Abstract
BACKGROUND
The sperm chromatin of fertile men retains a small number of nucleosomes that are enriched at developmental gene promoters and imprinted gene loci. This unique chromatin packaging at certain gene promoters provides these genomic loci the ability to convey instructive epigenetic information to the zygote, potentially expanding the role and significance of the sperm epigenome in embryogenesis. We hypothesize that changes in chromatin packaging may be associated with poor reproductive outcome.
METHODS
Seven patients with reproductive dysfunction were recruited: three had unexplained poor embryogenesis during IVF and four were diagnosed with male infertility and previously shown to have altered protamination. Genome-wide analysis of the location of histones and histone modifications was analyzed by isolation and purification of DNA bound to histones and protamines. The histone-bound fraction of DNA was analyzed using high-throughput sequencing, both initially and following chromatin immunoprecipitation. The protamine-bound fraction was hybridized to agilent arrays. DNA methylation was examined using bisulfite sequencing.
RESULTS
Unlike fertile men, five of seven infertile men had non-programmatic (randomly distributed) histone retention genome-wide. Interestingly, in contrast to the total histone pool, the localization of H3 Lysine 4 methylation (H3K4me) or H3 Lysine 27 methylation (H3K27me) was highly similar in the gametes of infertile men compared with fertile men. However, there was a reduction in the amount of H3K4me or H3K27me retained at developmental transcription factors and certain imprinted genes. Finally, the methylation status of candidate developmental promoters and imprinted loci were altered in a subset of the infertile men.
CONCLUSIONS
This initial genome-wide analysis of epigenetic markings in the sperm of infertile men demonstrates differences in composition and epigenetic markings compared with fertile men, especially at certain imprinted and developmental loci. Although no single locus displays a complete change in chromatin packaging or DNA modification, the data suggest that moderate changes throughout the genome exist and may have a cumulative detrimental effect on fecundity.
Keywords: epigenetic, DNA methylation, ART, infertility, histone modifications, sperm
Introduction
The incidence of infertility is rising and estimated to affect one of six couples. As a result, the use of assisted reproductive technologies (ARTs) is rising and it is now estimated that ART accounts for 1–2% of all births in developed countries (Manipalviratn et al., 2009). Although ART has become a widely accepted and implemented therapy for many forms of infertility, there have been concerns about safety of ART for resulting offspring. Recently, a series of reports raised the concern that ART is associated with an increased incidence of major congenital malformations, chromosomal aberrations, miscarriage rates, intrauterine growth restriction, early childhood cancers and imprinting disorders (such as Angelmann's syndrome, Beckwith–Weidmann's syndrome and Retinoblastoma) (Hansen et al., 2002; Place and Englert, 2003; Rebar and DeCherney, 2004; Reefhuis et al., 2009). These reports suggest that many of the effects of ART on offspring are genetic or epigenetic in nature (Bourque et al., 2010; Nelissen et al., 2011).
The precedence for epigenetic abnormalities in ART stems from earlier work on animal models that suggests an increased incidence of imprinting errors in offspring conceived by ART compared with natural conception (Maher, 2005). This rise in imprinting errors was attributed to embryo or gamete manipulation, in vitro culture conditions, hormonal stimulation or ovulation induction (Khosla et al., 2001; Cox et al., 2002; DeBaun et al., 2003; Mann et al., 2004; Ludwig et al., 2005). However, a currently accepted alternative notion is that the increased incidence of imprinting disorders may be due to facilitation of conception using gametes of infertile couples that may have an elevated risk of epigenomic errors. This view aligns with the limited number of reports showing abnormal methylation patterns at imprinted loci in the gametes of infertile men (Marques et al., 2004, 2008, 2010; Kagami et al., 2007; Kobayashi et al., 2007, 2009; Hammoud et al., 2009b).
Recently, the understanding of the germline epigenome has expanded. We, and others, have shown that the sperm genome is hypomethylated with respect to a differentiated somatic cell (Weber et al., 2007; Hammoud et al., 2009a), and that the small percentage of nucleosomes retained in the mature sperm of human and mouse are enriched at developmental promoters (i.e. HOX clusters), miRNA genes and imprinted loci (Arpanahi et al., 2009; Hammoud et al., 2009a; Brykczynska et al., 2010). Furthermore, the promoters of genes involved in spermatogenesis, cell cycle and cell metabolism were associated with H3 Lysine 4 methylation (H3K4me3: an activating histone modification), while lacking H3 Lysine 27 methylation (H3K27me3: a repressive histone modification), in keeping with their activation during gametogenesis. In contrast, many of the promoters for genes encoding transcription factors important for embryonic development and morphogenesis bear two marks with antagonistic roles: H3K4me3 and H3K27me3—termed ‘bivalent’ chromatin. Here, large regions of H3K27me3 often overlap with smaller regions of H3K4me3. This bivalent chromatin observed in sperm was first described in embryonic stem cell, and is believed to help poise genes for either activation or repression later in development (Bernstein et al., 2006). These findings suggest that the sperm genome may be packaged and poised for two programs: the first is a reminiscent gametogenesis program (active chromatin marks), and the second is a future embryonic program (bivalent domains).
The conservation in epigenomic packaging in both the mouse and humans suggests a potential role for paternal histones in early developing embryos (Arpanahi et al., 2009; Hammoud et al., 2009a; Brykczynska et al., 2010). It is understood that some cases of IVF failure appear to be due to paternal factors, and one current focus of research is to determine whether abnormal epigenetics in gametes could account for failure of embryogenesis (Tesarik, 2005; Carrell, 2008) and subsequent infertility. We hypothesized that the retained nucleosomes in the male gamete exhibit a functional role in early embryogenesis and that changes in histone retention and histone modifications may have important clinical ramifications on embryo outcome and/or fertility potential. Therefore, we studied the epigenome of two classes of infertility: men with abnormal semen parameters [also previously shown to have altered protamine 1-to-protamine 2 (P1/P2 ratios)] and men with unexplained, poor embryogenesis during IVF therapy. This is the first report to show alterations genome-wide in the epigenomic landscape of infertile men.
Materials and Methods
Study participants
Sperm samples were obtained from seven infertility patients and four donors of known fertility attending the University of Utah Andrology and IVF Laboratories. All patients were consented under an IRB-approved protocol. The first four patients (Patients 1–4) had presumed male factor infertility who had an abnormal semen analysis and an abnormal P1/P2 ratio [two had lower than normal P1/P2 ratio (<0.8) and two with an elevated P1/P2 ratio (>1.2)]. Two of the patients had previously undergone IVF without obtaining a pregnancy, although morphologically normal embryos were transferred. The other two patients had undergone intrauterine inseminations without obtaining a pregnancy.
The remaining three patients had unexplained embryo arrest during IVF. Two of the poor embryo patients had good quality embryos on Day 2 of embryo culture, but all embryos arrested on Day 3. The third patient had pronuclear arrest of all embryos. Electron microscopy revealed normal sperm centrosome morphology in this patient. Sperm aneuploidy testing for five chromosomes was also performed on this patient and the aneuploidy rate was not elevated.
Semen sample collection and clinical parameters
Semen samples were collected after 2–5 days of abstinence and subjected to somatic cell lysis (0.1% SDS, 0.5% Triton-X in DEPC H2O) for 20 min on ice and washed with 1X phosphate-buffered saline to eliminate white blood cell contamination. Semen parameters were measured following WHO criteria (World Health Organization., 1999). Sperm count, motility, morphology, DNA damage, P1/P2 ratio and percentage of histone retention are summarized in Table I. DNA damage was assessed using the terminal deoxynucleotidyl transferase-mediated dUDP nick-end labeling assay (Aoki et al., 2005; Zini et al., 2008), and P1/P2 ratio was determined as previously described (Carrell and Liu, 2001).
Table I.
Patient population description and summary of semen parameters.
| Patient code | Female age | Male age | Progressive motility | Morphology | Sperm count | % TUNEL positive | P1/P2 ratio | % histone retention | Number Of IUI | Number Of IVF | Embryo quality |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor 1 | NA | 26 | 62 | 49 | 132 | 5 | 0.923 | Donor pool 3–5% | NA | NA | Many pregnancies |
| Donor 2 | NA | 28 | 55 | 35 | 100 | 9 | 1.0 | Donor pool 3–5% | NA | NA | Many pregnancies |
| Donor 3 | NA | 28 | 58 | 40 | 75 | 3 | 0.99 | Donor pool 3–5% | NA | NA | Many pregnancies |
| Donor 4 | NA | 35 | 50 | 46 | 60 | 14 | 0.78 | Donor pool 3–5% | NA | NA | Many pregnancies |
| Patient 1 | 31 | 31 | 28 | 14 | 119 | 52 | 1.73 | 25% | 0 | 2 | Day 3 embryos-No pregnancy twice |
| Patient 2 | 33 | 33 | 37 | 15 | 75 | 40 | 0.732 | 5% | 2 | 0 | No IVF data |
| Patient 3 | 29 | 32 | 23 | 10 | 10.7 | 2 | 1.539 | 13% | 0 | 1 | Day No pregnancy |
| Patient 4 | 30 | 30 | 37 | 15 | 75.5 | 30 | 0.739 | 8% | 3 | 0 | No IVF data |
| Patient 5 | 27 | 29 | 7 | 11 | 2.45 | 42 | 0.8 | 32% | 3 | 1 | Pronuclear arrest (twice) |
| Patient 6 | 26 | 29 | 42 | 35 | 49 | 19 | 0.96 | 16% | 1 | 3 | All embryos arrested on Day 3 (twice) |
| Patient 7 | 27 | 32 | 50 | 39 | 36 | 15 | 0.72 | 16.6% | 0 | 2 | All embryos arrested Day 3 (twice) |
Partitioning of histone and protamine-associated DNA
Partitioning of histone and protamine-associated DNA was performed as previously described (Hammoud et al., 2009a). Briefly, 10–20 million sperm cells were treated with an increasing concentration of micrococcal nuclease (MNase 10–240 U). The histone fraction was either used directly for chromatin immunoprecipitation (ChIP) or the DNA was purified and mononucleosomal length fragments (∼140–155 bp) were gel-purified and subjected to Illumina GAIIx, ∼10–20 million reads were mapped for each sample. The protamine-associated DNA was hybridized to the agilent expanded promoter arrays, a two-slide set (244 k X2) (Hammoud et al., 2009a).
Chromatin IP and preparation for genomics methods
ChIP and sequencing methods were as described previously (Hammoud et al., 2009a). ChIPs used the following antibodies: anti-H3K27me3 (Upstate 07–449) and H3K4me3 (Abcam 8580). For each modification, ∼10–20 million reads were aligned.
Methylation profiling using bisulfite sequencing
Sperm DNA was extracted as previously described (Hammoud et al., 2009b). One microgram of genomic DNA was converted using the epitect bisulfite kit (Qiagen, Valencia, CA). PCR products were cloned into TOPO 2.1 vector (Invitrogen), and bacterial cells were grown on kanamycin (50 µg/ml) and X-gal (25 µg/ml) plates overnight. Colony PCR was used to screen 10–20 colonies; however, 8–10 positive clones were submitted to sequencing. All genomics data sets will be deposited in GEO. Raw and processed data is available for manual download from https://bioserver.hci.utah.edu/gnomex/gnomex.html. See following Analysis IDs: A246 (GI36), A247(PTO4), A248(X7FC), A249(WA1V), A250(IXO1), A251(JQ81), A252(ODN2), A253 (TotalGenomicDNAInput-withProcessingNotes). The q-Value FDR tracks can also be directly visualized in the Integrated Genome Browser (http://bioserver.hci.utah.edu/BioInfo/index.php/Software:IGB) under the Data Access Tab → Human → H_sapiens_Mar_2006 → UofUtahBioinfoCore → Cairns Lab → SpermEpiProject→ Infertile Patients.
Computational data analysis
Low- and high-level agilent array and Illumina GAIIx sequencing data analysis were performed as previously described (Hammoud et al., 2009a). Both Timat2 and Useq analysis packages are available at the SourceForge projects (http://useq.sourceforge.net/, http://timat2.sourceforge.net/). For the array data, the relative difference pseudo-median scores were converted to log2 ratio values and a log2 value of 1 (2-fold enrichment) was the selected threshold for all analysis. For the sequencing data, 36 bp reads were generated using Illumina's Genome Analyser IIx and processed according to their standard software pipeline. Reads were mapped to the March 2006 NCBI Build 36.1 human genome and analyzed using the USeq ChIP-Seq application where a q-value threshold of 20 [false discovery rate (FDR) <0.01] and log2 ratio 1 was used to select regions for analysis.
Results
Histone localization is altered in the gametes of infertile men
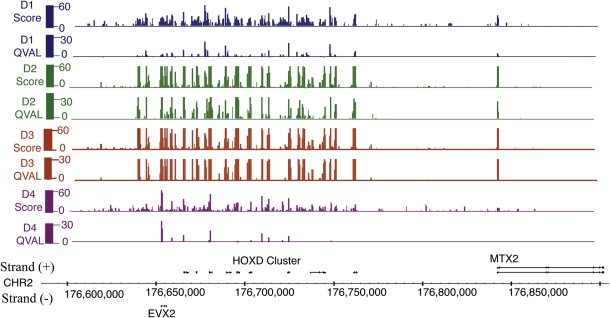
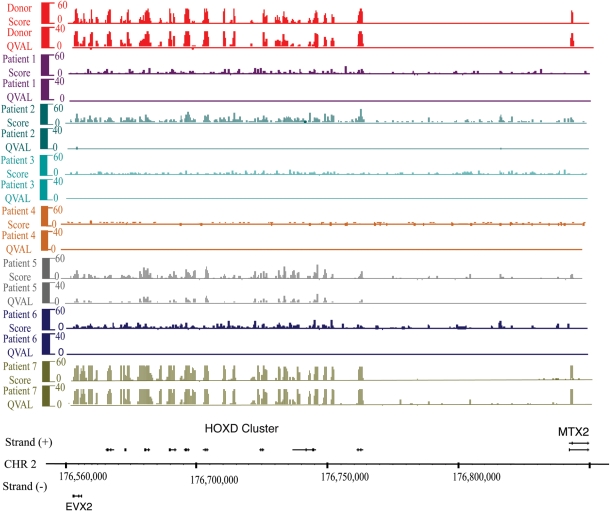
To better understand the relationship between the sperm epigenome and infertility, we localized nucleosomes in the gametes of seven men with infertility. All sperm samples were subjected to sequential increasing amounts of micrococcal nuclease digestion, and mononucleosomal length fragments were gel-purified and submitted to high-throughput sequencing to determine histone localization. Interestingly, unlike the consistent and repeated pattern of histone localization noted in fertile men (Fig. 1: note that each donor has ∼5 million reads compared with 20 million reads in patients), three men with abnormal semen parameters/protamine ratios and two of the three men from couples with embryo failure couples (five of seven infertile) had dispersed histone localization patterns genome-wide, suggesting that histone retention in infertile men maybe random (Fig. 2). For fertile donors and all infertile patients, we depict both sequencing read scores and the results of a statistical windowing program that quantifies the significance of read/histone enrichment (Qval, see Materials and Methods section). This distribution of nucleosomes is illustrated across the HOXD cluster (Figs 1 and 2); however, the observation of random histone localization for five of the seven patients extended genome-wide.
Figure 1.
Ac depiction of histone localization in each of the four donors in the HOXD cluster. Approximately, each donor has 5 million reads. The background reads in histone localization is minimal compared with patients (Fig. 2). Mapped sequencing reads were scored for enrichment (score) and for significance. Due to a low number of reads for each donor, statistical levels are much lower than the pooled (Fig. 2).
Figure 2.
High-throughput sequencing of mononucleosomes reveals histone retention at the HOX loci, and relatively random histone association in most infertile patients. Mapped sequencing reads were scored for enrichment (score) and for significance. Regions significantly enriched for histone relative to the input control (sheared total sperm DNA) were identified using a 300-bp window metric. For display, we depict score and FDR window scores [–10 log(FDR)]. Note that an FDR of 20 is equal to <0.01 and FDR 40 <0.001. Each patient sample has ∼20 million reads.
An altered histone localization profile in infertile men might be related to the percentage of the genome packaged in histone, as the failure to efficiently remove histone from the bulk genome will obscure the observation of relative retention patterns at developmental loci. Previous work from this laboratory showed that the histone retention in normal subjects was estimated to be in the range of 3–5% (Table I, Hammoud et al., 2009a,b). In contrast to fertile men, three of the four men with abnormal semen parameters/abnormal protamine ratios, and all of the men from couples with embryonic failure, had elevated histone retention in their bulk genome. Histone levels in infertile men were variable and ranged from 5 to 32% (Table I). This supports the notion that greater histone retention overall obscures the observation of specific enrichment at developmental loci. However, one of the embryo failure subjects (Patient 7), despite a markedly elevated histone retention (16%: Fig. 2, Table I), retained relative histone enrichment at expected promoters. Furthermore, a patient with an abnormal protamine ratio (Patient 2), although retaining a normal range of histone (5%: Table I, Fig. 2), did not display a normal retention profile. Therefore, measuring bulk nucleosome retention is insufficient to conclude nucleosomes are properly placed—one must determine the localization.
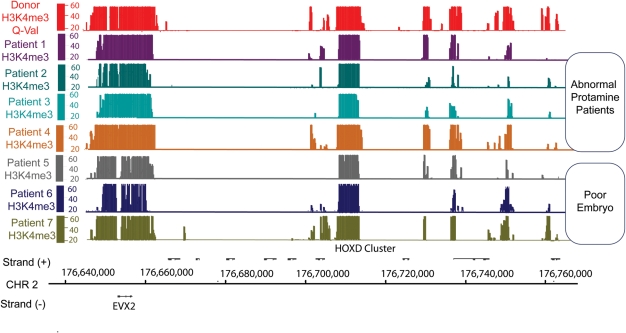
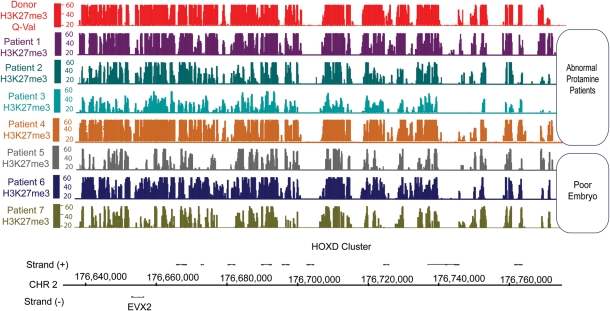
The loss of apparent histone enrichment at particular promoters may be due to excess histone across the genome, or instead due to the loss of histone from promoters at developmental and gametogenesis genes. To test this, we examined the level and placement of histone modifications, reasoning that if these attributes are retained, then properly marked histones remain. Here, we performed ChIP experiments for two histone modifications, H3K4me3 and H3K27me3, in our seven study subjects. The ChIP data were normalized to each subject's mononucleosome input, and a 300 bp window was used to identify enriched regions. We found that H3K4me and H3K27me methylation patterns in infertile men were generally similar to that of fertile donors (Figs 3 and 4 and Table II). Therefore, it seems likely that the abnormal histone localization pattern in infertile men is because of an incomplete replacement of histone by protamine in the genome as a whole, resulting in an increased background of abnormally retained histone that obscures the pattern of appropriately retained histones seen in normal sperm.
Figure 3.
H3K4me3 localization patterns are generally normal in the gametes of infertile men. This figure illustrates H3K4me3 retention aligned to the physical map of the HOXD locus. The y-axis depicts Storey q-value false discovery rate [–10log(FDR)]. Note that an FDR of 20 is equal to <0.01 and FDR 40 <0.001.
Figure 4.
H3K27me3 localization patterns are generally normal in the gametes of infertile men. This figure illustrates H3K27me3 retention aligned to the physical map of the HOXD locus. The y-axis depicts Storey q-value false discovery rate [–10log (FDR)]. Note that an FDR of 20 is equal to <0.01 and FDR 40 <0.001.
Table II.
Summary of fertile versus infertile men data set correlations.
| Comparison | r |
|---|---|
| Donor H3K4me3 versus Patient 1 H3K4me3 | 0.786 |
| Donor H3K4me3 versus Patient 2 H3K4me3 | 0.826 |
| Donor H3K4me3 versus Patient 3 H3K4me3 | 0.788 |
| Donor H3K4me3 versus Patient 4H3K4me3 | 0.868 |
| Donor H3K4me3 versus Patient 5 H3K4me3 | 0.698 |
| Donor H3K4me3 versus Patient 6 H3K4me3 | 0.749 |
| Donor H3K4me3 versus Patient 7 H3K4me3 | 0.749 |
| Donor H3K27me3 versus Patient 1 H3K27me3 | 0.807 |
| Donor H3K27me3 versus Patient 2 H3K27me3 | 0.771 |
| Donor H3K27me3 versus Patient 3 H3K27me3 | 0.818 |
| Donor H3K27me3 versus Patient 4 H3K27me3 | 0.787 |
| Donor H3K27me3 versus Patient 5 H3K27me3 | 0.666 |
| Donor H3K27me3 versus Patient 6 H3K27me3 | 0.775 |
| Donor H3K27me3 versus Patient 7 H3K27me3 | 0.785 |
Correlations for sequencing data were calculated using a 500-bp sliding window, moving every 100 bp. Windows with ≥5 reads were included in the analysis.
The sperm epigenome of infertile men
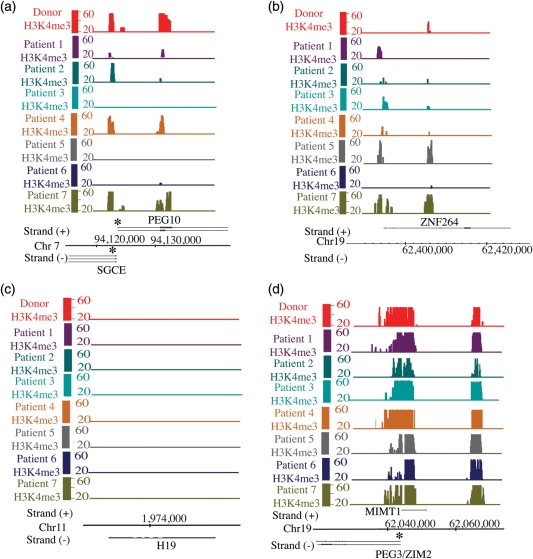
This genome-wide similarity in H3K4me3 and H3K27me3 data sets observed in fertile subjects and in subjects with evidence for reproductive dysfunction (Figs 3 and 4) was substantiated using gene ontology analysis (data not shown) and Pearson correlations (Table II). Similar to fertile controls, H3K4me gene ontology analysis revealed that H3K4me localization in subjects with sperm/protamine abnormalities or embryonic failure was enriched at the promoters of cell cycle genes, cellular metabolic processes, RNA processing and splicing, spermatogenesis gene promoters and developmental transcription factors (FDR 40 ≤ 1/1000). In contrast to H3K4me localization, H3K27me localization in control and abnormal subjects was similar, showing enrichment only at developmental gene promoters (FDR 40 ≤ 1/1000). These results imply that as in normal fertile men, sperm from men with reproductive disorders exhibit H3K4me enrichment at the gametogenesis program and H3K27me represses loci in the future embryonic program. The similarity in modification data sets for the two groups of subjects was also confirmed by the Pearson correlation coefficients on raw data sets (Table II). The correlation coefficients (r) for many of the data sets were estimated 0.7 or greater. These findings confirm that modification profiles for the fertile subjects and subjects with reproductive dysfunction are highly correlated, but not completely so, such that differences exist between individuals and classes (Table II). An illustrative gene-specific difference for H3K4me comparing normal and abnormal subjects can be observed at imprinted loci (Fig. 5). In fertile subjects, nearly all paternally expressed imprinted genes were enriched with H3K4me, while maternally imprinted loci lacked H3K4me3. In contrast, among the seven subjects with evidence for reproductive dysfunction, three (Patients 3, 5 and 6) showed absence of H3K4me3 at the promotors of three paternally expressed imprinted loci (PEG10, SGCE and NAP1L5), and six showed a gain of H3K4me3 at a maternally imprinted gene (ZNF264) (Fig. 5).
Figure 5.
A few maternally or paternally imprinted loci are improperly modified in the gametes of infertile men. (a) A depiction of two paternally imprinted genes (PEG10 and SGCE) that lack or have reduced levels of H3K4me3 in patients. (b) Representation of a maternally imprinted gene (ZNF264) that lacks K4me in fertile donors but acquires K4me in infertile men. (c, d) Represention of two properly marked maternally imprinted (H19) or paternally imprinted gene (PEG3). Asterisk represents the bisulfite amplicon.
Although the localization of modified nucleosomes was similar to controls in all seven subjects with evidence for reproductive abnormalities, a differential analysis was performed to identify loci that bear differences in the amount of H3K4me/H3K27me enrichment (read count at promoters). The analysis outputs (i) enriched regions: which are regions enriched in the patient and reduced in controls and/or (ii) reduced regions: which are regions deficient in patients and elevated in controls. These regions are subsequently submitted for gene ontology analysis to identify altered programs or pathways in infertile men. Interestingly, the top 300 genes with reduced H3K4me3 were predominately genes of bivalent promoters involved in embryonic processes such as multicellular oragnismal development, system development, organ development, pattern specification and not the promoters of cell cycle or spermatogenesis (FDR 40 < 1/1000). Similarly, the top 300 genes with reduced H3K27me3 promoters in the subjects were developmental promoters. This latter finding is expected because H3K27me3 is enriched only at developmental promoters in patients and controls (FDR 40 < 1/1000).
Changes in chromatin modifications do not generally result in marked changes in DNA methylation
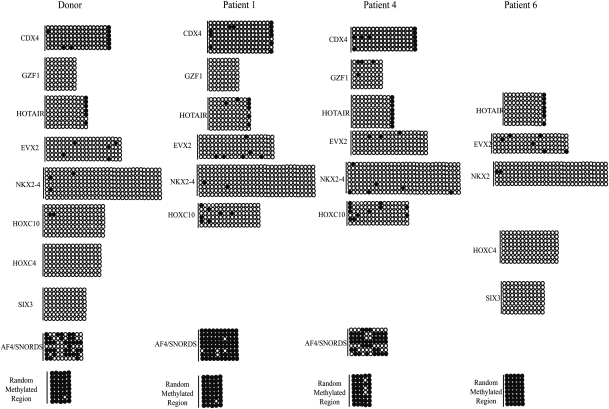
Recently, structural and in vitro data showed that H3K4 methylation deters DNA methylation by DNMT3A2 and DNMT3L in mice (Ooi et al., 2007). To examine whether a change in chromatin modifications might result in a change in DNA methylation, bisulfite sequencing of several candidate developmental loci that lacked or had significantly reduced levels of H3K4me3 were assessed in three infertile men (Patients 1, 4 and 6; Table III). As seen in fertile control subjects, the developmental promoters tested in the three infertile men were generally unmethylated with very few changes in methylation observed relative to controls (Fig. 6). Expanding the DNA methylation analysis to imprinted loci and including all subjects with evidence for reproductive dysfunction revealed more pronounced changes at the imprinted loci of Patients 3 and 5. However, the remainder of the subjects showed very limited changes in methylation (Fig. 7). The gain of DNA methylation did not always correlate with a loss of H3K4me. Subjects 3 and 5 lost K4me at PEG10, whereas 30–70% of their alleles gained methylation (Figs 5 and 7). In contrast, Patient 6 lost H3K4me at PEG10 but did not acquire methylation (Figs 5 and 7). Therefore, although we could not always correlate a loss of H3K4me with a gain in DNA methylation, infertile men appear more susceptible to changes in DNA methylation independent of the changes in chromatin in the mature sperm.
Table III.
Bisulfite primer coordinates for imprinted loci.
| Chr-start–stop (Hg18) | Forward primer | Reverse primer | Reference | |
|---|---|---|---|---|
| NAP1L5 | Chr4: 89,837,427–89,837,932 | FTTGGYGAGTAGGGGTTTATAGATGTTATTATATT NF-GTTTTTGTAGTTTTTTGAGGGTTAGGATT | AAAACCAAAAACCTACRAAACCTAACCAAA | In house |
| SGCE | Chr7: 94,122,796–94,123,031 | F-AAAAGATGTGTTTGTGTAGYGGGGA NF-TAGYGGGGAYGTAGTTTGATATTTATTTTT TTTG | AAATTCCATCTCCAATTCCTATTCCCAACAT | In house |
| PEG3 | Chr19: 62,043,540–62,043,862 | AAAAGGTATTAATTATTTATAGTTTGGT | AAAAATATCCACCCTAAACTAATAA | El-Maarri (2003) |
| ZAC | Chr6: 144,370,897–144,371,056 | GGGGTAGTYGTGTTTATAGTTTAGTA | CRAACACCCAAACACCTACCCTA | Kamikihara (2005) |
| PEG10 | Chr7: 94,123,573–94,123,828 | GGTGTAATTTATATAAGGTTTATAGTTTG | AACAAAAAAAATAAAATCCCACAC | Kim (2007) |
Figure 6.
DNA hypomethylation is largely maintained at developmental promoters of infertile men. Bisulfite sequencing of developmental promoters that lack or have reduced levels of K4me. CpGs are represented as open dots (if unmethylated) or filled dots (if methylated). Lines represent the number of alleles sequenced per patient.
Figure 7.
DNA methylation patterns are altered at a subset of paternally imprinted loci in the gametes of infertile men. CpGs are represented as open dots (if unmethylated) or filled dots (if methylated).
Discussion
This paper provides several lines of evidence that the genome packaging and epigenomic modifications are altered in the gametes of some infertile men. A few reports have shown that the methylation at several imprinted loci were altered in the gametes of infertile men (Marques et al., 2004, 2008, 2010; Kobayashi et al., 2007; Hammoud et al., 2009b; Boissonnas et al., 2010; Poplinski et al., 2010). We add that the alteration in the epigenome extends beyond DNA methylation at imprinted loci and entails changes in histone modifications and localization genome-wide at a large number of loci.
Altered histone localization patterns identified in the sperm of infertile men generally reflect a more random pattern of nucleosome retention rather than the localized retention seen in all fertile donors. This incomplete removal of nucleosomes might be attributed to problems with the chromatin remodeling machinery or improper histone acetyl transferase activity during the histone-to-protamine exchange; however, the effect of improper histone retention in early embryos is currently not well understood. One can speculate that if histones are replaced following fertilization, then non-programmatic histone retention may not necessarily affect the embryo. In contrast, if nucleosomes are retained but genome-wide reprogramming, e.g. DNA demethylation, requires naked DNA, then this non-programmatic histone retention may affect the efficiency and extent of reprogramming of the paternal pronucleus, though the perdurance of sperm nucleosomes in the embryo has not been established.
While nucleosomes were randomly retained genome-wide in infertile men, the localization of modified nucleosomes was similar to fertile controls suggesting that the establishment of epigenetic marks in the spermatogonial stem cell remain largely intact in infertile men. However, subtle but potentially important differences were identified. For example, developmental promoters enriched with H3K4 and H3K27me in known fertile men were lost or significantly reduced in most infertile men. The most striking observation is the loss of H3K4me3 from three paternally imprinted loci, a mark normally highly correlated with hypomethylation, in keeping with structural and functional studies of DNMT3L and DNMT3A in mice (Ooi et al., 2007). However, the loss or reduction of K4me3 in the mature sperm correlated with a gain in DNA methylation in only one of the seven patients. This shows that the loss of H3K4me3 is likely insufficient to cause DNA methylation, suggesting that additional mechanisms are likely involved in preventing DNA methylation at these imprinted loci.
The clinical ramifications of an altered H3K4me/H3K27me bound promoters in the gametes of infertile men are not yet known. However, clinical case studies have established that imprinting diseases in particular are not binary (presence or absence), but instead can result from moderate changes in DNA methylation status, e.g. in Beckwith–Weidmann's syndrome a 20% reduction in the number of methylated CpGs in a paternally methylated DMR results in disease onset. Therefore, the fidelity of the epigenome may need to be quite high to ensure embryo viability and health. Future studies that determine the relevance of chromatin marks in the germ-line are essential.
Although the number of patients assessed in this study is small (necessarily so due to the extreme costs associated with genome-wide analysis), the results are intriguing. Previously we, and others, have shown that all fertile men tested programmatically retain nucleosomes genome-wide. In contrast to fertile controls, six of seven infertile men had abnormal histone retention patterns, suggesting that histone to protamine exchange is defective in many infertile men. Furthermore, a subset of the patients displayed abnormalities in histone modifications (H3K4me and H3K27me) and DNA methylation at imprinted and developmental loci, which was absent in the fertile control pooled sample. This study demonstrates differences in the chromatin composition and epigenetic marking in the sperm of infertile men, especially at imprinted and developmental loci. Although no single locus displays a complete change in chromatin packaging or DNA modification, these moderate changes at many loci may have a cumulative detrimental effect on fecundity. This is aligned with current notions, since infertility is a multifactorial disease, and like other complex diseases, numerous etiologies likely contribute to male infertility or poor embryo outcome, with epigenetics being just one potential contributor.
In summary, this is the first report examining the sperm epigenome (histone localization and histone modifications) genome-wide in men with abnormal spermatogenesis or repeated poor embryo outcome. The risks associated with germ-line epigenetic abnormalities for the embryo is unclear, but we know that subtle differences in chromatin packaging were observed in the gametes of infertile men, and the penetrance of epigenetic abnormalities according to the literature is low; therefore, it is not entirely surprising that the frequency of epigenetic anomalies from ART is rare. The low disease penetrance can be attributed to several selected defense mechanism acquired through evolution and natural selection to ensure the formation of a viable conceptus.
Authors’ roles
B.R.C., D.T.C. and S.S.H. played a role in overall design. D.T.C. and S.S.H. were involved in the acquisition of samples, clinical logistics, patient consenting and Institutional Review Board documents. B.R.C., S.S.H. and D.A.N. detailed molecular and genomics approaches. S.S.H. and D.A.N. took part in data processing, array and sequencing analysis; S.S.H. carried out experiments and the figures. S.S.H. wrote the manuscript; D.T.C., B.R.C. (an investigator with HHMI), A.H. and M.G. played a role in manuscript review and input.
Funding
Financial support was provided by the Andrology and IVF laboratories clinical funds and Howard Hughes Medical Institute funds.
Acknowledgements
We thank Brian Dalley for microarray and sequencing expertise and Ken Boucher for statistical analysis. Financial support provided from the Department of Urology and the Howard Hughes Medical Institute.
References
- Aoki VW, Moskovtsev SI, Willis J, Liu L, Mullen JB, Carrell DT. DNA integrity is compromised in protamine-deficient human sperm. J Androl. 2005;26:741–748. doi: 10.2164/jandrol.05063. doi:10.2164/jandrol.05063. [DOI] [PubMed] [Google Scholar]
- Arpanahi A, Brinkworth M, Iles D, Krawetz SA, Paradowska A, Platts AE, Saida M, Steger K, Tedder P, Miller D. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res. 2009;19:1338–1349. doi: 10.1101/gr.094953.109. doi:10.1101/gr.094953.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. doi:10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boissonnas CC, Abdalaoui HE, Haelewyn V, Fauque P, Dupont JM, Gut I, Vaiman D, Jouannet P, Tost J, Jammes H. Specific epigenetic alterations of IGF2-H19 locus in spermatozoa from infertile men. Eur J Hum Genet. 2010;18:73–80. doi: 10.1038/ejhg.2009.117. doi:10.1038/ejhg.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque DK, Avila L, Penaherrera M, von Dadelszen P, Robinson WP. Decreased placental methylation at the H19/IGF2 imprinting control region is associated with normotensive intrauterine growth restriction but not preeclampsia. Placenta. 2010;31:197–202. doi: 10.1016/j.placenta.2009.12.003. doi:10.1016/j.placenta.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schubeler D, Stadler MB, Peters AH. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol. 2010;17:679–687. doi: 10.1038/nsmb.1821. doi:10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- Carrell DT. Contributions of spermatozoa to embryogenesis: assays to evaluate their genetic and epigenetic fitness. Reprod Biomed Online. 2008;16:474–484. doi: 10.1016/s1472-6483(10)60454-3. doi:10.1016/S1472-6483(10)60454-3. [DOI] [PubMed] [Google Scholar]
- Carrell DT, Liu L. Altered protamine 2 expression is uncommon in donors of known fertility, but common among men with poor fertilizing capacity, and may reflect other abnormalities of spermiogenesis. J Androl. 2001;22:604–610. [PubMed] [Google Scholar]
- Cox GF, Burger J, Lip V, Mau UA, Sperling K, Wu BL, Horsthemke B. Intracytoplasmic sperm injection may increase the risk of imprinting defects. Am J Hum Genet. 2002;71:162–164. doi: 10.1086/341096. doi:10.1086/341096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBaun MR, Niemitz EL, Feinberg AP. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet. 2003;72:156–160. doi: 10.1086/346031. doi:10.1086/346031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Maarri O, Buiting K, Peery EG, Kroisel PM, Balaban B, Wagner K, Urman B, Heyd J, Lich C, Brannan CI, et al. Maternal methylation imprints on human chromosome 15 are established during or after fertilization. Nat Genet. 2001;27:341–344. doi: 10.1038/85927. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009a;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Purwar J, Pflueger C, Cairns BR, Carrell DT. Alterations in sperm DNA methylation patterns at imprinted loci in two classes of infertility. Fertil Steril. 2009b;94:1728–1733. doi: 10.1016/j.fertnstert.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Hansen M, Kurinczuk JJ, Bower C, Webb S. The risk of major birth defects after intracytoplasmic sperm injection and in vitro fertilization. N Engl J Med. 2002;346:725–730. doi: 10.1056/NEJMoa010035. doi:10.1056/NEJMoa010035. [DOI] [PubMed] [Google Scholar]
- Kagami M, Nagai T, Fukami M, Yamazawa K, Ogata T. Silver-Russell syndrome in a girl born after in vitro fertilization: partial hypermethylation at the differentially methylated region of PEG1/MEST. J Assist Reprod Genet. 2007;24:131–136. doi: 10.1007/s10815-006-9096-3. doi:10.1007/s10815-006-9096-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamikihara T, Arima T, Kato K, Matsuda T, Kato H, Douchi T, Nagata Y, Nakao M, Wake N. Epigenetic silencing of the imprinted gene ZAC by DNA methylation is an early event in the progression of human ovarian cancer. Int J Cancer. 2005;115:690–700. doi: 10.1002/ijc.20971. [DOI] [PubMed] [Google Scholar]
- Khosla S, Dean W, Brown D, Reik W, Feil R. Culture of preimplantation mouse embryos affects fetal development and the expression of imprinted genes. Biol Reprod. 2001;64:918–926. doi: 10.1095/biolreprod64.3.918. doi:10.1095/biolreprod64.3.918. [DOI] [PubMed] [Google Scholar]
- Kim KP, Thurston A, Mummery C, Ward-van Oostwaard D, Priddle H, Allegrucci C, Denning C, Young L. Gene-specific vulnerability to imprinting variability in human embryonic stem cell lines. Genome Res. 2007;17:1731–1742. doi: 10.1101/gr.6609207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Hiura H, John RM, Sato A, Otsu E, Kobayashi N, Suzuki R, Suzuki F, Hayashi C, Utsunomiya T, et al. DNA methylation errors at imprinted loci after assisted conception originate in the parental sperm. Eur J Hum Genet. 2009;17:1582–1591. doi: 10.1038/ejhg.2009.68. doi:10.1038/ejhg.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Sato A, Otsu E, Hiura H, Tomatsu C, Utsunomiya T, Sasaki H, Yaegashi N, Arima T. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum Mol Genet. 2007;16:2542–2551. doi: 10.1093/hmg/ddm187. doi:10.1093/hmg/ddm187. [DOI] [PubMed] [Google Scholar]
- Ludwig M, Katalinic A, Gross S, Sutcliffe A, Varon R, Horsthemke B. Increased prevalence of imprinting defects in patients with Angelman syndrome born to subfertile couples. J Med Genet. 2005;42:289–291. doi: 10.1136/jmg.2004.026930. doi:10.1136/jmg.2004.026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher ER. Imprinting and assisted reproductive technology. Hum Mol Genet. 2005;14((1)):R133–138. doi: 10.1093/hmg/ddi107. doi:10.1093/hmg/ddi107. [DOI] [PubMed] [Google Scholar]
- Manipalviratn S, DeCherney A, Segars J. Imprinting disorders and assisted reproductive technology. Fertil Steril. 2009;91:305–315. doi: 10.1016/j.fertnstert.2009.01.002. doi:10.1016/j.fertnstert.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann MR, Lee SS, Doherty AS, Verona RI, Nolen LD, Schultz RM, Bartolomei MS. Selective loss of imprinting in the placenta following preimplantation development in culture. Development. 2004;131:3727–3735. doi: 10.1242/dev.01241. doi:10.1242/dev.01241. [DOI] [PubMed] [Google Scholar]
- Marques CJ, Carvalho F, Sousa M, Barros A. Genomic imprinting in disruptive spermatogenesis. Lancet. 2004;363:1700–1702. doi: 10.1016/S0140-6736(04)16256-9. doi:10.1016/S0140-6736(04)16256-9. [DOI] [PubMed] [Google Scholar]
- Marques CJ, Costa P, Vaz B, Carvalho F, Fernandes S, Barros A, Sousa M. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol Hum Reprod. 2008;14:67–74. doi: 10.1093/molehr/gam093. doi:10.1093/molehr/gam093. [DOI] [PubMed] [Google Scholar]
- Marques CJ, Francisco T, Sousa S, Carvalho F, Barros A, Sousa M. Methylation defects of imprinted genes in human testicular spermatozoa. Fertil Steril. 2010;94:585–594. doi: 10.1016/j.fertnstert.2009.02.051. doi:10.1016/j.fertnstert.2009.02.051. [DOI] [PubMed] [Google Scholar]
- Nelissen EC, van Montfoort AP, Dumoulin JC, Evers JL. Epigenetics and the placenta. Hum Reprod Update. 2010 doi: 10.1093/humupd/dmq052. [DOI] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. doi:10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place I, Englert Y. A prospective longitudinal study of the physical, psychomotor, and intellectual development of singleton children up to 5 years who were conceived by intracytoplasmic sperm injection compared with children conceived spontaneously and by in vitro fertilization. Fertil Steril. 2003;80:1388–1397. doi: 10.1016/j.fertnstert.2003.06.004. doi:10.1016/j.fertnstert.2003.06.004. [DOI] [PubMed] [Google Scholar]
- Poplinski A, Tuttelmann F, Kanber D, Horsthemke B, Gromoll J. Idiopathic male infertility is strongly associated with aberrant methylation of MEST and IGF2/H19 ICR1. Int J Androl. 2010;33:642–649. doi: 10.1111/j.1365-2605.2009.01000.x. [DOI] [PubMed] [Google Scholar]
- Rebar RW, DeCherney AH. Assisted reproductive technology in the United States. N Engl J Med. 2004;350:1603–1604. doi: 10.1056/NEJMp048046. doi:10.1056/NEJMp048046. [DOI] [PubMed] [Google Scholar]
- Reefhuis J, Honein MA, Schieve LA, Correa A, Hobbs CA, Rasmussen SA. Assisted reproductive technology and major structural birth defects in the United States. Hum Reprod. 2009;24:360–366. doi: 10.1093/humrep/den387. doi:10.1093/humrep/den387. [DOI] [PubMed] [Google Scholar]
- Tesarik J. Paternal effects on cell division in the human preimplantation embryo. Reprod Biomed Online. 2005;10:370–375. doi: 10.1016/s1472-6483(10)61798-1. doi:10.1016/S1472-6483(10)61798-1. [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. doi:10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- World Health Organization. WHO Laboratory Manual for the Examination of Human Semen and Sperm-Cervical Mucus Interaction. 4th edn. Cambridge, UK; New York, NY: Published on behalf of the World Health Organization Cambridge University Press,; 1999. [Google Scholar]
- Zini A, Boman JM, Belzile E, Ciampi A. Sperm DNA damage is associated with an increased risk of pregnancy loss after IVF and ICSI: systematic review and meta-analysis. Hum Reprod. 2008;23:2663–2668. doi: 10.1093/humrep/den321. doi:10.1093/humrep/den321. [DOI] [PubMed] [Google Scholar]