Abstract
The V600E mutation in the BRAF oncogene is associated with colorectal carcinomas, with mismatch-repair deficiency and, recently, with nonresponse to epidermal growth factor receptor inhibitor therapy. The use of reliable techniques for its detection is important. The aim of our study was to compare the performance characteristics in V600E detection of denaturing high-performance liquid chromatography (dHPLC) and high-resolution melting (HRM) with TaqMan allelic discrimination as well as direct-sequencing methods in a series of 195 colorectal paraffin-embedded specimens up to the age of 15 years. The effectiveness for obtaining results on mutation status was best using TaqMan (96.9%), followed by dHPLC (93.3%), HRM (88.7%), and sequencing (88.2%). In general, TaqMan was best for analyzing older tissues, whereas sequencing was the least efficient. Heterozygotic V600E was detected in 11.6%, 9.9%, 11.6%, and 9.9% of tissues using TaqMan, dHPLC, HRM, and sequencing, respectively. Result concordances between dHPLC and TaqMan or sequencing were excellent (κ = 0.9411 and κ = 0.8988, respectively); for HRM, the concordances were good (κ = 0.7973 and κ = 0.7488, respectively). By using DNA dilutions from tumor tissue, a minimum of 10% of V600E harboring cancer content was required for the analysis by dHPLC and HRM. dHPLC could detect four non-V600E mutations, whereas HRM detected one. Our results indicate that dHPLC and HRM are techniques that can be reliably used for the detection of the BRAFV600E mutation in archival paraffin-embedded tissues.
Recent advances in DNA sequencing technology have made it possible to use high-throughput sequencing on the entire human colorectal cancer (CRC) spectrum, finding 848 genes that may be somatically mutated.1,2 The subsequent study of these mutations in CRC specimens worldwide would be facilitated by the use of less expensive and more efficient screening methods for mutation detection. Because most of these mutations are found in heterozygosis, screening techniques based on heteroduplex detection, such as denaturing high-performance liquid chromatography (dHPLC) or high-resolution melting (HRM) analysis, are strongly recommended. The V600E mutation in the BRAF oncogene has been found in CRCs with mismatch-repair gene deficiency. Familial CRCs with mismatch-repair gene deficiency do not harbor this mutation; rather, V600E is associated with the sporadic form of CRC associated with mismatch-repair gene deficiency. In addition to the diagnostic value of the BRAF mutation assessment, recent evidence3,4 has demonstrated that stage IV CRCs with the BRAF V600E mutation do not respond to epidermal growth factor receptor inhibitor therapy, thus extending its value to predictive grounds. In previous articles,5 V600E detection using TaqMan chemistry (Applied Biosystem, Foster City, CA) saved cost, time, and manual labor over direct sequencing, with the former showing 100% sensitivity and 100% specificity when the latter is considered as the reference method. Also, HRM has proved to be a reliable technique for V600E detection when compared with dHPLC, allele-specific PCR, and direct sequencing.6 The aim of our work was to compare the performance characteristics of BRAF V600E detection by dHPLC and HRM with the previously described TaqMan allelic discrimination method and direct sequencing.
Materials and Methods
Study Cases and DNA Extraction
A total of 195 colorectal paraffin blocks (172 CRCs and 23 specimens of normal mucosa from surgical margins) were collected from a single institution (Santa María del Rosell University Hospital, Cartagena, Spain). The study cases come from a previously described series of serrated and conventional CRCs from January 1995 to December 2009 (mean, 2003; SD, ±3.0 years).7 All patient identifiers were deleted to protect patient confidentiality, and the study was approved by the local ethical board.
Areas selected by two pathologists (J.G.-S. and M.P.-G.) were cut in five sections (4-μm thick) from the original paraffin blocks. A brand new microtome blade was used for step sectioning each paraffin block to avoid DNA cross contamination. Histological sections were deparaffinized with xylol, and genomic DNA extraction was performed using the QIAamp DNA minikit (catalogue no. 51306) and the QiaCube automatic nucleic acid extractor (Qiagen, Hilden, Germany), according to the instruction manuals. Genomic DNA was quantified by UV absorbance using the Biophotometer (Eppendorf AG, Hamburg, Germany). The average DNA concentration was 80 ng/μL. DNA samples were distributed in two 96-well plates. One nontemplate control was included per plate. Melanoma cell lines HBL, WR2, and BEU, harboring wild-type (WT) BRAF and the heterozygous and homozygous BRAF V600E mutations, respectively, were used as positive controls (a gift from Prof. Ghanem E. Ghanem, Oncology and Experimental Surgery Laboratory (LOCE) Faculty of Medicine, Université Libre de Bruxelles, Brussels, Belgium).
TaqMan Allelic Discrimination
DNA samples were diluted to 5 ng/μL and subjected to allelic discrimination using TaqMan probes for BRAF V600E detection and following the protocol described in the study by Benlloch et al.5 Primers from set 1 (125-bp amplicon size) and probes from set 2 were used (Applied Biosystems, Foster City, CA), as suggested by the researchers. The only modifications introduced by us compared with the previously mentioned procedure were the use of TaqMan GTXpress Master Mix onto a 7500F platform for PCR and fluorescent analysis (Applied Biosystems for both). Genotype assignment was performed using SDS version 1.4 software (Applied Biosystems).
dHPLC Analysis
A DNA sequence fragment of 173 bp containing the BRAFV600E mutation was amplified twice for each sample, with 50 ng DNA, 200 μmol/L deoxyribonucleotide triphosphates, 1 U Taq polymerase (AmpliTaq Gold; Applied Biosystems), and 1 μmol/L primers [5′-TGCTTGCTCTGATAGGAAAATG-3′ (forward) and 5′-CCACAAAATGGATCCAGACA-3′ (reverse)], in a final volume of 25 μL. To improve the detection of sequence changes in the amplified product, a 40-base GC clamp was anchored to the 5′ end of the reverse primer. PCR cycling conditions were as follows: 96°C for 9 minutes; 40 cycles of 95°C for 15 seconds, 60°C for 15 seconds, and 72°C for 15 seconds; and a final incubation at 72°C for 5 minutes. Before dHPLC analysis, PCR products were heated to 95°C for 10 minutes and then slowly cooled to room temperature to allow heteroduplex formation.
The PCR product, 5 μL, was then injected into a preheated reverse-phase column (Helix DVB; Varian Analytical Instruments, Walnut Creek, CA) equilibrated by triethyl ammonium acetate, 0.1 mol/L, in a Helix ProStar dHPLC instrument (Varian Analytical Instruments). DNA was removed from the column at a constant flow rate of 0.45 mL/minute by a linear acetonitrile gradient, achieved by mixing buffer A (0.1 mol/L triethyl ammonium acetate) with buffer B (0.1 mol/L triethyl ammonium acetate and 25% acetonitrile), with a final buffer B concentration of 63%. The temperature for separation of the V600E heteroduplex from the homoduplex was 57.5°C. In addition, the dHPLC assay was also performed at 58.0°C and 58.5°C for discarding false positives and identifying other non-V600E mutations.
The eluted DNA was detected at 260 nm. All results were analyzed using Star Reviewer software (Varian Analytical Instruments). When a heteroduplex curve similar to a positive V600E control was observed in both amplifications of the same sample, the case was considered as V600E positive.
Sequencing was performed using the same amplicon on an independent PCR for all cases of study (sequencing conditions are described later).
HRM Analysis
HRM analysis was set up using DNA from the HBL and WR2 melanoma cell lines, which are homozygotic V600 and heterozygotic V600E, respectively. After testing different combinations of primers and template DNA concentrations, the reaction was settled at 20 ng of DNA, 100 nmol/L of each of the primers, and 10 μL of MeltDoctor HRM Master Mix (reference 4415440; Applied Biosystems), in a total volume of 20 μL, according to instructions given by the HRM master mix purveyor. Primer sequences were the same as described by Benlloch et al5 for the TaqMan assays: BRAF-51F, 5′-CTACTGTTTTCCTTTACTTACTACACCTCAGA-3′; and BRAF-176R, 5′-ATCCAGACAACTGTTCAAACTGATG-3′. For DNA extracted from paraffin-embedded tissue, 50 ng was the preferred amount for optimal amplification. As suggested by the manufacturer, the amplification cycle was initially performed in two steps (denaturation at 95°C for 15 seconds and annealing/extension at 60°C for 1 minute). A three-step amplification cycle was tested, obtaining better results for amplification signal and V600E discrimination. Therefore, the PCR program was as follows: 95°C for 10 minutes; 40 cycles of 95°C for 15 seconds, 64°C for 30 seconds, and 72°C for 1 minute; and a dissociation cycle consisting of 95°C for 10 seconds, 60°C for 1 minute, and 95°C for 15 seconds (ramp rate, 1%). A PCR was performed in a 7500F real-time thermocycler, and HRM curves were analyzed using HRM software version 2.0 (Applied Biosystems for both). The use of a shorter amplicon (97 bp) and of 0.5 mmol/L increments in MgCl2 did not render better discrimination of BRAF mutations (data not shown).
Sensitivity Tests
To test the sensitivity of dHPLC and HRM in BRAF V600E mutation detection, serial dilutions from 50% to 0.5% of mutant allele (WR2 cell line) into WT DNA (HBL cell line) were performed. The fragment of interest was amplified twice in every dilution and analyzed separately. To estimate the minimum cancer content suitable for analysis, another sensitivity test was performed. The percentage of tumor cells harboring the V600E mutation was estimated by counting 25 microscopic fields (at ×20 magnification). Half of this percentage was assigned to the percentage of V600E alleles in the specimen. DNA extracted from this case was serially diluted in DNA from a WT case with a similar concentration. The limit of sensitivity was established by the dilution with the lower percentage of mutant allele in which the heteroduplex could be detected.
Direct Sequencing
Sequencing was performed on the same amplicon, with the same PCR conditions, as used for dHPLC. Before sequence analysis, 5 μL of PCR products was purified with the enzyme ExoSap-IT (USB, High Wycombe, UK) for 15 minutes at 37°C, followed by 15 minutes at 80°C. A sequencing reaction was performed with 1 μL of purified PCR product and the BigDye Terminator version 1.1 Cycle Sequencing Kit (Applied Biosystems), according to the standard protocol. The internal forward sequence, 5′-TGATAGGAAAATGAGATCTAC-3′, was used as the sequencing primer.
Before analysis, purification of the sequencing reaction product was performed with Performa DTR Gel Filtration Cartridges (EdgeBio, Gaithersburg, MD). Analysis was performed using a four-capillary automated sequencer (ABI Prism 3130 Genetic Analyzer; Applied Biosystems).
Statistical Analysis
Statistical analysis was performed using Epidat software version 3.1. (Xunta de Galicia, Spain). Cohen's κ coefficient was used for intertechnique concordance. Performance indexes (sensitivity and specificity) were used for dHPLC and HRM test evaluations, considering TaqMan allelic discrimination and direct sequencing as the references.
Results
Of 195 colorectal specimens, 189 (96.9%) could be genotyped by TaqMan allelic discrimination. Heterozygotic V600E was detected in 22 cases (11.6%), and the WT genotype was found in the rest of the cases.
PCR before dHPLC was able to amplify 93.3% of colorectal specimens. In 18 (9.9%) of these 182 cases, the V600E mutation was detected. All duplicate amplifications render the same result for each specimen. None of these techniques detected the V600E mutation in normal mucosa specimens.
Concerning HRM analysis, V600E heterozygote samples rendered a double-peak curve or a wide peak comprising the melting temperatures 75.4°C and 74.9°C, corresponding to WT and V600E alleles, respectively (Figure 1). Amplification products were obtained from 88.7% of the specimens. In 20 (11.6%) of 173 cases, V600E was detected; and in five cases, non-V600E variants were assigned by HRM software. One of these variants was identified as R603X by sequencing, and another one was detected as V600E by TaqMan, dHPLC, and sequencing.
Figure 1.
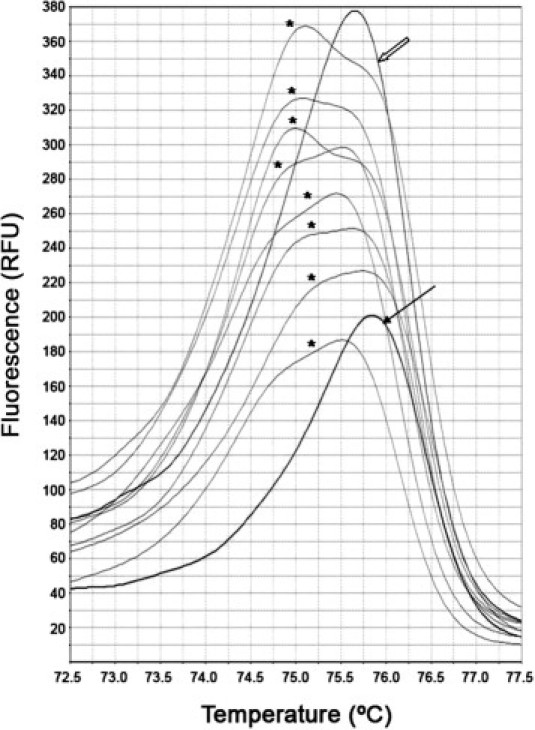
HRM analysis curves of specimens harboring heterozygous V600E (asterisk), WT V600 (empty arrow), and heterozygous R603X (solid arrow) detected by HRM as a non-V600E variant. RFU indicates relative fluorescence units.
A DNA sequence was obtained from 172 specimens (88.2%) by direct sequencing. A V600E mutation was detected in 17 cases (9.9%). In addition, nine non-V600E mutations were detected by this technique. Four of these mutations (ie, G581D, G596D, G596S, and A598T+W604X) (Figure 2) were detected by dHPLC, and one (R603X) was detected by HRM (Figure 1 and Table 1).
Figure 2.
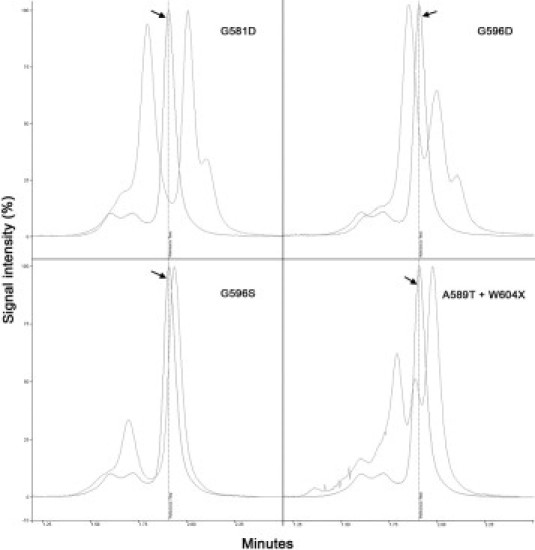
dHPLC curves obtained for different non-V600E mutations compared with the WT curve (arrows).
Table 1.
BRAF Mutations Found Using TaqMan, dHPLC, HRM, and Direct Sequencing
| Method (n = 195) |
||||
|---|---|---|---|---|
| Variable | TaqMan | dHPLC | HRM | Direct sequencing |
| Suitable for evaluation [no. (%)]⁎ | 189 (96.9) | 182 (93.3) | 173 (88.7) | 172 (88.2) |
| V600 WT | 167 | 160 | 152† | 146 |
| V600E [no. (%)]‡ | 22 (11.6) | 18 (9.9) | 20 (11.6) | 17 (9.9) |
| Mutation | ||||
| c.1800G>A | NAD | NAD | NAD | 1 |
| G581D | NAD | 1 | NAD | 1 |
| G596D | NAD | 1 | NAD | 1 |
| G596S | NAD | 1 | NAD | 3 |
| A598T+W604X | NAD | 1 | NAD | 1 |
| S602F | NAD | NAD | NAD | 1 |
| R603X | NAD | NAD | 1 | 1 |
| H608Y§ | NAD | NAD | NAD | 1 |
| Unsuitable for evaluation | 6 | 13 | 22 | 23 |
NAD, not able to be detected by this technique.
Referred to the total number of cases (n=195).
Including four non-V600E assigned mutations.
Referred to the number of cases suitable for evaluation.
H608Y was found in a specimen carrying a V600E mutation.
The amplification rate decreases with the age of paraffin-embedded tissues. In general, the TaqMan technique was the best when analyzing older tissues, whereas sequencing was the least efficient. dHPLC and HRM showed similar amplification rates (Figure 3).
Figure 3.
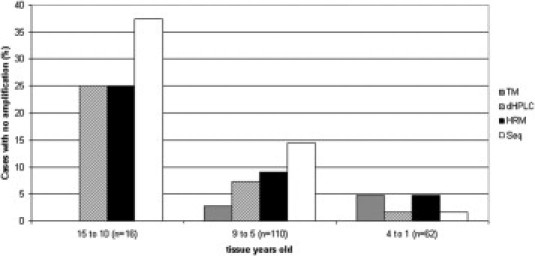
Comparison of the percentage of cases rendering no amplification for TaqMan (TM), dHPLC, HRM, and sequencing (Seq) techniques, according to the age of paraffin-embedded tissue.
Concordances between dHPLC and HRM versus TaqMan and direct sequencing in V600E detection are shown in Table 2. Cohen's κ coefficient rendered an excellent concordance between dHPLC and TaqMan for V600E detection (κ = 0.9411; 95% CI, 0.8602 to 1; P < 0.0001) and between dHPLC and sequencing (κ = 0.8988; 95% CI, 0.7857 to 1; P < 0.0001). The sensitivity of dHPLC in detecting V600E was 90% (95% CI, 74.4% to 100%), and the specificity was 100% (95% CI, 99.7% to 100%), when using TaqMan as the reference test. When direct sequencing was considered as the reference test, the sensitivity of dHPLC was 93.8% (95% CI, 78.8% to 100%) and the specificity was 98.6% (95% CI, 96.4% to 100%) (Table 2).
Table 2.
Concordance and Performance Indexes between dHPLC and HRM versus TaqMan and Direct Sequencing in V600E Detection
| dHPLC |
HRM |
|||||||
|---|---|---|---|---|---|---|---|---|
| Method | Positive (no.) | Negative (no.) | Sensitivity [% (95% CI)] | Specificity [% (95% CI)] | Positive (no.) | Negative (no.) | Sensitivity [% (95% CI)] | Specificity [% (95% CI)] |
| TaqMan | ||||||||
| Positive | 13 | 2 | 90 (77.4–100) | 100 (99.7–100) | 16 | 3 | 84.2 (65.2–100) | 97.4 (94.5–100) |
| Negative | 0 | 151 | 4 | 147 | ||||
| Direct sequencing | ||||||||
| Positive | 12 | 1 | 93.8 (78.8–100) | 98.6 (96.4–100) | 14 | 2 | 87.5 (68.2–100) | 95.7 (91.9–99.4) |
| Negative | 1 | 136 | 6 | 132 | ||||
Result concordances between HRM and TaqMan (κ = 0.7973; 95% CI, 0.6521 to 0.9424) and between HRM and sequencing (κ = 0.7488; 95% CI, 0.5831 to 0.9144) were good (P < 0.0001 for both). The sensitivity of HRM analysis was 84.2% (95% CI, 65.2% to 100%), and the specificity was 97.4% (95% CI, 94.5% to 100%), when using TaqMan as the reference. When direct sequencing was considered as the reference test, HRM sensitivity was 87.5% (95% CI, 68.2% to 100%) and specificity was 95.7% (95% CI, 91.9% to 99.4%).
In four cases, curve patterns that resembled the curve associated with V600E were observed by dHPLC. When these cases were injected at 58.5°C in the presence of positive and negative controls, it could be confirmed that these cases were not V600E carriers.
The limit of sensitivity for heteroduplex detection for dHPLC and HRM was established between 1% of mutated allele in 99% of WT allele and 5% of mutated allele in 95% of WT allele for both techniques (Figure 4). For the estimation of minimum cancer content, percentages of 10% to 20% of V600E allele in tumor sample for dHPLC and 5% to 10% of V600E allele in tumor sample for HRM were estimated (Figure 5).
Figure 4.
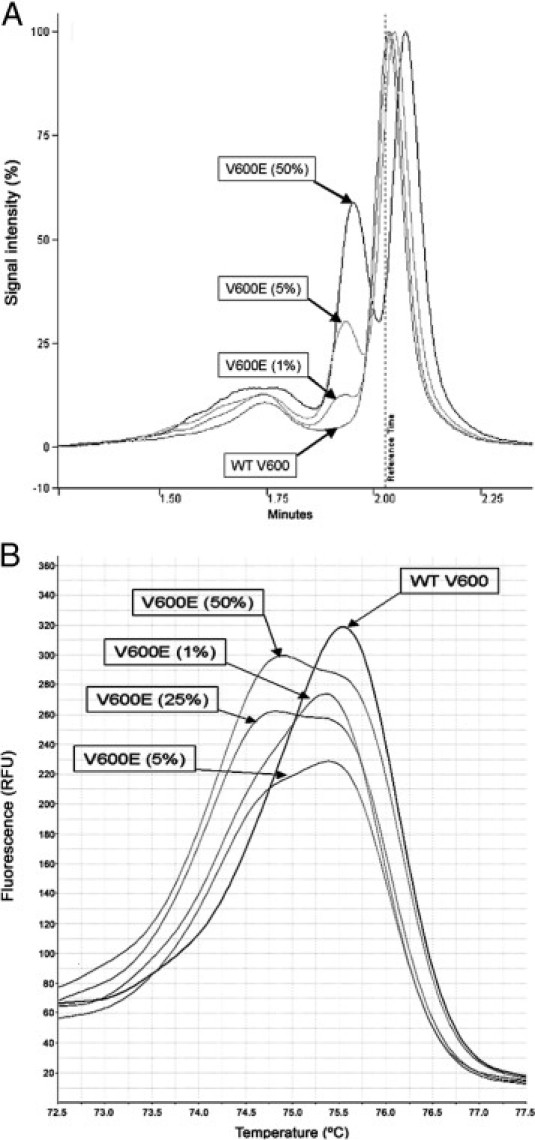
Sensitivity test for the detection of the V600E mutation by dHPLC (A) and HRM (B). DNA from the V600E heterozygote cell line was serial diluted in DNA from the WT cell line. Percentages indicate the proportion of the V600E allele.
Figure 5.
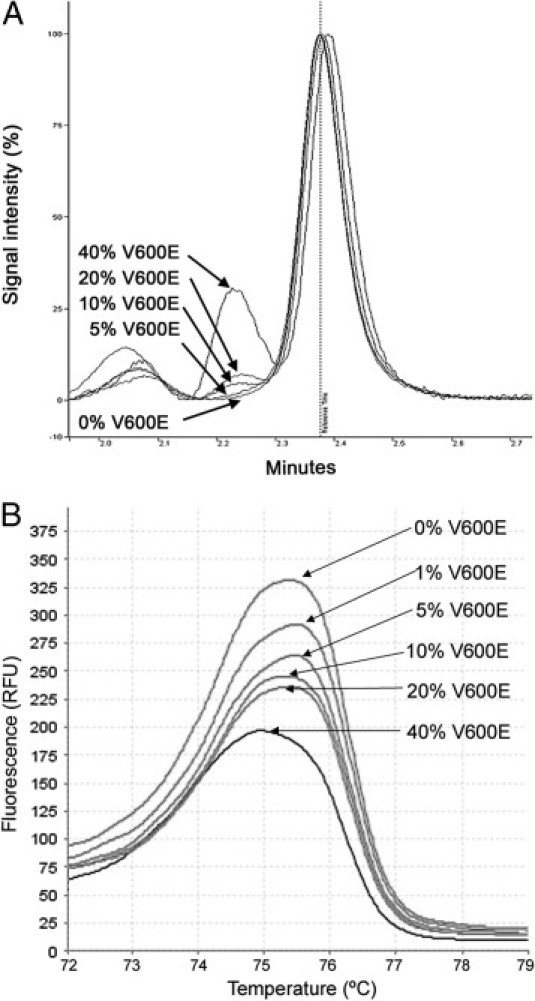
Estimation of minimum cancer content harboring the V600E mutation by dHPLC and HRM. Percentages indicated are the estimated proportion of the V600E allele versus WT in the DNA extracted from paraffin-embedded tissue, as measured by dHPLC (A) and HRM (B). RFU indicates relative fluorescence units.
Discussion
Screening techniques, such as dHPLC or HRM, will help to define the clinical importance of less frequent mutations in cancer. This finding is crucial given the fact that massive sequencing of CRCs has revealed that somatically mutated colorectal candidate genes harbor an average of 3.56 mutations per gene and, apart from APC, KRAS, and TP53, colorectal candidate genes are mutated in <21% of CRCs.1 For this reason, and given the diagnostic and predictive value attributed to the BRAF V600E mutation in CRC, we decided to assess the efficiencies of dHPLC and HRM for V600E detection and compare them with those of allelic discrimination by TaqMan and direct sequencing.
The V600E mutation is mostly found in heterozygosis in tissue specimens. In addition, tumor cells are accompanied by nontumoral WT BRAF cells. For this reason, a detection technique with high sensitivity is required. We performed a sensitivity test for dHPLC and HRM techniques. The test was designed by diluting DNA from cell lines harboring the V600E mutation with DNA from a WT cell line. Results for dHPLC and HRM suggest that these techniques are adequately sensitive for detecting the mutation when present in ratios as low as 1% to 5% (of V600E allele in the background of WT allele). Pichler et al6 obtained similar results performing HRM with reagents, real-time PCR equipment, and HRM analysis software (Roche Diagnostics, Vienna, Austria).
Our study cases come from a described series of CRCs,7 including specimens up to the age of 15 years, that are useful for testing sample requirements, given the fact that, in clinical practice, mutation detection experiments are usually performed on archival material. DNA from paraffin-embedded samples is seriously degraded after formalin treatment. The percentages of samples suitable for mutation analysis were comparable among TaqMan, dHPLC, and HRM (96.8%, 93.0%, and 88.7%, respectively). Therefore, amplification sensitivity was better for TaqMan, followed by dHPLC and HRM, whereas direct sequencing showed the poorest sensitivity. One reason for the slightly higher amplification rate for TaqMan could be that this technique combines both amplification and hybridization with specific fluorescent probes, thus making it more sensitive than amplification alone. Also, TaqMan is the technique that detects more instances of V600E. We also compared the four techniques looking at the relationship between tissue age and percentage of nonamplification (Figure 3). The TaqMan assay is especially convenient when handling older tissue specimens. In contrast, HRM and dHPLC assays increase their effectiveness when the samples are more recent, whereas sequencing is especially inadvisable with older samples. V600E detection rates suggest comparable sensitivities for dHPLC, HRM, and TaqMan; the rate is slightly lower for dHPLC when handling cases >9 years. Intriguingly, the only two cases that were WT by dHPLC and V600E by TaqMan came from paraffin blocks >9 years. In addition, one of them carried a double mutation (V600E + H608Y). This case was the one that was judged as WT by dHPLC and as V600E by sequencing (Table 1). The possible effect of heteroduplex destabilization because of the second mutation might influence the result; however, for those specimens that were adequate for evaluation with both techniques, excellent agreement was observed. In four cases, curve patterns that were similar (but not identical) to that of V600E were observed by dHPLC. When the temperature for injection was increased up to 58.5°C, these cases were confirmed as non-V600E carriers.
Four cases were V600E positive by HRM but WT by TaqMan, dHPLC, and sequencing. A possible explanation for this finding was given by Pichler et al,6 who also observed a noticeable rate of false-positive cases with HRM and attributed it to the failure of Taq polymerase to recognize modified bases and, therefore, amplify more artifacts existing in DNA extracted from paraffin-embedded tissue than true mutations.
By using direct sequencing, eight non-V600E mutations, which cannot be identified by TaqMan, have also been detected. Seven of them were previously reported in CRC, whereas A598T was reported in melanoma.8 dHPLC could detect more of these non-V600E mutations than HRM, thus adding support to the use of the former technique. However, the main objective of this study was to compare the efficiency of TaqMan, dHPLC, and HRM for detecting the V600E mutation in the BRAF gene, which has proven diagnostic and predictive value. Therefore, although it is possible that at least some of these non-V600E mutations (with uncertain clinical implications) could be detected by dHPLC at different analysis temperatures, the identification of other mutations in exon 15 of BRAF is out of the scope of this study.
The amplicon size for dHPLC and sequencing (173 bp) was similar to that for TaqMan and HRM (125 bp), indicating that the reason for differences in sensitivity is mainly because of the techniques themselves. The estimation of minimum cancer content harboring V600E mutation was slightly better for HRM (5% to 10%) than for dHPLC (10% to 20%), probably because of the higher sensitivity of fluorescent techniques.
In conclusion, the diagnostic and recent predictive values attributable to BRAF V600E justify the use of optimized techniques for its detection. The dHPLC and HRM screening methods for the detection of such a mutation are comparable to TaqMan allelic discrimination for sensitivity and specificity. dHPLC and HRM are useful techniques for detecting less frequent mutations, such as BRAF V600E, that also have an oncogenic role and might have clinical value in CRC patient management.
Acknowledgments
We thank Prof. Ghanem E. Ghanem (Université Libre de Bruxelles, Brussels, Belgium) and Prof. José C. García-Borrón (Murcia University, Murcia, Spain) for their help in the validation of these techniques and Dr. Diego Arcas for reviewing the English version of the manuscript.
Footnotes
Supported by grants from Carlos III Health Institute, Spanish Ministry of Health, Spain, and European regional development fund (FEDER), European Union (P1081210 and 127990).
P.C. and M.C.T. contributed equally to this work.
References
- 1.Sjöblom T., Jones S., Wood L.D., Parsons D.W., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., Szabo S., Buckhaults P., Farrell C., Meeh P., Markowitz S.D., Willis J., Dawson D., Willson J.K., Gazdar A.F., Hartigan J., Wu L., Liu C., Parmigiani G., Park B.H., Bachman K.E., Papadopoulos N., Vogelstein B., Kinzler K.W., Velculescu V.E. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 2.Wood L.D., Parsons D.W., Jones S., Lin J., Sjöblom T., Leary R.J. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 3.Di Nicolantonio F., Martini M., Molinari F., Sartore-Bianchi A., Arena S., Saletti P., De Dosso S., Mazzucchelli L., Frattini M., Siena S., Bardelli A. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 4.Markowitz S.D., Bertagnolli M.M. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benlloch S., Payá A., Alenda C., Bessa X., Andreu M., Jover R., Castells A., Llor X., Aranda F.I., Massutí B. Detection of BRAF V600E mutation in colorectal cancer: comparison of automatic sequencing and real-time chemistry methodology. J Mol Diagn. 2006;8:540–543. doi: 10.2353/jmoldx.2006.060070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pichler M., Balic M., Stadelmeyer E., Ausch C., Wild M., Guelly C., Bauernhofer T., Samonigg H., Hoefler G., Dandachi N. Evaluation of high-resolution melting analysis as a diagnostic tool to detect the BRAF V600E mutation in colorectal tumors. J Mol Diagn. 2009;11:140–147. doi: 10.2353/jmoldx.2009.080100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.García-Solano J., Pérez-Guillermo M., Conesa-Zamora P., Acosta-Ortega J., Trujillo-Santos J., Cerezuela-Fuentes P., Mäkinen M.J. Clinicopathological study of 85 colorectal serrated adenocarcinomas: further insights into the full recognition of a new subset of colorectal carcinoma. Hum Pathol. 2010;41:1359–1368. doi: 10.1016/j.humpath.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Deichmann M., Krahl D., Thome M., Wüst K., Hassanzadeh J., Helmke B. The oncogenic B-raf V599E mutation occurs more frequently in melanomas at sun-protected body sites. Int J Oncol. 2006;29:139–145. [PubMed] [Google Scholar]


