for the development of novel ovarian cancer therapies.
Materials and Methods
Cell culture
Normal ovaries were obtained from the operating room under an Institutional Review Board (IRB) exemption at Montefiore Medical Center, NY and human ovarian surface epithelial (HOSE) cells were cultured following a published protocol (12). Immortalized normal ovarian surface epithelial (IOSE) cells including IOSE 144, 523 and 385 were provided by the Canadian Ovarian Tissue Bank and cultured in M199:MCDB105 (1:1) containing 5% fetal bovine serum (FBS). Human ovarian cancer cell lines including ES2, SKOV3 and OV90 were purchased from the American Type Culture Collection (Manassas, VA). ES2 and SKOV3 were cultured in McCoy’s 5a containing 10% FBS and OV90 was cultured in M199:MCDB105 (1:1) containing 10% FBS. OCC1 was purchased from MD Anderson Cancer Center, TX and was cultured in RPMI 1640 containing 10% FBS. OVCAR8 was cultured in RPMI 1640 containing 10% FBS. OVCA429 was provided by Dr. Barbara Vanderhyden, University of Ottawa, Ontario and was cultured in αMEM containing 10% FBS and 1% nonessential amino acids.
Clinical specimens
Formalin-fixed paraffin-embedded (FFPE) tissue blocks of ovarian serous borderline tumor (n=10) and ovarian serous carcinoma (n=17) were obtained from Pathology at Montefiore Medical Center, NY under our IRB exemption. The criteria for non-invasive serous borderline tumor were tumor composed of arborizing papillary structures lined by a proliferating population of serous epithelial cells, which show stratification, cytologic atypia, mitotic activity and tufting of cells (destructive invasion is not present). The criteria for invasive serous carcinoma were tumors that showed all of the above with destructive or frank stromal invasion and usually with marked cytologic atypia. Three different regions containing more than 90% tumor cells were scraped from the tissue section of each specimen (7 μm). Genomic DNA and total RNA were purified from the same scraped tissue using the AllPrep DNA/RNA FFPE kit (Qiagen). Real-time RT-PCR and Sequenom® EpiTYPER Mass Array were performed as described in the following sections.
5-Aza-2′-deoxycytidine and Trichostatin A treatment
Ovarian cancer cells were seeded in 6-well plates at a density of 1 × 105 cells per well 16 h before treatment. Cells were treated with 5-aza-2′-deoxycytidine (DAC; Sigma-Aldrich) daily for 72 h or with Trichostatin A (TSA; Sigma-Aldrich) once for 24 h.
Transfection of Cells with FILIP1L plasmids
Cloning of FILIP1LΔC103 (amino acid 1–790) was described previously (5). Plasmids were purified using Endo-free maxiprep kit (Qiagen). ES2 and SKOV3 cells were transfected with equimolar amounts of either control empty plasmid or plasmid encoding FILIP1LΔC103 using X-fect solution following the manufacturer’s protocols (Clontech). 24 h after transfection, the cells were subjected to cell invasion assay and western blot analysis.
Quantitative real-time RT-PCR
HOSE cells, IOSE cells and ovarian cancer cells, either untreated or DAC- and TSA-treated, were cultured and harvested at ~80% confluence. Total RNA was prepared by RNeasy kit (Qiagen), and cDNA was prepared by Superscript II reverse transcriptase (Invitrogen). qPCR was performed using ABI 7900HT SDS real-time PCR instrument as per manufacturer (Applied Biosystems). Expression of the FILIP1L gene was normalized to hRPL7 gene expression. The primers used were 5′-AACGCTGGTATCATGGCTGAA-3′ and 5′-ATCTCTGCACTGCTCCTCCATT-3′ for FILIP1L (probe A); 5′-AAGAAGCGAATTGCTTTGACAGA-3′ and 5′-CAAATCCTCCATGCAGATGATG-3′ for hRPL7. For FFPE tissue specimens, another set of FILIP1L probe (probe B; 5′-GGCAGTTCAGATTAAAGAAGCTAATTG-3′ a n d 5′-TGGTGACCCTTTTCTCCTTTTC-3′) was also used.
Western blot
Whole cell lysates were prepared from radioimmunoprecipitation assay (RIPA) buffer, separated on SDS-PAGE and transferred to nitrocellulose membrane. The membranes were blotted with antibodies against FILIP1L (5) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Chemicon) followed by incubation with anti-mouse antibody conjugated to horseradish peroxidase. The signal was detected using chemiluminescence (Millipore).
Pull-down assay
Nuclear extracts were prepared from IOSE523 cells using NE-PER nuclear protein extraction kit (Thermo Scientific) as recommended by the manufacturer. Custom-synthesized biotinylated oligos were purchased from Operon. Nuclear extracts (200 μg) were incubated with or without biotinylated oligo (4 μg) in the presence of Streptavidin-sepharose (Cell Signaling Technology) at 4ºC for 2h. The precipitated immune complex was subject to immunoblot analysis with anti-CREB antibody (86B10; Cell Signaling Technology).
Methylation analysis
Genomic DNA was extracted using Wizard SV Genomic DNA purification kit (Promega). Bisulphite modification was performed using EZ DNA Methylation kit (Zymo Research) following manufacturer’s instructions. Bisulphite-modified DNA was subjected to nested PCR using HotStar Taq DNA Polymerase kit (Qiagen) following manufacturer’s instructions. Primers were selected with MethPrimer software (http://www.urogene.org/methprimer/). The nested PCR primers used were 5′-aggaagagagGGATGTGTATTGAAGTTTTTGAAGTAA-3′ and 5′-cagtaatacgactcactatagggagaaggctCAACCACCCACAAACTTACTACCTA-3′ for the first reaction; 5′-aggaagagagAGTTTTAGGGATTTAGAGGGAAAAA-3′and 5′-cagtaatacgactcactatagggagaaggctTCTAACAACAATACCCCTTAATAAAA-3′ for the second reaction (Forward primer sequences contain a 10bp tag at their 5′ ends (aggaagagag); Reverse primer sequences contain a 31bp tag at their 5′ ends (cagtaatacgactcactatagggagaaggct)). Quantitative DNA methylation was analyzed by Sequenom® EpiTYPER Mass Array (13–16). The assays were performed using the company’s standard protocol through Genomics Shared Facility at Albert Einstein College of Medicine, NY. Matched peak data was exported using EpiTYPER software and analyzed quantitatively.
Immunohistochemistry
FFPE tissue blocks of ovarian serous borderline tumor and ovarian serous carcinoma were obtained as described above. FFPE tissue sections (n=10 each) were deparaffinized and hydrated in xylene and serial alcohol solutions, respectively. Endogenous peroxidase was blocked by incubation in 3% H2O2 for 10 min. Antigen retrieval was performed in a steam pressure cooker with pre-warmed antigen retrieval buffer pH 9 (DakoCytomation) at 95ºC for 20 min. To minimize non-specific staining, the section was incubated with protein block (DakoCytomation) for 10 min. After washing with a buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl and 0.05% Tween-20, the specimen was incubated with 7 μg/mL anti-FILIP1L antibodies at room temperature for 2h. Antigen-antibody reactions were detected with DAKO Flex+ linker (Dako). The staining was visualized using 3,3′-diaminobenzidine plus (Dako). The staining was lightly counterstained with hematoxylin, dehydrated in ethanol and cleared in xylene. Images were acquired by AxioImager microscope (Zeiss). A second pathologist scored the staining under blinded conditions. FILIP1L cytoplasmic staining was scored according to the staining intensity [categorized as 0 (absent), 1 (weak), 2 (moderate), or 3 (strong)].
Cell invasion assay
IOSE cells and ovarian cancer cells either untransfected or FILIP1LΔC103-transfected were cultured at ~80% confluence. Cells were starved in basal medium containing 0.2% bovine serum albumin for 16 h. Matrigel invasion was measured using the BD BioCoat Tumor Invasion System (BD Biosciences #354165) as recommended by the manufacturer. It consists of a BD FluoroBlok™ 24-multiwell insert plate with an 8.0 micron pore size PET membrane that is coated with BD Matrigel Matrix. 4.5 × 104 of starved cells were seeded into the apical chambers and a chemoattractant (10% FBS) was added to the basal chambers. After 20 h incubation, quantification of cell invasion was achieved by post-cell invasion labeling with a fluorescent dye, calcein AM (BD Biosciences), and measuring the fluorescence of invading cells of the underside of the membrane at 494/517 nm (excitation/emission). Since the BD FluoroBlok membrane effectively blocks the passage of light from 490–700 nm at >99% efficiency, fluorescently-labeled cells that have not invaded are not detected by a bottom-reading fluorescence plate reader. Synergy Mx microplate reader (BioTek) was used to measure fluorescence and Gen5 software (BioTek) was used to analyzed the data.
Statistical analysis
Statistical analyses were performed using a two-tailed Student’s t test (GraphPad Prism 3.0), and differences were considered statistically significant at a value of P < 0.05. The correlation of the FILIP1L mRNA expression with DNA methylation status of the CpG island in the FILIP1L promoter as well as that with invasiveness of the cells was estimated by Spearman’s rank correlation method (GraphPad Prism 3.0).
Results
Down-regulation of FILIP1L in ovarian cancer cell lines
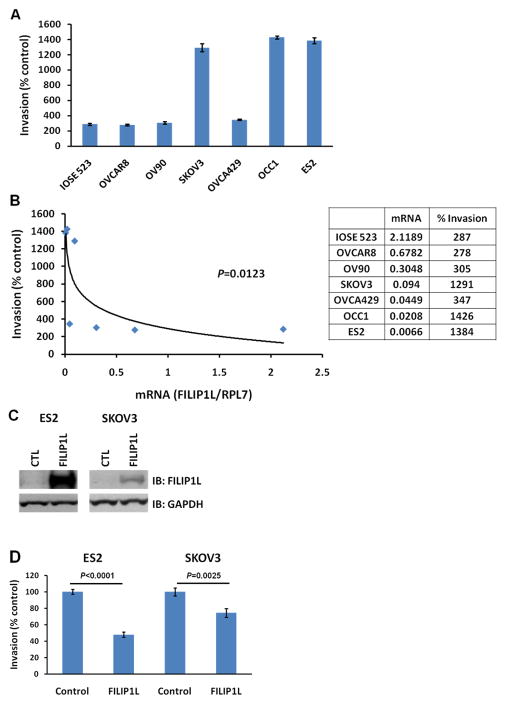
Although FILIP1L mRNA has been shown to be preferentially expressed in human ovarian surface epithelial (HOSE) cells compared to ovarian carcinoma cells derived from patients, the quantitative expression of FILIP1L mRNA has not been previously investigated. To test the expression levels of FILIP1L in ovarian cells, we measured FILIP1L mRNA expression in HOSE cells as well as in ovarian cell lines by real-time RT-PCR. Three immortalized ovarian surface epithelial (IOSE) cell lines as well as various ovarian cancer lines were used. As shown in Figure 1A, mRNA expression of FILIP1L was highest in HOSE cells and higher in IOSE cells compared to ovarian cancer cell lines. In order to test if the protein expression of FILIP1L is correlated with its mRNA expression in ovarian cells, we measured the FILIP1L protein in these cells by western blot analysis using anti-FILIP1L antibody. As shown in Figure 1B, FILIP1L protein expression was much higher in HOSE and IOSE cells compared to ovarian cancer cell lines. In particular, FILIP1L protein was undetectable in ovarian cancer cell lines SKOV3, OVCA429, OCC1 and ES2. These data demonstrate that the protein expression of FILIP1L is correlated with its mRNA expression in ovarian cells. Interestingly, while the amount of the full-length FILIP1L protein was correlated with its mRNA expression, FILIP1L-high expressing cells also showed a substantial amount of detection of smaller sized FILIP1L proteins as shown in Supplementary Figure S1. Thus, these findings suggest that FILIP1L expression is down-regulated in ovarian cancer cell lines compared with normal ovarian cells.
Figure 1. Down-regulation of FILIP1L in ovarian cancer cell lines.
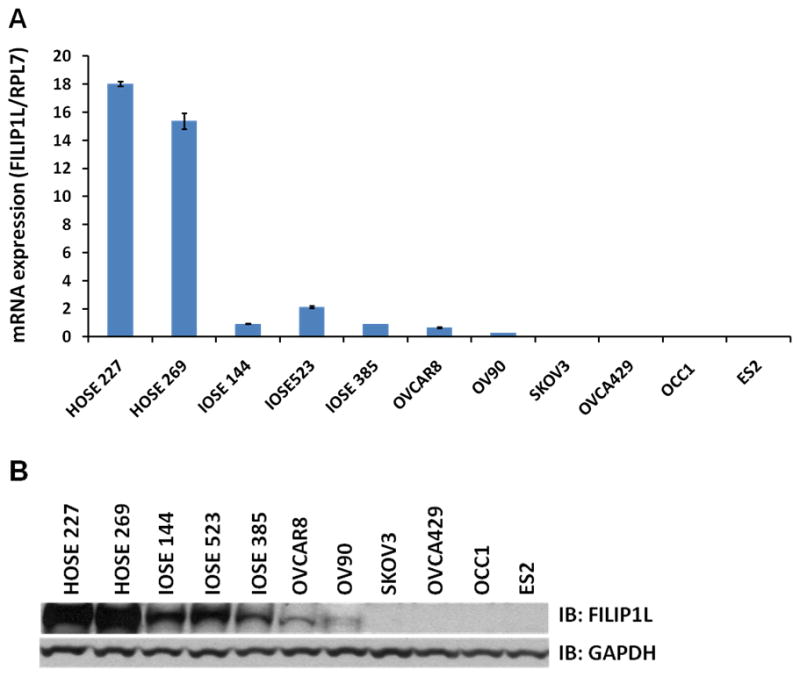
A, Real-time RT-PCR analysis for FILIP1L on cDNA from normal HOSE cells, immortalized normal IOSE cell lines and ovarian cancer cell lines. The y axis represents FILIP1L mRNA expression which was standardized with the housekeeping gene hRPL7. Error bars indicate SEM (n = 3). The result is an average of three independent experiments. B, Immunoblot analysis for FILIP1L in the same cells used in section A. GAPDH blot is shown as the loading control. The result is representative of three independent experiments. A full-length blot is presented in Supplementary Figure S1.
Inverse correlation of FILIP1L expression with the invasive potential of ovarian cell lines
We tested whether FILIP1L expression is inversely correlated with the invasive potential of ovarian cell lines. We examined the invasive activity for the same cell lines utilized in Figure 1 by Matrigel invasion assay. As shown in Figure 2A, all the cell lines that demonstrated low FILIP1L expression invaded Matrigel significantly more than those that demonstrated high FILIP1L expression. In addition, the FILIP1L mRNA expression demonstrated a significant inverse correlation with invasiveness of the cells (Figure 2B), suggesting that FILIP1L expression is inversely correlated with the invasive potential of ovarian cell lines. We then tested if overexpression of FILIP1L in FILIP1L-low expressing, highly-invasive ovarian cancer cell lines resulted in inhibition of cell invasion. Our previous studies have shown that FILIP1LΔC103 (COOH terminal truncation mutant 1–790) was more potent than wild-type FILIP1L in mediating antiproliferative activity in human umbilical vein endothelial cells (HUVEC) (5). In addition, overexpression of FILIP1LΔC103 in HUVECs as well as DU145 prostate cancer cells, lead to inhibition of cell migration (5). Thus, we transfected ES2 and SKOV3 cells with a plasmid encoding either control or FILIP1LΔC103 cDNA, and measured invasion. FILIP1LΔC103 protein expression was confirmed by western blot analysis using anti-FILIP1L antibody (Figure 2C). As shown in Figure 2D, both cell lines transfected with FILIP1LΔC103 cDNA invaded Matrigel significantly less than those transfected with control. We also transfected ES2 cells with a plasmid encoding wild-type FILIP1L cDNA, and measured invasion. As shown in Supplementary Figure S3, ES2 cells transfected with wild-type FILIP1L cDNA invaded Matrigel significantly less than those transfected with control. Collectively, these data suggest that down-regulation of FILIP1L is associated with an invasive phenotype in ovarian cancer cell lines and that this phenotype can be reversed by overexpression of FILIP1L.
Figure 2. Inverse correlation of FILIP1L expression with the invasive potential of ovarian cell lines.
A, Matrigel cell invasion assay for IOSE and ovarian cancer cell lines. Matrigel invasion was measured using the BD BioCoat Tumor Invasion System as described in Materials and Methods. The y axis represents a percent change over serum-free control. Error bars indicate SEM (n = 4). P value comparison between each cell line and IOSE523 were: NS for OVCAR8 and OV90; p<0.0001 for SKOV3, OCC1 and ES2; p=0.0028 for OVCA429. The result is representative of two independent experiments. B, A significant inverse correlation of the FILIP1L mRNA expression with invasiveness of the cells (p=0.0123 by Spearman’s rank correlation method). y axis; Invasiveness of the cells of a percent change over serum-free control shown in section A was used. x axis; standardized FILIP1L mRNA expression shown in Figure 1A was used. C, Immunoblot analysis for FILIP1L in ES2 and SKOV3 cells transfected with either control or FILIP1LΔC103 cDNA. GAPDH blot is shown as the loading control. The result is representative of three independent experiments. D, Matrigel cell invasion assay for the same cells used in section C. Transfected cells were subject to invasion assay at 24 h after transfection. The same experimental procedures were followed as described in section A. P value comparison was: p<0.0001 for ES2 cells; p=0.0025 for SKOV3 cells. The result is representative of three independent experiments. Representative image of an area scan is shown in Supplementary Figure S2.
Reduced expression of FILIP1L mRNA in invasive serous carcinomas compared with non-invasive serous borderline tumors
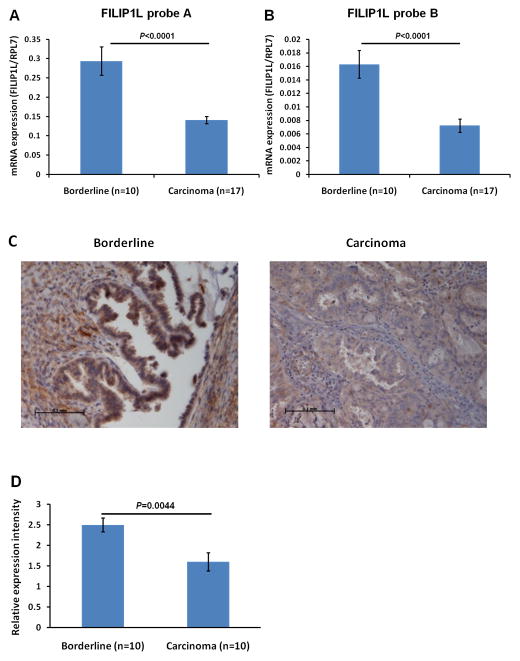
Having observed that FILIP1L expression is inversely correlated with invasive potential in cell lines, we examined if down-regulation of FILIP1L expression is also associated with the invasive potential in clinical ovarian specimens. We chose to compare serous borderline tumors (low malignant potential tumors) with invasive serous carcinomas since the former are distinguished from carcinomas by their lack of stromal invasion (17). Total RNA was purified from formalin-fixed paraffin-embedded (FFPE) tissue sections of ovarian cancer patient specimens and real-time RT-PCR was performed. As shown in Figure 3A, FILIP1L mRNA expression was significantly lower in invasive serous carcinoma than in non-invasive serous borderline tumors. We confirmed that these data were not produced by artifacts resulting from the handling of FFPE tissues, by real-time RT-PCR using another probe spanning an inter-exon region of the FILIP1L genome. We observed that FILIP1L mRNA expression was also significantly lower in invasive serous carcinoma than in non-invasive serous borderline tumors using this second probe (Figure 3B). We then examined FILIP1L protein expression in these tissues using immunohistochemical staining. As shown in Figure 3C and D, FILIP1L expression was significantly lower in invasive serous carcinoma than in non-invasive serous borderline tumors. These findings further suggest that down-regulation of FILIP1L is associated with an invasive phenotype in ovarian cancer.
Figure 3. Reduced expression of FILIP1L mRNA in invasive serous carcinomas compared with non-invasive serous borderline tumors.
Real-time RT-PCR analysis for FILIP1L on cDNA from ovarian serous carcinomas and ovarian serous borderline tumors. The y axis represents FILIP1L mRNA expression which was standardized with the housekeeping gene hRPL7. Error bars indicate SEM (n = 10 for serous borderline tumors and n = 17 for serous carcinoma). P value comparison was: p<0.0001 for TaqMan probe A (A); p<0.0001 for TaqMan probe B (B). C, Immunohistochemical staining of FILIP1L in ovarian serous carcinomas and ovarian serous borderline tumors is shown. Scale bar shown indicates 0.1 mm. The result is a representative image from ten independent tissue specimens each. D, FILIP1L expression in invasive serous carcinomas was significantly less than that in non-invasive serous borderline tumors (p = 0.0044). Ten specimens from each group were immunohistochemically stained and the staining was scored in a blinded fashion. Error bars indicate SEM. Expression score was performed as described in Materials and Methods.
Methylation in the CpG island of the FILIP1L promoter in ovarian cancer cells
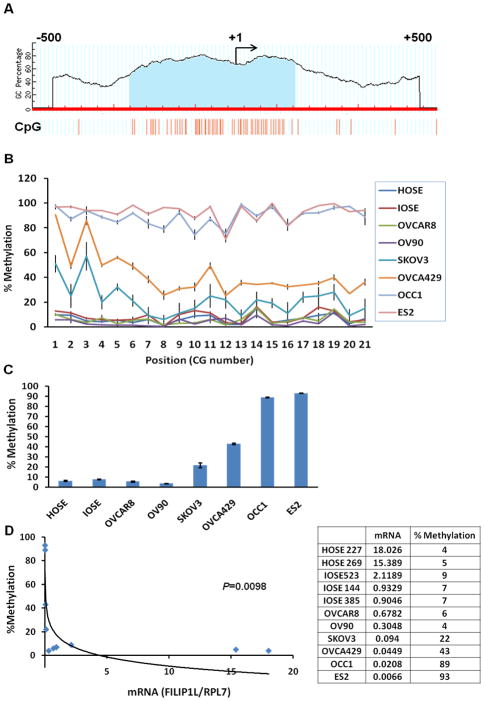
Epigenetic modifications have been shown to be associated with ovarian tumorigenesis (10, 11). A number of genes including the tumor suppressor BRCA1 have been shown to be hypermethylated and down-regulated in ovarian cancer. Using MethPrimer software (http://www.urogene.org/methprimer/), we found that the promoter region of FILIP1L has a CpG island of 407 base pairs (Figure 4A). In order to investigate whether or not DNA methylation mediates FILIP1L down-regulation in ovarian cancer cells, we tested whether the DNA methylation status at the FILIP1L promoter inversely correlates with FILIP1L expression. The percent methylation for each CG site in the CpG island of the FILIP1L promoter is shown in Figure 4B. The average overall methylation for all 21 CG sites is shown in Figure 4C. The analyzed CG sites out of total 59 CG sites in the CpG island is shown in supplementary Table 1. Most cytosines analyzed in the CpG island of the FILIP1L promoter demonstrated similar percent methylation in each cell type (Figure 4B). The average overall methylation demonstrated that these CG sites were highly methylated in ovarian cancer cells such as ES2 (93.1±0.3%) and OCC1 (88.9±0.5%) whereas these sites were partially methylated in OVCA429 (43.0±0.9%) and SKOV3 (21.7±2.3%) (Figure 4C). In contrast, these sites were unmethylated in normal HOSE (average of 10 HOSE cells; 6.4 ± 0.5%) and IOSE (average of 3 IOSE cell lines; 7.7±0.5%) cells as well as some ovarian cancer cell lines such as OVCAR8 (5.7±0.6%) and OV90 (3.6±0.2%). We tested if the DNA methylation status of the CpG island of the FILIP1L promoter (Figure 4C) inversely correlated with FILIP1L mRNA expression (Figure 1A). As shown in Figure 4D, the DNA methylation status of the CpG island of the FILIP1L promoter displayed a significant inverse correlation with FILIP1L mRNA expression, suggesting that DNA methylation in the FILIP1L promoter may mediate FILIP1L down-regulation in ovarian cancer cells.
Figure 4. Methylation in the CpG island of the FILIP1L promoter in ovarian cancer cells.
A, The schematic diagram of the putative CpG island in the FILIP1L promoter identified by a bioinformatics analysis. The axis is the percentage of the dinucleotide, guanine and cytosine. The genomic DNA sequences (−500 to +500 nt) were analyzed by MethPrimer software. Transcription start site (+1) is indicated by the arrow. The location of the putative CpG island of 407 base pairs is displayed in the shaded area. B, DNA methylation status of the CpG island in the FILIP1L promoter from HOSE, IOSE and ovarian cancer cell lines was analyzed by Sequenom® EpiTYPER Mass Array. Mass Array results are shown as group-median percent methylation values with SEM (y axis; n = 3 except for HOSE cells where 10 independent cultures were used) for each individual CG site (in sequential order along the x axis). The result is average of three independent experiments. C, Mass Array results are shown as the average overall methylation for all 21 CG sites from the same cells used in section A. Error bars indicate SEM (n = 3 except for HOSE cells where 10 independent cultures were used). D, A significant inverse correlation of the DNA methylation status of the CpG island in the FILIP1L promoter with FILIP1L mRNA expression (p=0.0098 by Spearman’s rank correlation method). y axis; Percent methylation of the average overall methylation for all 21 CG sites shown in section C was used. x axis; standardized FILIP1L mRNA expression shown in Figure 1A was used.
Association of reduced methylation in the FILIP1L promoter with restoration of FILIP1L expression in ovarian cancer cells following treatment with a DNA demethylating agent
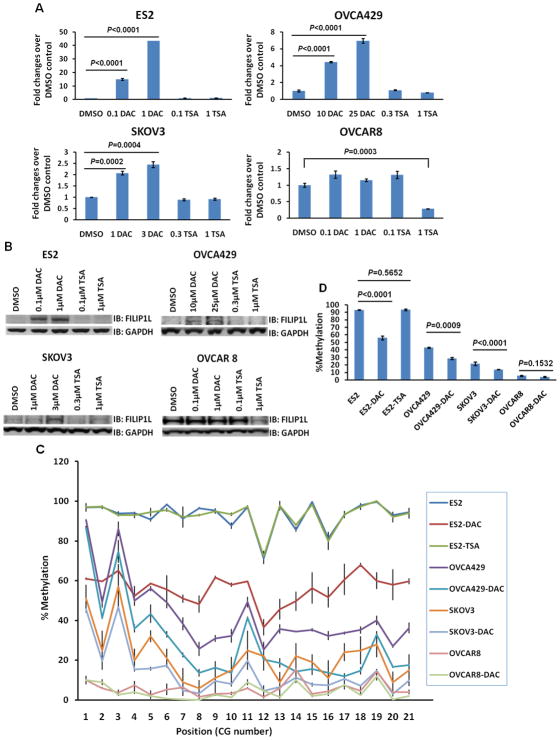
To further test if epigenetic regulation results in re-expression of FILIP1L, we treated ovarian cancer cells with either a DNA demethylating agent or histone deacetylase inhibitor. We chose to use ES2, OVCA429 and SKOV3 cells because they showed lower amounts of FILIP1L expression (Figure 1A and B). Treatment of these cells with a DNA demethylating agent, 5-aza-2′-deoxycytidine (DAC), but not with the histone deacetylase inhibitor, Trichostatin A (TSA) resulted in increased mRNA (Figure 5A) and protein expression (Figure 5B) of FILIP1L. FILIP1L mRNA expression in DAC- or TSA-treated cells was compared with that in DMSO control-treated cells. Significant increase in FILIP1L mRNA was observed in all three cell lines following DAC treatment. Induction of FILIP1L protein expression was observed in all three cell lines following DAC treatment. However, significant increases in FILIP1L mRNA and protein expression following TSA treatment were not observed. When a FILIP1L-high expressing cell such as OVCAR8 was treated with these reagents, we did not observe a significant increase in mRNA and protein expression (Figure 5A and B). In order to examine whether reduced methylation in the FILIP1L promoter is associated with restoration of FILIP1L expression, we analyzed the methylation status of the FILIP1L promoter in ovarian cancer cells following DAC treatment. The same cell lines used in Figure 5A and B were tested. Most CG sites analyzed in the CpG island of the FILIP1L promoter demonstrated similar reduction in percent methylation in each cell type (Figure 5C). As shown in Figure 5D, ES2, OVCA429 and SKOV3 cells treated with DAC demonstrated a significant decrease in methylation (from 93.1±0.3% to 56.1±2.6% in ES2 cells, from 43.0±0.9% to 28.5±1.4% in OVCA429 and from 21.7±2.3% to 13.9±0.3% in SKOV3 cells). However, DAC-treated OVCAR8 cells demonstrated no changes in methylation throughout the cytosines in the CpG island (from 5.7±0.6% to 4.2±0.6%). These data demonstrate that DNA methylation in the FILIP1L promoter is a mechanism by which FILIP1L is down-regulated in ovarian cancer.
Figure 5. Association of reduced methylation in the FILIP1L promoter with restoration of FILIP1L expression in ovarian cancer cells following treatment with a DNA demethylating agent.
A, Real-time RT-PCR analysis for FILIP1L on cDNA from ES2, OVCA429, SKOV3 and OVCAR8 cells treated with either 5-aza-2′-deoxycytidine (DAC) or Trichostatin A (TSA). The y axis represents a fold change over DMSO-treated control cells where each value was standardized with the housekeeping gene hRPL7. Error bars indicate SEM (n = 3). P value comparison between DMSO-treated control and either DAC- or TSA-treated experiments were: both 0.1 and 1 μM DAC (p<0.0001) and both 0.1 and 1 μM TSA (not significant (NS)) for ES2; both 10 and 25 μM DAC (p<0.0001) and both 0.3 and 1 μM TSA (NS) for OVCA429; 1 μM DAC (p=0.0002), 3 μM DAC (p=0.0004) and both 0.3 and 1 μM TSA (NS) for SKOV3; 0.1 μM DAC, 1 μM DAC and 0.1 μM TSA (NS), and 1 μM TSA (p=0.0003) for OVCAR8. The result is an average of three independent experiments. B, Immunoblot analysis for FILIP1L in the same cells used in section A. GAPDH blot is shown as the loading control. Note that in FILIP1L-high expressing OVCAR8 cells, DAC treatment did not result in increased FILIP1L expression whereas TSA treatment decreased FILIP1L expression. The result is representative of three independent experiments. C, DNA methylation status of the CpG island in the FILIP1L promoter from the same cells used in section A was analyzed by Sequenom® EpiTYPER Mass Array. Mass Array results are shown as described in Figure 4B. The result is an average of three independent experiments. D, Mass Array results are shown as described in Figure 4C from the same cells used in section A. P value comparison between DMSO-treated control and DAC- or TSA-treated experiments were: 1 μM DAC (p<0.0001) and 1 μM TSA (NS) for ES2; 25 μM DAC (p=0.0009) for OVCA429; 3 μM DAC (p<0.0001) for SKOV3; 1 μM DAC (NS) for OVCAR8. Note that either TSA-treated ES2 or DAC-treated OVCAR8 did not decrease DNA methylation compared with DMSO-treated control.
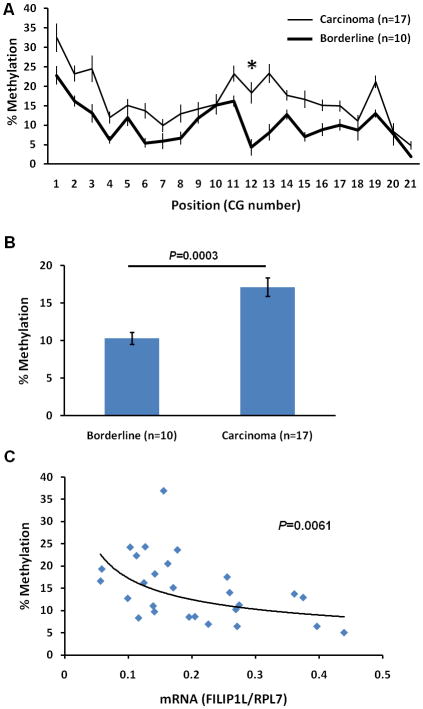
Increased methylation in the CpG island of the FILIP1L promoter is seen in invasive serous carcinomas compared with non-invasive serous borderline tumors
Having observed that the down-regulation of FILIP1L expression associated with DNA methylation in the FILIP1L promoter is related with invasive potential in ovarian cancer cell lines, we examined if down-regulation of FILIP1L expression is associated with the degree of DNA methylation in the FILIP1L promoter in clinical ovarian tissue specimens. Genomic DNA was purified from the same scraped tissues used in Figure 3A and B for mRNA expression analysis, and DNA methylation was analyzed. 13 out of 21 CG sites analyzed in the CpG island of the FILIP1L promoter demonstrated a significantly higher degree of methylation in invasive serous carcinomas than in non-invasive serous borderline tumors (Figure 6A; p=0.0204 for CG site 2; p=0.0268 for CG site 3; p=0.0063 for CG site 4; p=0.0063 for CG site 6; p=0.0204 for CG site 11; p=0.0039 for CG site 12; p<0.0001 for CG site 13; p=0.0281 for CG site 14; p=0.0053 for CG site 15; p=0.0147 for CG site 16; p=0.0426 for CG site 17; p=0.0009 for CG site 19; p=0.0254 for CG site 21). In Figure 6B, the average overall methylation for all 21 CG sites also demonstrated a significantly higher methylation in invasive serous carcinomas than that in non-invasive serous borderline tumors. We then tested if the DNA methylation status of the CpG island of the FILIP1L promoter (Figure 6B) inversely correlated with FILIP1L mRNA expression (Figure 3A) in these clinical ovarian tissue specimens. As shown in Figure 6C, the DNA methylation status of the CpG island of the FILIP1L promoter displayed a significant inverse correlation with FILIP1L mRNA expression. These findings suggest that down-regulation of FILIP1L associated with CpG island methylation of the FILIP1L promoter is related with the invasive phenotype of ovarian cancer.
Figure 6. Increased methylation in the CpG island of the FILIP1L promoter in invasive serous carcinomas compared with non-invasive serous borderline tumors.
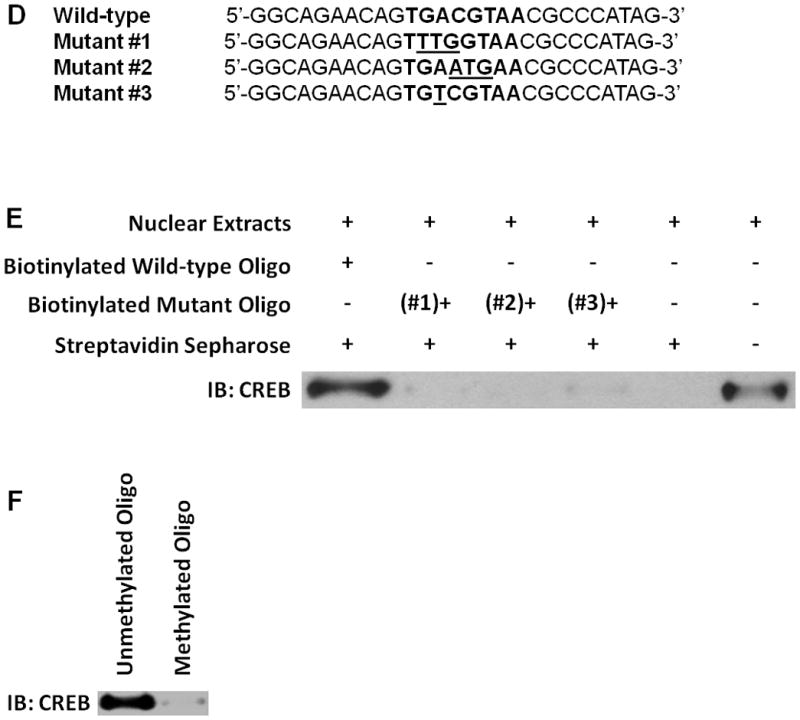
A, DNA methylation status of the CpG island in the FILIP1L promoter from ovarian serous carcinomas and ovarian serous borderline tumors was analyzed by Sequenom® EpiTYPER Mass Array. Mass Array results are shown as described in Figure 4B (y axis; n = 10 for serous borderline tumors and n = 17 for serous carcinoma). CG site 12 (*), the CG site in the putative CREB/ATF site is shown. B, Mass Array results are shown as described in Figure 4C from the same cells used in section A. Error bars indicate SEM (n = 10 for serous borderline tumors and n = 17 for serous carcinoma; p=0.0003). C, A significant inverse correlation of the DNA methylation status of the CpG island in the FILIP1L promoter with FILIP1L mRNA expression in clinical ovarian tissue specimens (p=0.0061 by Spearman’s rank correlation method). y axis; Percent methylation of the average overall methylation for all 21 CG sites shown in section B was used. x axis; standardized FILIP1L mRNA expression shown in Figure 3A was used. Data to plot the graph is presented in Supplementary Table 2. D, Sequence of the oligos used in pull-down assay. The sequence from the FILIP1L promoter including CREB/ATF site is shown as wild-type. Three mutant oligos containing the mutated sequence in CREB/ATF site are shown as mutant #1–3. The CREB/ATF site is shown in bold. Mutated nucleotides are shown as the underlined. E, A pull-down assay for CREB binding was performed as described in Materials and Methods, and is shown as an immunoblot analysis for CREB. The far right lane demonstrates the presence of CREB in the nuclear extracts (2.5 μg) from IOSE523 cells. The result is representative of three independent experiments. F, A pull-down assay for CREB binding was performed as described in section E except the oligo methylated at the cytosine in the CREB/ATF site was used. The result is representative of three independent experiments.
Binding of a transcription activator, CREB to the CREB/ATF site in the CpG island of the FILIP1L promoter
Inspection of the FILIP1L genomic sequence reveals that it has a cyclic-AMP response element binding protein (CREB)/activating transcription factor (ATF) site (TGACGTAA) 10 base pairs upstream of the transcription initiation site (−10) in the CpG island of its promoter region. CREB and a family of ATFs are transcription activators (18). Methylation of the CREB/ATF site in the CpG island of a promoter region results in suppression of CREB binding to the CREB/ATF site, which in turn suppresses gene expression (19–25). As shown in Figure 4B, the CpG in the CREB/ATF site in the CpG island of the FILIP1L promoter, CG site 12, is methylated in ovarian cancer cells (71.1±2.8% for ES2, 75.3±4.2% for OCC1, 25.3±2.3% for OVCA429 and 22.2±7.2% for SKOV3). In addition, the same CG site demonstrated a significantly higher degree of methylation in invasive serous carcinomas than in the non-invasive serous borderline tumors (Figure 6A; 18.3±2.8% for carcinomas; 4.3±2.1% for borderline tumors; p=0.0039). Thus, we questioned whether the putative CREB/ATF site plays a role in mediating down-regulation of FILIP1L in ovarian cancer. To determine if the CREB protein binds to this site, we performed a pull-down assay as previously described (26). Biotinylated DNA oligos spanning the CREB/ATF site at −10 from the wild-type sequence were used to pull-down the CREB complex. Three mutant biotinylated DNA oligos containing mutated nucleotides for the CREB/ATF binding site were constructed (Figure 6D). As shown in Figure 6E, the wild-type oligo, but not mutant oligos, demonstrated a pull-down of the CREB complex. To directly test the effect of DNA methylation on CREB binding activity, we performed a pull-down assay using methylated oligo. The oligo methylated at the cytosine in the CREB/ATF site failed to pull down the CREB complex (Figure 6F). Thus, these data suggest that methylation of the CREB/ATF site in the FILIP1L promoter plays a role in mediating down-regulation of FILIP1L in ovarian cancer.
Discussion
Epigenetic aberrations have been shown to be intimately associated with ovarian tumorigenesis (10, 11). A number of genes including the classic tumor suppressor, BRCA1, p16, MLH1, RASSF1A, ANGPTL2, ARH1, LOT1, ICAM1, HSulf-1, PALB2 and TUBB3 have been shown to be hypermethylated and down-regulated in ovarian cancer (11). In the present study, we demonstrated that down-regulation of FILIP1L (previously termed DOC1) is associated with DNA methylation in ovarian cancer. We have shown that both the mRNA and protein expression of FILIP1L are down-regulated in ovarian cancer cells. FILIP1L expression is significantly lower in invasive serous carcinoma than in non-invasive serous borderline tumors. FILIP1L expression is inversely correlated with the invasive potential of ovarian cell lines as well as clinical ovarian specimens. The CpG island in the FILIP1L promoter is heavily methylated in ovarian cancer cells and almost completely non-methylated in normal HOSE cells. The DNA methylation status of the FILIP1L promoter is inversely correlated with FILIP1L expression in ovarian cancer. Reduced methylation in the FILIP1L promoter following treatment with a DNA demethylating agent was associated with restoration of FILIP1L expression. A transcription activator, CREB, binds to the CREB/ATF site in the CpG island of the FILIP1L promoter. Overall, these findings suggest that down-regulation of FILIP1L associated with DNA methylation is related with the invasive phenotype in ovarian cancer and indicates that FILIP1L has the potential to be a biomarker for invasive potential and a target for novel ovarian cancer therapy.
In order to identify common intracellular mediators of proliferation, migration and apoptosis, we previously analyzed gene expression profiles of endothelial cells after treatment with angiogenesis inhibitors such as endostatin, fumagillin and EMAP-II (3, 4). FILIP1L was up-regulated in endothelial cells in response to these inhibitors. We subsequently demonstrated that overexpression of FILIP1L resulted in inhibition of cell proliferation and migration and increased apoptosis (5). In addition, targeted expression of FILIP1L in tumors inhibited tumor growth in vivo (5). These findings suggested that FILIP1L may be an important inhibitor of cell proliferation and migration as well as an inducer of apoptosis. Our previous work suggested that agents, which increase the expression of FILIP1L in cells have the effect of inhibiting the proliferation and migration of those cells. The findings from the present study suggest that down-regulation of FILIP1L is associated with the invasive phenotype in ovarian cancer and that modulation of FILIP1L expression has the potential to be a target for ovarian cancer therapy.
Ovarian borderline tumors are epithelial ovarian cancers with histologic and biologic features intermediate between benign and malignant neoplasms. They display evidence of epithelial proliferation with frequent high grade nuclear features but rare to no invasive implants. Approximately 15% of ovarian cancers are borderline tumors (ovarian tumors of low malignant potential) (17). We have shown that FILIP1L expression is significantly lower in invasive serous carcinoma than in non-invasive serous borderline tumors (Figure 3). The methylation of the CpG island in the FILIP1L promoter was significantly higher in invasive serous carcinomas than that in non-invasive serous borderline tumors (Figure 6A and B). Others have shown that borderline tumors demonstrate a lower degree of methylation than malignant ovarian cancers. Genes such as IGFBP-3, ER-α, p16, BRCA1, hMLH1 and TGFBI are less methylated in borderline tumors than in invasive ovarian cancers (27–31). Serous is the most frequent subtype of epithelial ovarian cancer (17). Although we have shown that FILIP1L down-regulation is implicated in serous ovarian cancers, it will be important to expand our study to examine if the same mechanism is also involved in other epithelial ovarian cancer subtypes.
We have shown that a DNA demethylating agent DAC, but not the histone deacetylase inhibitor TSA, resulted in increased mRNA (Figure 5A) and protein expression (Figure 5B) of FILIP1L in FILIP1L-low expressing ovarian cancer cells, suggesting that DNA methylation, but not histone modification, is associated with the down-regulation of FILIP1L in these cells. However, while FILIP1L mRNA was down-regulated, percent DNA methylation was low in ovarian cancer cell lines such as SKOV3 and OVCA429 (Figure 4D) as well as in ovarian serous carcinomas (Figure 6A). Whether low/moderate level of DNA methylation can down-regulate FILIP1L in these specimens needs to be further investigated. It is also possible that DNA methylation is not the only mechanism by which FILIP1L is down-regulated.
Standard therapy for ovarian cancer is surgical cyto-reduction followed by combination chemotherapy using a taxane (paclitaxel) and platinum (carboplatin) (32). The majority of ovarian cancers will unfortunately recur despite optimal front line therapy (17). Recurrent cancer is often resistant to conventional agents. Platinum resistance has been shown to be associated with DNA methylation, specifically with methylation of the CpG units of the MLH1 mismatch repair gene (33, 34). Studies have shown a clinical effect of a combination of carboplatin and DNA demethylating agents (35, 36). Epigenetic-based therapies that lead to the re-expression of tumor suppressor genes may have a role in resensitizing tumors to cytotoxic chemotherapies. In patients whose ovarian cancers have low expression of FILIP1L, there may be utility to the addition of a demethylating agent in order to increase FILIP1L expression. This therapy may revive the tumor suppressor-like function of FILIP1L. In conjunction with cytotoxic therapies, agents targeted to increase the expression of FILIP1L may have the potential to improve progression-free and overall survival in a disease with a dismal outcome. Determining the level of expression of FILIP1L in ovarian cancer may also be useful in predicting the invasive potential of those tumors and assist in staging, grading and management.
Differential expression of the FILIP1L gene has been demonstrated in other histologies. cDNA microarray analysis was used to identify FILIP1L as one of the genes whose transcription is induced in senescent human prostate epithelial cells but significantly repressed in immortalized prostate epithelial cells (37, 38). FILIP1L mRNA expression was down-regulated in microvascular endothelial cells infected with Kaposi’s sarcoma-associated herpes virus but not in uninfected microvascular endothelial cells (39). Furthermore, mRNA expression was down-regulated in B cells transformed with the oncogene TaxBLV (bovine leukemia virus Tax) but not in untransformed B cells (40). Therefore, future studies to test whether DNA methylation in the FILIP1L promoter plays a role in mediating down-regulation of FILIP1L in other tumor histologies will be of interest.
In summary, we have demonstrated that down-regulation of FILIP1L is associated with the invasive phenotype in ovarian cancer. Down-regulation of FILIP1L is mediated by promoter methylation. Further characterization of the mechanism of FILIP1L down-regulation may improve the understanding of the role played by FILIP1L in ovarian carcinogenesis and lead to the development of more effective anticancer agents.
Supplementary Material
Acknowledgments
This research was supported in part by a generous gift from Linda and Earle Altman, the Albert Einstein College of Medicine and the NCI Intramural Program.
This work was supported in part by a generous research gift from Linda and Earle Altman. We thank the Canadian Ovarian Tissue Bank for providing IOSE 144, 523 and 385 and Dr. Barbara Vanderhyden for providing OVCA429. We thank Drs. John Greally and Silvia Gravina for helpful discussion and Kris Ylaya for technical assistance.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Yap TA, Carden CP, Kaye SB. Beyond chemotherapy: targeted therapies in ovarian cancer. Nat Rev Cancer. 2009;9:167–81. doi: 10.1038/nrc2583. [DOI] [PubMed] [Google Scholar]
- 3.Mazzanti CM, Tandle A, Lorang D, Costouros N, Roberts D, Bevilacqua G, et al. Early genetic mechanisms underlying the inhibitory effects of endostatin and fumagillin on human endothelial cells. Genome Res. 2004;14:1585–93. doi: 10.1101/gr.2552804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tandle AT, Mazzanti C, Alexander HR, Roberts DD, Libutti SK. Endothelial monocyte activating polypeptide-II induced gene expression changes in endothelial cells. Cytokine. 2005;30:347–58. doi: 10.1016/j.cyto.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 5.Kwon M, Hanna E, Lorang D, He M, Quick JS, Adem A, et al. Functional characterization of filamin a interacting protein 1-like, a novel candidate for antivascular cancer therapy. Cancer Res. 2008;68:7332–41. doi: 10.1158/0008-5472.CAN-08-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mok SC, Wong KK, Chan RK, Lau CC, Tsao SW, Knapp RC, et al. Molecular cloning of differentially expressed genes in human epithelial ovarian cancer. Gynecol Oncol. 1994;52:247–52. doi: 10.1006/gyno.1994.1040. [DOI] [PubMed] [Google Scholar]
- 7.Matei D, Graeber TG, Baldwin RL, Karlan BY, Rao J, Chang DD. Gene expression in epithelial ovarian carcinoma. Oncogene. 2002;21:6289–98. doi: 10.1038/sj.onc.1205785. [DOI] [PubMed] [Google Scholar]
- 8.Quaye L, Dafou D, Ramus SJ, Song H, Gentry-Maharaj A, Notaridou M, et al. Functional complementation studies identify candidate genes and common genetic variants associated with ovarian cancer survival. Hum Mol Genet. 2009;18:1869–78. doi: 10.1093/hmg/ddp107. [DOI] [PubMed] [Google Scholar]
- 9.Notaridou M, Quaye L, Dafou D, Jones C, Song H, Hogdall E, et al. Common alleles in candidate susceptibility genes associated with risk and development of epithelial ovarian cancer. Int J Cancer. 2011;128:2063–74. doi: 10.1002/ijc.25554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barton CA, Hacker NF, Clark SJ, O’Brien PM. DNA methylation changes in ovarian cancer: implications for early diagnosis, prognosis and treatment. Gynecol Oncol. 2008;109:129–39. doi: 10.1016/j.ygyno.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 11.Matei DE, Nephew KP. Epigenetic therapies for chemoresensitization of epithelial ovarian cancer. Gynecol Oncol. 2010;116:195–201. doi: 10.1016/j.ygyno.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shepherd TG, Theriault BL, Campbell EJ, Nachtigal MW. Primary culture of ovarian surface epithelial cells and ascites-derived ovarian cancer cells from patients. Nat Protoc. 2006;1:2643–9. doi: 10.1038/nprot.2006.328. [DOI] [PubMed] [Google Scholar]
- 13.Ehrich M, Turner J, Gibbs P, Lipton L, Giovanneti M, Cantor C, et al. Cytosine methylation profiling of cancer cell lines. Proc Natl Acad Sci. 2008;105:4844–9. doi: 10.1073/pnas.0712251105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radpour R, Haghighi MM, Fan AX, Torbati PM, Hahn S, Holzgreve W, et al. High-throughput hacking of the methylation patterns in breast cancer by in vitro transcription and thymidine-specific cleavage mass array on MALDI-TOF silico-chip. Mol Cancer Res. 2008;6:1702–9. doi: 10.1158/1541-7786.MCR-08-0262. [DOI] [PubMed] [Google Scholar]
- 15.Radpour R, Kohler C, Haghighi MM, Fan AX, Holzgreve W, Zhong XY. Methylation profiles of 22 candidate genes in breast cancer using high-throughput MALDI-TOF mass array. Oncogene. 2009;20;28:2969–78. doi: 10.1038/onc.2009.149. [DOI] [PubMed] [Google Scholar]
- 16.Vanaja DK, Ehrich M, Van den Boom D, Cheville JC, Karnes RJ, Tindall DJ, et al. Hypermethylation of genes for diagnosis and risk stratification of prostate cancer. Cancer Invest. 2009;27:549–60. doi: 10.1080/07357900802620794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DiSaia PJ, Creasman WT. Clinical gynecologic oncology. 6. St. Louis, Mo: Mosby; 2002. [Google Scholar]
- 18.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 19.Mancini DN, Rodenhiser DI, Ainsworth PJ, O’Malley FP, Singh SM, Xing W, et al. CpG methylation within the 5′ regulatory region of the BRCA1 gene is tumor specific and includes a putative CREB binding site. Oncogene. 1998;16:1161–9. doi: 10.1038/sj.onc.1201630. [DOI] [PubMed] [Google Scholar]
- 20.DiNardo DN, Butcher DT, Robinson DP, Archer TK, Rodenhiser DI. Functional analysis of CpG methylation in the BRCA1 promoter region. Oncogene. 2001;20:5331–40. doi: 10.1038/sj.onc.1204697. [DOI] [PubMed] [Google Scholar]
- 21.Yossifoff M, Kisliouk T, Meiri N. Dynamic changes in DNA methylation during thermal control establishment affect CREB binding to the brain-derived neurotrophic factor promoter. Eur J Neurosci. 2008;28:2267–77. doi: 10.1111/j.1460-9568.2008.06532.x. [DOI] [PubMed] [Google Scholar]
- 22.Fox KE, Colton LA, Erickson PF, Friedman JE, Cha HC, Keller P, et al. Regulation of cyclin D1 and Wnt10b gene expression by cAMP-responsive element-binding protein during early adipogenesis involves differential promoter methylation. J Biol Chem. 2008;283:35096–105. doi: 10.1074/jbc.M806423200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuroda A, Rauch TA, Todorov I, Ku HT, Al-Abdullah IH, Kandeel F, et al. Insulin gene expression is regulated by DNA methylation. PLoS one. 2009;4:e6953. doi: 10.1371/journal.pone.0006953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sunahori K, Juang YT, Tsokos GC. Methylation status of CpG islands flanking a cAMP response element motif on the protein phosphatase 2Ac alpha promoter determines CREB binding and activity. J Immunol. 2009;182:1500–8. doi: 10.4049/jimmunol.182.3.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med. 2007;204:1543–51. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He A, Pu WT. Genome-wide location analysis by pull down of in vivo biotinylated transcription factors. Curr Protoc Mol Biol. 2010;Chapter 21(Unit 21):0. doi: 10.1002/0471142727.mb2120s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiley A, Katsaros D, Chen H, Rigault de la Longrais IA, Beeghly A, Puopolo M, et al. Aberrant promoter methylation of multiple genes in malignant ovarian tumors and in ovarian tumors with low malignant potential. Cancer. 2006;107:299–308. doi: 10.1002/cncr.21992. [DOI] [PubMed] [Google Scholar]
- 28.Wang C, Horiuchi A, Imai T, Ohira S, Itoh K, Nikaido T, et al. Expression of BRCA1 protein in benign, borderline, and malignant epithelial ovarian neoplasms and its relationship to methylation and allelic loss of the BRCA1 gene. J Pathol. 2004;202:215–23. doi: 10.1002/path.1507. [DOI] [PubMed] [Google Scholar]
- 29.Rathi A, Virmani AK, Schorge JO, Elias KJ, Maruyama R, Minna JD, et al. Methylation profiles of sporadic ovarian tumors and nonmalignant ovaries from high-risk women. Clin Cancer Res. 2002;8:3324–31. [PubMed] [Google Scholar]
- 30.Makarla PB, Saboorian MH, Ashfaq R, Toyooka KO, Toyooka S, Minna JD, et al. Promoter hypermethylation profile of ovarian epithelial neoplasms. Clin Cancer Res. 2005;11:5365–9. doi: 10.1158/1078-0432.CCR-04-2455. [DOI] [PubMed] [Google Scholar]
- 31.Kang S, Dong SM, Park NH. Frequent promoter hypermethylation of TGFBI in epithelial ovarian cancer. Gynecol Oncol. 2010;118:58–63. doi: 10.1016/j.ygyno.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 32.Morgan RJ., Jr Ovarian cancer guidelines: treatment progress and controversies. J Natl Compr Canc Netw. 2011;9:4–5. doi: 10.6004/jnccn.2011.0002. [DOI] [PubMed] [Google Scholar]
- 33.Asadollahi R, Hyde CA, Zhong XY. Epigenetics of ovarian cancer: from the lab to the clinic. Gynecol Oncol. 2010;118:81–7. doi: 10.1016/j.ygyno.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 34.Balch C, Huang TH, Brown R, Nephew KP. The epigenetics of ovarian cancer drug resistance and resensitization. Am J Obstet Gynecol. 2004;191:1552–72. doi: 10.1016/j.ajog.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 35.Fu S, Hu W, Iyer R, Kavanagh JJ, Coleman RL, Levenback CF, et al. Phase 1b-2a study to reverse platinum resistance through use of a hypomethylating agent, azacitidine, in patients with platinum-resistant or platinum-refractory epithelial ovarian cancer. Cancer. doi: 10.1002/cncr.25701. Epub 2010 Nov 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plumb JA, Strathdee G, Sludden J, Kaye SB, Brown R. Reversal of drug resistance in human tumor xenografts by 2′-deoxy-5-azacytidine-induced demethylation of the hMLH1 gene promoter. Cancer Res. 2000;60:6039–44. [PubMed] [Google Scholar]
- 37.Schwarze SR, DePrimo SE, Grabert LM, Fu VX, Brooks JD, Jarrard DF. Novel pathways associated with bypassing cellular senescence in human prostate epithelial cells. J Biol Chem. 2002;277:14877–83. doi: 10.1074/jbc.M200373200. [DOI] [PubMed] [Google Scholar]
- 38.Schwarze SR, Fu VX, Desotelle JA, Kenowski ML, Jarrard DF. The identification of senescence-specific genes during the induction of senescence in prostate cancer cells. Neoplasia. 2005;7:816–23. doi: 10.1593/neo.05250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poole LJ, Yu Y, Kim PS, Zheng QZ, Pevsner J, Hayward GS. Altered patterns of cellular gene expression in dermal microvascular endothelial cells infected with Kaposi’s sarcoma-associated herpesvirus. J Virol. 2002;76:3395–420. doi: 10.1128/JVI.76.7.3395-3420.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klener P, Szynal M, Cleuter Y, Merimi M, Duvillier H, Lallemand F, et al. Insights into gene expression changes impacting B-cell transformation: cross-species microarray analysis of bovine leukemia virus tax-responsive genes in ovine B cells. J Virol. 2006;80:1922–38. doi: 10.1128/JVI.80.4.1922-1938.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.