Abstract
The Src/FAK complex is involved in many signaling pathways and plays crucial roles in cell adhesion/migration. It becomes clear that the subcellular localization of Src and FAK is crucial for their activities and functions. In this article, we first overview the molecular mechanisms and functions of Src and FAK involved in cell adhesion/migration. We then introduce the development of genetically encoded biosensors based on fluorescence resonance energy transfer (FRET) to visualize the activities of Src and FAK in live cells with high spatiotemporal resolutions. Different kinds of signal peptides targeting subcellular compartments are also discussed. FRET-based biosensors fused with these targeting signals peptides are further introduced to provide an overview on how these targeting signals can facilitate the localization of biosensors to continuously monitor the local activity of Src and FAK at subcellular compartments. In summary, genetically-encoded FRET biosensors integrated with subcellular compartment-targeting signals can provide powerful tools for the visualization of subcellular Src and FAK activities in live cells and advance our in-depth understanding of Src/FAK functions at different subcellular compartments.
Keywords: Focal adhesion kinase, Src, FRET, Lipid rafts, Live cell imaging
INTRODUCTION
Non-receptor tyrosine kinases Src and FAK play important roles in cell differentiation, adhesion, migration and invasion6,37,45,49 The gene encoding the mutated and active form of Src, v-Src, was isolated as the first oncogene from the Rous sarcoma virus7 The cellular homologue, c-Src, was identified in 1978 as the first proto-oncogene and its product protein is the first tyrosine kinase.13,31,36 Focal adhesion kinase (FAK) was later discovered as one of the Src substrates at focal adhesions, the contact sites of a cell on extracellular matrix (ECM).33 The structure and function of Src and FAK, the interaction of Src/FAK and their clinical implications have been extensively reviewed.4,6,8,37,49 Interested readers are encouraged to pursue the details from these articles. In this review, we will specifically focus on the imaging study of Src/FAK enzymatic activities in live cells.
Src contains SH4, a SH3, a SH2, a tyrosine kinase domain, and a regulatory C-terminal tail.49 In its resting, Src exists as a closed conformation mainly through the intramolecular binding between the phosphorylated C-terminal Tyr527 and SH2 domain. This closed conformation is also facilitated by the interaction between SH3 domain and the linker region.56 When the closed conformation is disrupted, the kinase domain of Src can be exposed to phosphorylate its substrates. Therefore, Src activation mechanism includes the de-phosphorylation of Tyr527 by phosphatases, and the competitive bindings of phosphorylated tyrosines or proline-rich domains from other molecules with the Src SH2 or SH3 domain, respectively.56 For example, the auto-phosphorylated Tyr397 of FAK can recruit and bind to SH2 domain of Src, causing the activation of Src. Similar scenario exists for the FAK activation mechanism. FAK contains N-terminal FERM (protein 4.1, ezrin, radixin and moesin homology) domain, kinase domain, and C-terminal focal adhesion targeting (FAT) domain.38 When FAK is inactive, the N-terminal FERM domain directly binds to and masks the kinase domain. At the same time, the FERM domain also sequesters the linker region containing the major auto-phosphorylation site Tyr397.32 When this auto-inhibited conformation of FAK is destabilized, Tyr397 can be phosphorylated, creating a binding site for the SH2 domain of Src. Src can be subsequently recruited to phosphorylate Tyr516 and Tyr517 in the kinase domain of FAK, causing the further activation of FAK. Thus, Src and FAK work closely to form a molecular complex, which regulates numerous signaling pathways, in particular, in cell adhesion/migration.38
It becomes clear that the functions of Src and FAK are largely affected by their subcellular locations.38,49 Indeed, Src can be attracted to the plasma membrane via its N-terminal myristoylation signal and polybasic amino acids to become functional.49 FAK also contains the focal adhesion targeting domain in its C-terminus, which can interact with integrin-binding proteins, e.g. paxillin and talin, to lead FAK toward the membrane regions.38 As such, Src and FAK can be activated at the plasma membrane in response to various stimulations, e.g. the integrin clustering and growth factors. Genetically encoded biosensors based on fluorescence resonance energy transfer (FRET) have been designed to detect the FRET changes induced by the molecular interactions upon Src/FAK activation, allowing the visualization of the Src/FAK activities in live cells. These biosensors can be further targeted to the plasma membrane where the Src/FAK complex functions to continuously monitor the Src/FAK activities in these local sites with high spatiotemporal resolution.
In this review, we will introduce the Src/FAK signaling pathways in regulating the focal adhesion and cytoskeleton dynamics during cell migration. We will then discuss recent advances in the development of Src/FAK biosensors based on FRET. Specific targeting motifs for subcellular localization will also be summarized. Finally, we will introduce recent studies visualizing the Src/FAK activities at different microdomains of plasma membrane by FRET biosensors with specific compartment-targeting signals.
Src/FAK SIGNALING IN CELL MIGRATION
Cell migration is a cyclic process consisting of cell polarization, protrusion and adhesion formation at the leading edge, and retraction at the rear.46 This highly integrated process requires the coordinated regulation of focal adhesion (FA) dynamics and cytoskeleton reorganization, which are largely controlled by Src/FAK signaling pathways. For example, Src and FAK can be rapidly phosphorylated upon cell adhesion to participate in the FAs formation.21,33,49 Other studies showed that fibroblasts deficient in FAK26 or expressing kinase-defective Src16 displayed increased sizes in FAs and decreased speeds in migration. These evidences suggest that the Src/FAK complex is involved in both FA assembly and disassembly. The Src/FAK complex has also been shown to regulate the reorganization of cytoskeleton.38,49 The FAK-mediated stabilization of local microtubules can result in cell polarization,42 which is a crucial step for the directional migration.46 The Src/FAK complex can also regulate actin reorganizations through Rho GTPases.3 Therefore, the Src/FAK complex plays crucial roles in cell adhesion/migration by regulation of FA dynamics and cytoskeleton reorganization.
Focal Adhesion Dynamics
Focal adhesion is composed of structural and regulatory protein complexes coupling the membrane receptor integrins and extracellular matrix (ECM), providing an anchor on which the cell can push or pull over ECM.19 It becomes clear that the integrin signaling involves a complex and robust molecular network.30,38 The Src/FAK complex is part of this network and can serve as a positive regulator for the FA assembly by recruiting related signaling molecules. FAK can localize at FAs via its C-terminal FAT domain, which can bind integrin-associated adaptor proteins, paxillin and talin.19 Integrin clustering upon cell adhesion causes FAK autophosphorylation at Tyr397, creating a binding site for the SH2-domain of Src.24 The recruited Src and the subsequent formation of Src/FAK complex can cause the activation of the type I phosphatidylinositol phosphate kinase γ (PIPKIγ) to increase the production of phosphatidylinolsitol-(4,5)-biphosphate (PIP2).12 PIP2 can function as binding sites for focal adhesion molecules such as α-actinin, vinculin, and talin,10 to promote the FA assembly. Thus, the Src/FAK complex can facilitate the formation of FA by modulating lipid composition of the plasma membrane and creating docking sites to recruit FA proteins.
The FAs near the leading edge are disassembled as new FAs assemble nearby in the migration direction. The disassembly of FAs at the rear and the retraction of the trailing edge of the cell can facilitate the complete of the migration cycle and enable the cell translocation. It becomes clear that the Src/FAK complex also plays important roles in these FA turnovers via various routes. The Src/FAK complex can recruit Grb2 into FAs and the subsequent ERK activation can displace FAK from paxillin, causing the disassembly of focal adhesion complexes. The autophosphorylation of FAK at Tyr397 can recruit Src, which in turn phosphorylates Tyr925 in the FAT domain of FAK. This phosphorylated Tyr925 creates the binding site for the SH2 domain of Grb2. Because the binding site of Grb2 in the FAK FAT domain partially overlaps with that of paxillin, FAK can be decoupled from paxillin and hence from focal adhesions.28 The recruited Grb2 can also lead to the activation of Ras and Erk2, which causes the phosphorylation at serine 910 in the FAT domain of FAK. This Ser910 phosphorylation can further facilitate the dissociation of FAK from paxillin and cause the focal adhesion disassembly.25 In addition, the Src/FAK complex can influence the focal adhesion disassembly through proteolysis. Calpain is a protease which cleaves talin and FAK. Calpain can be up-regulated by the Src/FAK-mediated Erk2 activity14 as well as local Ca2+ flux20 to cause the disassembly of focal adhesions. Matrix metalloproteases (MMPs), which can be regulated by Ras/ERK2 signaling, can also cleave the binding between focal adhesions and ECM to facilitate the FA disassembly.11 Therefore, the Src/FAK complex can mediate the focal adhesion turnover via the ERK activation.38
Besides the ERK signaling pathway, the Src/FAK complex can facilitate FA turnover by affecting the linkage between FA and actin cytoskeleton. α-Actinin is an actin-associated protein which can bind to vinculin and zyxin, thus crosslinking actin stress fiber to the focal adhesion. The Src/FAK complex can phosphorylate α-actinin at tyrosine 12, which subsequently reduces the binding afinity of α-actinin toward actin.27 This reduced linkage between focal adhesions and actin cytoskeleton can facilitate the focal adhesion turnover. As such, the Src/FAK complex can promote the disassembly of focal adhesion coordinating multiple molecular signaling pathways.
Cytoskeleton Reorganization
It has been shown that integrins can cause the FAK activation which affects the stabilization of the dynamic microtubules via RhoA and its effector mDia.42 This local microtubule stabilization in the leading edge can facilitate the cell polarization by mediating asymmetric distribution of adhesion molecules and membrane components, which is required for the directional migration. Thus, FAK facilitates cell polarization through microtubule stabilization during cell migration.
The reorganization of actin filaments is also crucial for cell protrusion, which consists of two different types of actin organizations: lamellipodia and filopodia. Lamellipodia is composed of branched actin filaments whereas filopodia is a structure of fine, parallel actin bundles.23 The Src/FAK complex can regulate actin remodeling during cell migration through Rho small GTPases. For example, the Src/FAK complex is involved in the formation of lamellipodia through Rac1 signaling.38 The adapter protein p130CAS can bind to the proline-rich domain of FAK via its SH3 domain and be phosphorylated by Src kinase. The phosphorylated p130CAS can create a binding site for the SH2 domain of Crk. Crk can then recruit DOCK180, a Rac guanine exchange factor (GEF), which subsequently activates Rac1. The phosphorylated Tyr397 of FAK can also recruit the SH2 domain of PI3K. The local accumulation of PIP3 caused by the PI3K activity can recruit another Rac GEF Vav2, which provides the second pathway for Rac1 activation.38 Activated Rac1 can then promote the formation of lamellipodia through the activation of WAVE and Arp2/3 proteins. Thus, the Src/FAK-mediated Rac1 activation can induce the lamellipodia protrusion in the leading edge, which is crucial for cell migration. Filopodia, a fine actin bundle structure in the leading edge, can sense and explore the extracellular environment of a migrating cell to guide cell migration. FAK can influence Cdc42 and its effecter N-WASP to promote the formation of filopodia.55 In fact, FAK can phosphorylate N-WASP at tyrosine 256, which can influence the subcellular localization of N-WASP to regulate filopodia. Therefore, the Src/FAK complex can regulate actin reorganization during cell migration through Rho GTPases.
LIVE CELL IMAGING OF THE Src/FAK SIGNALING BY FRET
Live cell imaging can provide tremendously more insights than traditional biochemical assays,1,22 which usually require the cells to be killed.53 The discovery of green fluorescence protein (GFP) and recent advances in fluorescence proteins (FPs) enabled visualizing of the distribution of a protein as well as tracking of its locations in live cells.34,58 Genetically-encoded bio-sensors based on FPs and fluorescence resonance energy transfer (FRET) can further allow the visualization and quantification of the molecular activity at subcellular levels with high spatiotemporal resolution. Fluorescence proteins for the FRET pair of biosensors can be chosen when the emission wavelength of a donor FP overlaps with the excitation wavelength of an acceptor FP. If these FRET pair FPs are in proximal with appropriate orientation, fluorescence resonance energy can be transferred to the acceptor when the donor is excited. Substrate-based kinase FRET biosensors can be developed accordingly by connecting a specific substrate peptide and its binding domain between a donor and an acceptor FP. When the target kinase is active in cells, it can phosphorylate the specific substrate in the biosensor, and the phosphorylated substrate can then bind to the intramolecular binding domain, e.g. SH2 domain. This intramolecular interaction can cause the conformation change of the bio-sensor, which results in the FRET change (Fig. 1). Thus, the molecular activity of the target kinase can be visualized in live cells by monitoring the FRET change. In this section, we will discuss the development of FRET biosensors for the visualization of Src and FAK activities in live cells.
FIGURE 1.
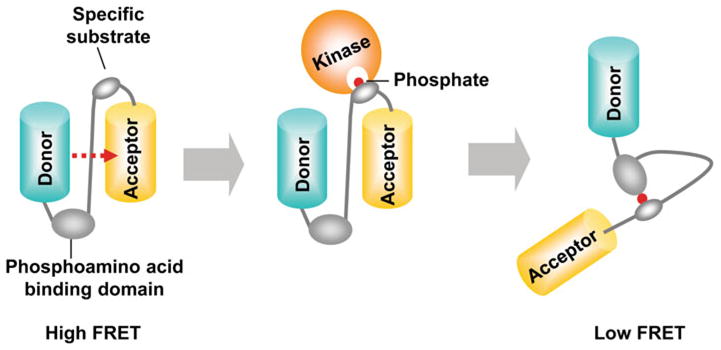
General design of a tyrosine kinase biosensor based on FRET. A typical kinase biosensor based on FRET is composed of a specific substrate peptide and its binding domain between a donor and an acceptor FPs. When the specific substrate in the biosensor is phosphorylated by active kinase, it can bind to the nearby binding domain, e.g. SH2 domain. This intramolecular interaction can cause the conformation change of the biosensor, which results in the FRET change.
Src FRET Biosensors
A FRET-based Src biosensor was initially developed using the strategy of substrate-based kinase biosensors described above.50 When the active Src phosphorylates the tyrosine of the substrate, the phosphotyrosine binds to the SH2 domain in the biosensor, causing the FRET change between a FRET pair FPs, enhanced cyan fluorescence protein (ECFP) and enhanced yellow fluorescence protein (EYFP). This Src biosensor showed around 30% of FRET changes in vitro when subjected to kinase assay in the presence of Src. However, the substrate of the biosensor, selected from in vitro library screening, also responded to EGFR, Abl and Lck.50 Because the specific FRET response is one of the essential features for the successful biosensor, the second version of Src biosensor was subsequently developed to improve the specificity.52 This Src bio-sensor has the substrate peptide derived from a Src substrate p130CAS, which can be specifically phosphorylated by Src. In addition, the intermolecular FRET, which can be caused by the dimerization tendency of FPs, was eliminated by the mutation of Ala206Lys in FPs. This improved biosensor displayed a high sensitivity and specificity toward Src kinase, and allowed the successful visualization of a wave propagation of Src activation upon mechanical stimulation induced by a laser-tweezer-traction of a bead seeded on top of human umbilical vein endothelial cell (HUVECs).5,52 Recently, ECFP and YPet were shown to serve as an improved FRET pair to allow a significantly enhanced dynamic range for various FRET biosensors, including the Src FRET biosensor.41 This third version of Src biosensor with enhanced sensitivity can allow the detection of subtle, but physiologically crucial signals.41 For example, this Src biosensor has allowed the visualization of a transient Src activation in bovine aortic endothelial cells (BAECs) upon vascular endothelial growth factor (VEGF) stimulation.41 With this bio-sensor, a rapid activation of Src upon mechanical stretch was also observed to have a speed 40 times faster than chemical stimulation.39
FAK FRET Biosensors
Several FAK biosensors based on FRET were recently developed. One of the FAK biosensors consists of a donor FP and an acceptor FP separately fused to FAK and a double-SH2 domain, respectively.9 This system is designed to report the intermolecular FRET increase upon the interaction between the autophosphorylated Tyr397 and SH2 domains. However, this intermolecular FRET system has some limitations compared to intramolecular FRET system. First, it is difficult to determine whether the FRET signals are engendered from the binding of the double-SH2 domain to the phosphorylated Tyr397 or other tyrosine sites in FAK. In addition, the acceptor/donor ratio at various subcellular locations is not uniform due to the different expression and localization of donors and acceptors. Thus, this approach requires more sophisticated mathematical tools to quantify the signals than a simple donor/acceptor ratio in live cells. Another two FAK biosensors were developed to detect the intramolecular FRET upon the conformational change of FAK.9,43 It has been shown that, in the auto-inhibited state of FAK, the N-terminal FERM domain binds to the kinase domain to block the phosphorylation of the active loop of FAK. This closed conformation can be open when FAK is activated.32 Accordingly, ECFP was fused at the N-terminus of FAK and EYFP inserted at the linker region between the FERM and the kinase domain of FAK to develop these FAK biosensors. Therefore, the conformational change of FAK, representing FAK activation, can cause the FRET change of these biosensors. Despite their similar design strategies, the FRET responses of these two biosensors upon FAK activation were opposite to each other: increasing in one case43 while decreasing in the other,9 suggesting a complex FRET mechanism of these reporters. The inserted FP at the linker regions may also partially expose the kinase domain, causing an elevated enzymatic activity of the biosensors comparing to the endogenous FAK.9 Also, the expression of this full-length FAK-containing bio-sensors may lead to the perturbation of the endogenous downstream signaling pathways, possibly by introducing extra enzymatic activities of FAK in host cells.
Recently, a substrate-based FAK FRET biosensor has been developed. This FAK biosensor is composed of a specific substrate and Src SH2 domain between an enhanced FRET pair ECFP and YPet. The substrate sequence is derived from the linker region encompassing Tyr397 of FAK. When Tyr397 in the substrate of the biosensor is phosphorylated by the active FAK (trans-activation), the phosphorylated substrate can then bind to the intramolecular SH2 domain, causing a conformation change and a decrease in FRET. This FAK biosensor has a design strategy similar to the Src FRET biosensor, with the only difference on the substrate peptide. In vitro kinase assay showed no FRET change of this FAK biosensor by addition of the active Src, indicating the specificity of the biosensor toward FAK, but not Src. In addition, the over-expression of this FAK biosensor did not affect the phosphorylation of ERK, suggesting the biosensor did not affect endogenous downstream signaling pathways (Seong et al., manuscript under review).
SUBCELLULAR VISUALIZATION OF THE Src/FAK SIGNALING
It is clear that the Src/FAK complex is involved in numerous crucial signaling pathways. To precisely control these various signaling events, the tight spatiotemporal regulation of the Src/FAK activity is important. In particular, proper subcellular localization is crucial because molecular interactions are largely dependent on the subcellular environment due to different sets and concentrations of molecular intermediates at different subcellular regions. Both Src and FAK can interact with membrane proteins such as integrins and growth factor receptors, it is hence expected that the activities of Src and FAK are largely determined by their localization at the plasma membrane. However, the plasma membrane is not uniform in structure and has different microdomains, e.g. lipid rafts, which are rich in cholesterol, sphingomyelin, and saturated fatty acids, and known to function as segregated signaling platforms. Therefore, the subcellular targeting of the Src or FAK biosensor is essential to visualize and study the activities of Src and FAK with high precision in space and time. In this section, we will discuss several targeting signals, and examples of their application in the subcellular targeting of FRET-based biosensors.
Protein Targeting Signals
Cells contain separate compartments including nucleus, mitochondria, endoplasmic reticulum (ER), and plasma membrane. Unlike diffusive dyes, e.g. Fura-2, one of the advantages of genetically encoded FRET biosensors is that they can be engineered to localize at the specific subcellular position and report the local activity of the target molecules. Many protein targeting signals have been discovered, e.g. nuclear localization signal (NLS),17 nuclear export signal (NES),51 the endoplasmic reticulum (ER)29 and plasma membrane targeting signals18,47 (Table 1). These short peptide sequences are identified from the endogenous proteins that are localized at the specific subcellular compartments. The fusion of these specific tags to the engineered proteins, such as FRET-based biosensors, can enable the targeting of the engineered proteins to the desired subcellular locations in a live cell. For example, a FRET-based Ca2+ biosensor was targeted into the ER, a major calcium store, to confirm the Ca2+ oscillation in human mesenchymal stem cells.29 The subcellular activity of JNK was also studied by a FRET-based JNK biosensor targeted to nucleus, mitochondria, and plasma membrane.17 The FRET biosensor detecting the activity of membrane type 1 matrix metalloproteinase (MT1-MMP) was tethered outside of the plasma membrane using the targeting sequence adopted from the transmembrane domain of the platelet-derived growth factor (PDGF) receptor β.40 Thus, this biosensor can successfully visualize the MT1-MMP activity dissolving the extracellular matrix in response to epidermal growth factor (EGF). Therefore, FRET-based biosensors with specific targeting signals can be a powerful tool to study the molecular signaling events in live cells with high spatiotemporal precisions.
TABLE 1.
The sequences of various subcellular targeting signals.
| Target site | Amino acids sequences | Location | References |
|---|---|---|---|
| Nucleus localization signal | PKKKRKVEDA | C-terminal | [17] |
| Nucleus export signal | LPPLERLTL | C-terminal | [51] |
| Endoplasmic reticulum | MLLPVLLLGLLGAAAD | N-terminal | [29] |
| KDEL | C-terminal | ||
| Mitochondria | MAIQLRSLFPLALPGMLALLGWWWFFSRKK | N-terminal | [17] |
| Outer plasma membrane | AVGQDTQEVIVVPHSLPFKVVVISAILALVVLTIISLIILIMLWEKKPR | C-terminal | [40] |
| Lipid rafts inside plasma membrane | MGCIKSKRKDNLNDDE | N-terminal | [18,47] |
| Non-Lipid rafts inside plasma membrane | KKKKKKSKTKCVIM | C-terminal | [18,47] |
Targeting Signals to Microdomains of the Plasma Membrane
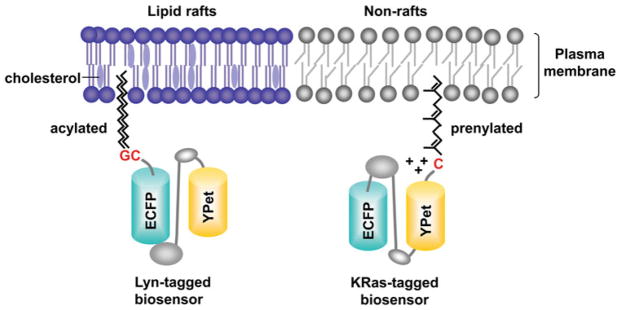
It has been shown that lipid modifications, e.g. acylation and prenylation, are sufficient to target a protein into different microdomains of plasma membrane.57 The FRET biosensor with these specific targeting signals can allow the live cell imaging of molecular activity in or outside of lipid rafts, whose size is beyond the resolution of the microscope. For example, the local activities of Src and Akt at lipid rafts of the plasma membrane were recently visualized to be different from those at the general membrane regions.18,47 The targeting signal for lipid rafts (Lyn-tag) contains the acylation substrate derived from Lyn kinase (Fig. 2). The N-terminal glycine and cysteine sites at this acylation sequence can be myristoylated and palmitoylated so that the tagged protein modified with these long saturated fatty acids can be incorporated into the lipid raft microdomains of the plasma membrane. Non-raft targeting signal (KRas-tag) was adopted from KRas which has C-terminal polybasic amino acids and a cysteine site for prenylation modification. The tagged protein with this long unsaturated fatty acids and positively charged amino acids can be attracted to the general membrane regions of the plasma membrane, but not partitioned into lipid rafts. As such, the Lyn- or KRas-tag can be added in the N- or C-terminus of the FRET-based biosensor, respectively, to localize the biosensor at different microdomains of the plasma membrane. These differentially localized Lyn- and KRas-tagged biosensors at the plasma membrane can be confirmed by modeling their movement in the cell geometry by two-dimensional diffusion simulation based on the finite element methods.35,48 Utilizing the images obtained by the fluorescent recovery after photobleach (FRAP) experiments, the finite element simulation and analysis have revealed that the Lyn-tagged biosensors are tightly attached to the membrane, with a slower motion than that of the KRas-tagged biosensors. The KRas-tagged biosensors appeared to be associated to the membrane with less stable binding state. The cytosolic biosensors, on the other hand, moved in 3D compartments between the nucleus and cytoplasm, which cannot be accurately described by a 2D diffusion model.35 These results confirmed that the Lyn- and KRas-tags can position the biosensors at different subcellular locations at the plasma membrane.
FIGURE 2.
Targeting of FRET biosensors into microdomains of plasma membrane. The targeting signal into lipid rafts (Lyn-tag) contains the acylation substrate derived from Lyn kinase. N-terminal glycine and cysteine can be myristoylated and palmitoylated. The tagged protein modified with these long saturated fatty acids can be incorporated into the lipid-raft microdomains of the plasma membrane. Non-raft targeting signal (KRas-tag) was adopted from KRas which has polybasic amino acids and a C-terminal cysteine site for prenylation modification. The tagged protein with this long unsaturated fatty acids and positively charged amino acids can be attracted to the general membrane regions of the plasma membrane, but not into lipid rafts.
Differential Src/FAK Activations at Different Microdomains of the Plasma Membrane
The Src activity at different microdomains of plasma membrane was studied utilizing the specific Src biosensor with Lyn- and KRas-tags described above.47 The Src biosensors tethered at different microdomains showed different FRET responses upon stimulations with pervanadate or growth factors: the FRET change of KRas-tagged Src biosensor was much faster and stronger than that of Lyn-tagged Src biosensor (Fig. 3). These results suggest that the Src activation outside of lipid rafts is faster and stronger than that within lipid rafts. In fact, Src has only a myristoylation motif whereas Lyn and Fyn, other Src family kinase members localized at lipid rafts, have the sites for both myristoylation and palmitoylation. Hence, this study supports the notion that a single acylation signal may not be sufficient to target the proteins inside lipid rafts. However, a small fraction of Src population can still be partitioned in lipid rafts, as evidenced by the observation of FRET response of the Lyn-tagged Src bio-sensor although it was much slower and weaker than that of the KRas-tagged Src biosensor. Computational analysis further revealed that growth factors can induce a Src activation at lipid rafts at clustered regions proximal to the cell periphery, possibly representing localized Src activation near the focal adhesion sites.35 It is possible that these different Src populations in and outside of lipid rafts can be involved in different downstream signaling events. Indeed, it has been shown that v-Src at lipid rafts regulates the PI3K/Akt signaling pathway, whereas Src at non-raft regions is involved in the MAPK/ERK signaling pathway.15 Therefore, lipid rafts function as segregated platforms of Src signaling pathways, allowing a fast and strong Src activation at non-raft regions and a slow and weak Src activation at lipid rafts.
FIGURE 3.
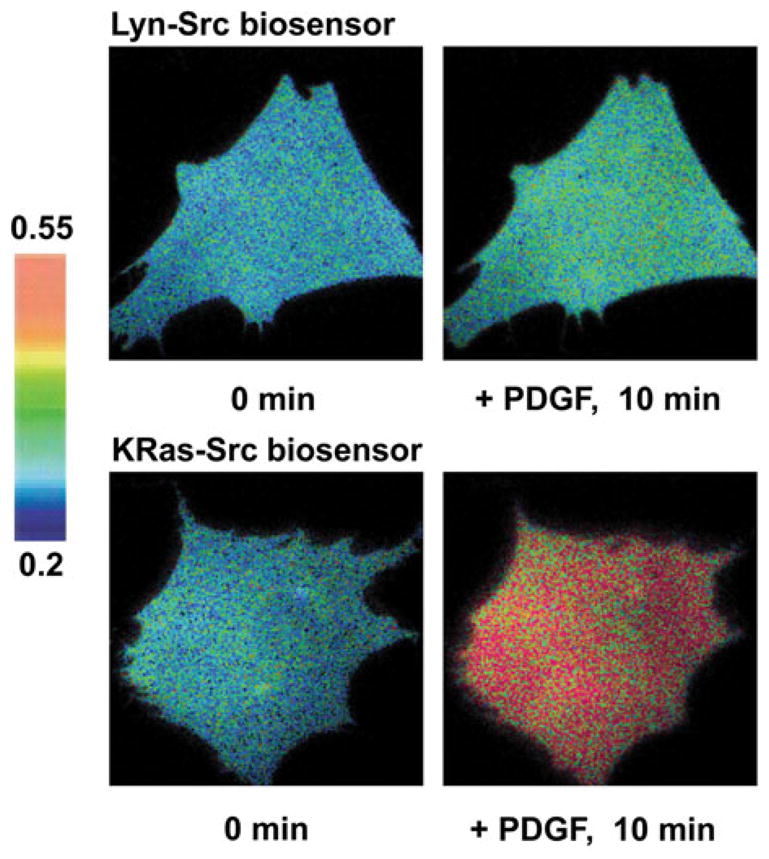
Differential Src activity in and outside of lipid rafts upon the PDGF stimulation. Mouse embryonic fibroblasts (MEFs) were transfected with Src biosensors containing microdomain targeting signals and stimulated with 10 ng/mL of PDGF. The ECFP/FRET images of Src biosensors before and after the PDGF stimulation are displayed. The color bars represent the ECFP/FRET ratio values.
The FAK activity at different microdomains of plasma membrane was recently studied with the FRET-based FAK biosensors with Lyn- and KRas-tags (Seong et al., manuscript under review). The FRET response of Lyn-FAK biosensor upon adhesion or PDGF stimulation was much stronger than that of KRas-FAK biosensor, suggesting that FAK activation might mainly occur at lipid rafts. This suggests a strong link between focal adhesions and lipid rafts. In fact, it has been previously shown that FAK can translocate to lipid rafts upon cell adhesion.2 The elimination of the raft-protein caveolin blocked the FAK phosphorylation upon integrin activation.54 The depletion of cholesterol and hence the disruption of rafts by methyl-β cyclodextrin (MβCD) also down-regulated FAK functions.44 These studies indicate that, although Src and FAK work closely as a molecular complex, they may be differentially localized at the plasma membrane: Src functions mainly outside lipid rafts, and only a minor fraction of Src co-localizes with FAK at lipid rafts. These studies also indicate that the integration of the FRET-based biosensors and specific targeting signals, i.e. acylation and prenylation, can provide power tools to study the activity of the Src/FAK complex at the subcellular compartments of the plasma membrane.
CONCLUSION AND PERSPECTIVES
In this review, we discussed the signaling pathways of the Src/FAK complex involved in cell adhesion/migration and the progress in the development of FRET-based biosensors to detect their subcellular activities. Recent studies utilizing specific Src/FAK FRET biosensors revealed the differential Src/FAK activities at different microdomains of the plasma membrane. It appears that Src has two distinct populations in and outside lipid rafts. It would be interesting to know further how different Src populations are tightly regulated to localize at different microdomains of the plasma membrane and to affect downstream pathophysiological consequences. In contrast to Src, FAK activity was mainly observed in lipid rafts of the plasma membrane. Since FAK is one of the major players in focal adhesions, it would be interesting to further delineate the relationship between lipid rafts and focal adhesions. These studies also showed that the subcellular targeting of the FRET biosensor provides a useful tool to visualize the local activity of proteins of interest in live cells. Therefore, utilizing this powerful method, more signaling pathways at the subcellular compartments can be studied in live cells. It is envisioned that FRET biosensors with further improved sensitivity and specificity will be developed. Biosensors with distinct colors will also become available in the future to allow the simultaneous visualization of multiple signals in the same live cell. All these should allow our in-depth understanding of the complex molecular signaling events in live cells.
Acknowledgments
This work is supported by grants from NIH HL098472, CA139272, NS063405, NSF CBET0846429, CMMI0800870 (Y.W.), and the Wallace H. Coulter Foundation and Beckman Laser Institute, Inc. (Y.W.).
References
- 1.Adachi T, Aonuma Y, Ito S, Tanaka M, Hojo M, Takano-Yamamoto T, Kamioka H. Osteocyte calcium signaling response to bone matrix deformation. J Biomech. 2009;42:2507–2512. doi: 10.1016/j.jbiomech.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 2.Baillat G, Siret C, Delamarre E, Luis J. Early adhesion induces interaction of FAK and Fyn in lipid domains and activates raft-dependent Akt signaling in SW480 colon cancer cells. Biochim Biophys Acta. 2008;1783:2323–2331. doi: 10.1016/j.bbamcr.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 3.Belsches AP, Haskell MD, Parsons SJ. Role of c-Src tyrosine kinase in EGF-induced mitogenesis. Front Biosci. 1997;2:d501–d518. doi: 10.2741/a208. [DOI] [PubMed] [Google Scholar]
- 4.Boggon TJ, Eck MJ. Structure and regulation of Src family kinases. Oncogene. 2004;23:7918–7927. doi: 10.1038/sj.onc.1208081. [DOI] [PubMed] [Google Scholar]
- 5.Botvinick EL, Wang Y. Laser tweezers in the study of mechanobiology in live cells. Methods Cell Biol. 2007;82:497–523. doi: 10.1016/S0091-679X(06)82018-2. [DOI] [PubMed] [Google Scholar]
- 6.Brown MT, Cooper JA. Regulation, substrates and functions of src. Biochim Biophys Acta. 1996;1287:121–149. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- 7.Brugge JS, Erikson RL. Identification of a transformation-specific antigen induced by an avian sarcoma virus. Nature. 1977;269:346–348. doi: 10.1038/269346a0. [DOI] [PubMed] [Google Scholar]
- 8.Brunton VG, Frame MC. Src and focal adhesion kinase as therapeutic targets in cancer. Curr Opin Pharmacol. 2008;8:427–432. doi: 10.1016/j.coph.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 9.Cai X, Lietha D, Ceccarelli DF, Karginov AV, Rajfur Z, Jacobson K, Hahn KM, Eck MJ, Schaller MD. Spatial and temporal regulation of focal adhesion kinase activity in living cells. Mol Cell Biol. 2008;28:201–214. doi: 10.1128/MCB.01324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calderwood DA. Integrin activation. J Cell Sci. 2004;117:657–666. doi: 10.1242/jcs.01014. [DOI] [PubMed] [Google Scholar]
- 11.Carragher NO, Levkau B, Ross R, Raines EW. Degraded collagen fragments promote rapid disassembly of smooth muscle focal adhesions that correlates with cleavage of pp125(FAK), paxillin, and talin. J Cell Biol. 1999;147:619–630. doi: 10.1083/jcb.147.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cary LA, Klinghoffer RA, Sachsenmaier C, Cooper JA. SRC catalytic but not scaffolding function is needed for integrin-regulated tyrosine phosphorylation, cell migration, and cell spreading. Mol Cell Biol. 2002;22:2427–2440. doi: 10.1128/MCB.22.8.2427-2440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collett MS, Brugge JS, Erikson RL. Characterization of a normal avian cell protein related to the avian sarcoma virus transforming gene product. Cell. 1978;15:1363–1369. doi: 10.1016/0092-8674(78)90061-2. [DOI] [PubMed] [Google Scholar]
- 14.Cuevas BD, Abell AN, Witowsky JA, Yujiri T, Johnson NL, Kesavan K, Ware M, Jones PL, Weed SA, DeBiasi RL, Oka Y, Tyler KL, Johnson GL. MEKK1 regulates calpain-dependent proteolysis of focal adhesion proteins for rear-end detachment of migrating fibroblasts. EMBO J. 2003;22:3346–3355. doi: 10.1093/emboj/cdg322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Diesbach P, Medts T, Carpentier S, D’Auria L, Van Der Smissen P, Platek A, Mettlen M, Caplanusi A, van den Hove MF, Tyteca D, Courtoy PJ. Differential subcellular membrane recruitment of Src may specify its downstream signalling. Exp Cell Res. 2008;314:1465–1479. doi: 10.1016/j.yexcr.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 16.Fincham VJ, Frame MC. The catalytic activity of Src is dispensable for translocation to focal adhesions but controls the turnover of these structures during cell motility. EMBO J. 1998;17:81–92. doi: 10.1093/emboj/17.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fosbrink M, Aye-Han NN, Cheong R, Levchenko A, Zhang J. Visualization of JNK activity dynamics with a genetically encoded fluorescent biosensor. Proc Natl Acad Sci USA. 2010;107:5459–5464. doi: 10.1073/pnas.0909671107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao X, Zhang J. Spatiotemporal analysis of differential Akt regulation in plasma membrane microdomains. Mol Biol Cell. 2008;19:4366–4373. doi: 10.1091/mbc.E08-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 20.Giannone G, Ronde P, Gaire M, Beaudouin J, Haiech J, Ellenberg J, Takeda K. Calcium rises locally trigger focal adhesion disassembly and enhance residency of focal adhesion kinase at focal adhesions. J Biol Chem. 2004;279:28715–28723. doi: 10.1074/jbc.M404054200. [DOI] [PubMed] [Google Scholar]
- 21.Guan JL, Shalloway D. Regulation of focal adhesion- associated protein tyrosine kinase by both cellular adhesion and oncogenic transformation. Nature. 1992;358:690–692. doi: 10.1038/358690a0. [DOI] [PubMed] [Google Scholar]
- 22.Guo XE, Takai E, Jiang X, Xu Q, Whitesides GM, Yardley JT, Hung CT, Chow EM, Hantschel T, Costa KD. Intracellular calcium waves in bone cell networks under single cell nanoindentation. Mol Cell Biomech. 2006;3:95–107. [PubMed] [Google Scholar]
- 23.Hall A, Paterson HF, Adamson P, Ridley AJ. Cellular responses regulated by rho-related small GTPbinding proteins. Philos Trans R Soc B-Biol Sci. 1993;340:267–271. doi: 10.1098/rstb.1993.0067. [DOI] [PubMed] [Google Scholar]
- 24.Hanks SK, Ryzhova L, Shin NY, Brabek J. Focal adhesion kinase signaling activities and their implications in the control of cell survival and motility. Front Biosci. 2003;8:d982–d996. doi: 10.2741/1114. [DOI] [PubMed] [Google Scholar]
- 25.Hunger-Glaser I, Fan RS, Perez-Salazar E, Rozengurt E. PDGF and FGF induce focal adhesion kinase (FAK) phosphorylation at Ser-910: dissociation from Tyr-397 phosphorylation and requirement for ERK activation. J Cell Physiol. 2004;200:213–222. doi: 10.1002/jcp.20018. [DOI] [PubMed] [Google Scholar]
- 26.Ilic D, Furuta Y, Kanazawa S, Takeda N, Sobue K, Nakatsuji N, Nomura S, Fujimoto J, Okada M, Yamamoto T. Reduced cell motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature. 1995;377:539–544. doi: 10.1038/377539a0. [DOI] [PubMed] [Google Scholar]
- 27.Izaguirre G, Aguirre L, Hu YP, Lee HY, Schlaepfer DD, Aneskievich BJ, Haimovich B. The cytoskeletal/non-muscle isoform of alpha-actinin is phosphorylated on its actin-binding domain by the focal adhesion kinase. J Biol Chem. 2001;276:28676–28685. doi: 10.1074/jbc.M101678200. [DOI] [PubMed] [Google Scholar]
- 28.Katz BZ, Miyamoto S, Teramoto H, Zohar M, Krylov D, Vinson C, Gutkind JS, Yamada KM. Direct transmembrane clustering and cytoplasmic dimerization of focal adhesion kinase initiates its tyrosine phosphorylation. Biochim Biophys Acta. 2002;1592:141–152. doi: 10.1016/s0167-4889(02)00308-7. [DOI] [PubMed] [Google Scholar]
- 29.Kim TJ, Seong J, Ouyang M, Sun J, Lu S, Hong JP, Wang N, Wang Y. Substrate rigidity regulates Ca2+ oscillation via RhoA pathway in stem cells. J Cell Physiol. 2009;218:285–293. doi: 10.1002/jcp.21598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kness M, Wang G, Zaman MH. Robustness of integrin signaling network. J Chem Phys. 2009;130:235103. doi: 10.1063/1.3149857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levinson AD, Oppermann H, Levintow L, Varmus HE, Bishop JM. Evidence that the transforming gene of avian sarcoma virus encodes a protein kinase associated with a phosphoprotein. Cell. 1978;15:561–572. doi: 10.1016/0092-8674(78)90024-7. [DOI] [PubMed] [Google Scholar]
- 32.Lietha D, Cai X, Ceccarelli DF, Li Y, Schaller MD, Eck MJ. Structural basis for the autoinhibition of focal adhesion kinase. Cell. 2007;129:1177–1187. doi: 10.1016/j.cell.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lipfert L, Haimovich B, Schaller MD, Cobb BS, Parsons JT, Brugge JS. Integrin-dependent phosphorylation and activation of the protein tyrosine kinase pp125FAK in platelets. J Cell Biol. 1992;119:905–912. doi: 10.1083/jcb.119.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lippincott-Schwartz J, Patterson GH. Development and use of fluorescent protein markers in living cells. Science. 2003;300:87–91. doi: 10.1126/science.1082520. [DOI] [PubMed] [Google Scholar]
- 35.Lu S, Ouyang M, Seong J, Zhang J, Chien S, Wang Y. The spatiotemporal pattern of Src activation at lipid rafts revealed by diffusion-corrected FRET imaging. PLoS Comput Biol. 2008;4:e1000127. doi: 10.1371/journal.pcbi.1000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin GS. The hunting of the Src. Nat Rev Mol Cell Biol. 2001;2:467–475. doi: 10.1038/35073094. [DOI] [PubMed] [Google Scholar]
- 37.Mitra SK, Schlaepfer DD. Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr Opin Cell Biol. 2006;18:516–523. doi: 10.1016/j.ceb.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 38.Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol. 2005;6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- 39.Na S, Collin O, Chowdhury F, Tay B, Ouyang M, Wang Y, Wang N. Rapid signal transduction in living cells is a unique feature of mechanotransduction. Proc Natl Acad Sci USA. 2008;105:6626–6631. doi: 10.1073/pnas.0711704105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ouyang M, Lu S, Li XY, Xu J, Seong J, Giepmans BN, Shyy JY, Weiss SJ, Wang Y. Visualization of polarized membrane type 1 matrix metalloproteinase activity in live cells by fluorescence resonance energy transfer imaging. J Biol Chem. 2008;283:17740–17748. doi: 10.1074/jbc.M709872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ouyang M, Sun J, Chien S, Wang Y. Determination of hierarchical relationship of Src and Rac at subcellular locations with FRET biosensors. Proc Natl Acad Sci USA. 2008;105:14353–14358. doi: 10.1073/pnas.0807537105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palazzo AF, Eng CH, Schlaepfer DD, Marcantonio EE, Gundersen GG. Localized stabilization of microtubules by integrin- and FAK-facilitated Rho signaling. Science. 2004;303:836–839. doi: 10.1126/science.1091325. [DOI] [PubMed] [Google Scholar]
- 43.Papusheva E, de Queiroz FM, Dalous J, Han Y, Esposito A, Jares-Erijmanxa EA, Jovin TM, Bunt G. Dynamic conformational changes in the FERM domain of FAK are involved in focal-adhesion behavior during cell spreading and motility. J Cell Sci. 2009;122:656–666. doi: 10.1242/jcs.028738. [DOI] [PubMed] [Google Scholar]
- 44.Park EK, Park MJ, Lee SH, Li YC, Kim J, Lee JS, Lee JW, Ye SK, Park JW, Kim CW, Park BK, Kim YN. Cholesterol depletion induces anoikis-like apoptosis via FAK down-regulation and caveolae internalization. J Pathol. 2009;218:337–349. doi: 10.1002/path.2531. [DOI] [PubMed] [Google Scholar]
- 45.Playford MP, Schaller MD. The interplay between Src and integrins in normal and tumor biology. Oncogene. 2004;23:7928–7946. doi: 10.1038/sj.onc.1208080. [DOI] [PubMed] [Google Scholar]
- 46.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 47.Seong J, Lu S, Ouyang M, Huang H, Zhang J, Frame MC, Wang Y. Visualization of Src activity at different compartments of the plasma membrane by FRET imaging. Chem Biol. 2009;16:48–57. doi: 10.1016/j.chembiol.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shvartsman DE, Donaldson JC, Diaz B, Gutman O, Martin GS, Henis YI. Src kinase activity and SH2 domain regulate the dynamics of Src association with lipid and protein targets. J Cell Biol. 2007;178:675–686. doi: 10.1083/jcb.200701133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas SM, Brugge JS. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- 50.Ting AY, Kain KH, Klemke RL, Tsien RY. Genetically encoded fluorescent reporters of protein tyrosine kinase activities in living cells. Proc Natl Acad Sci USA. 2001;98:15003–15008. doi: 10.1073/pnas.211564598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ullman KS, Powers MA, Forbes DJ. Nuclear export receptors: from importin to exportin. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- 52.Wang Y, Botvinick EL, Zhao Y, Berns MW, Usami S, Tsien RY, Chien S. Visualizing the mechanical activation of Src. Nature. 2005;434:1040–1045. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Shyy JY, Chien S. Fluorescence proteins, live-cell imaging, and mechanobiology: seeing is believing. Annu Rev Biomed Eng. 2008;10:1–38. doi: 10.1146/annurev.bioeng.010308.161731. [DOI] [PubMed] [Google Scholar]
- 54.Wei Y, Yang X, Liu Q, Wilkins JA, Chapman HA. A role for caveolin and the urokinase receptor in integrin-mediated adhesion and signaling. J Cell Biol. 1999;144:1285–1294. doi: 10.1083/jcb.144.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu X, Suetsugu S, Cooper LA, Takenawa T, Guan JL. Focal adhesion kinase regulation of N-WASP subcellular localization and function. J Biol Chem. 2004;279:9565–9576. doi: 10.1074/jbc.M310739200. [DOI] [PubMed] [Google Scholar]
- 56.Xu W, Harrison SC, Eck MJ. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 57.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 58.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]