Abstract
To elucidate the involvement of specific ultraviolet (UV) wavelengths in solar mutagenesis, we used a laser system to investigate the induction of DNA damage, both in the overall genome and at the nucleotide resolution level, in the genomic DNA of transgenic Big Blue mouse fibroblasts irradiated with a series of UV wavelengths, inclusive of UVC (λ<280 nm), UVB (λ=280–320 nm), and UVA (λ>320 nm). Subsequently, we sought correlation between the locations of UV-induced DNA lesions in the cII transgene of irradiated DNA samples and the frequency distribution and codon position of the induced cII mutations in counterpart mouse cells irradiated with simulated sunlight. Using a combination of enzymatic digestion assays coupled with gel electrophoresis, immunodot blot assays, and DNA footprinting assays, we demonstrated a unique wavelength-dependent formation of photodimeric lesions, i.e., cyclobutane pyrimidine dimers (CPDs) and (6–4) photoproducts [(6–4)PPs], based on direct UV absorption of DNA, in irradiated mouse genomic DNA, which could partially explain the induction of mutations in mouse cells irradiated with simulated sunlight. Most notably, there was a divergence of CPD and (6–4)PP formation at an irradiation wavelength of 296 nm in mouse genomic DNA. Whereas substantial formation of (6–4)PPs was detectable in samples irradiated at this wavelength, which intensified as the irradiation wavelength decreased, only small quantities of these lesions were found in samples irradiated at wavelengths of 300–305 nm, with no detectable level of (6–4)PPs in samples irradiated with longer wavelengths. Although CPD formation followed the same pattern of increase with decreasing wavelengths of irradiation, there were substantial levels of CPDs in samples irradiated with UVB wavelengths borderlined with UVA, and small but detectable levels of these lesions in samples irradiated with longer wavelengths. Because the terrestrial sunlight spectrum rolls off sharply at wavelengths ∼300 nm, our findings suggest that CPDs are the principal lesion responsible for most DNA damage-dependent biological effects of sunlight.—Besaratinia, A., Yoon, J. -I., Schroeder, C., Bradforth, S. E., Cockburn, M., Pfeifer, G. P. Wavelength dependence of ultraviolet radiation-induced DNA damage as determined by laser irradiation suggests that cyclobutane pyrimidine dimers are the principal DNA lesions produced by terrestrial sunlight.
Keywords: carcinogenesis, mutagenesis
Decades of epidemiologic studies have established an indisputable link between exposure to sunlight ultraviolet (UV) radiation and development of nonmelanoma skin cancers (i.e., basal and squamous cell carcinomas; refs. 1, 2). Conventionally, the UV spectrum of sunlight is divided into short wavelengths (UVC; λ<280 nm), middle wavelengths (UVB; λ=280–320 nm), and long wavelengths (UVA; λ=320–400 nm) (3, 4). The UVC fraction of sunlight is entirely absorbed by stratospheric oxygen (O2), which subsequently undergoes decomposition and recombination reactions, giving rise to ozone (O3) (5). The resulting O3 molecules can function as a filter to absorb the majority of sunlight UVB (5, 6). Consequently, the solar UV wavebands that reach the surface of the earth, and as such, are of relevance for photocarcinogenesis, are UVA and UVB, which comprise 95 and 5%, respectively, of the terrestrial sunlight UV (6, 7).
Earlier investigations of sunlight-induced carcinogenesis have solely focused on UVC and UVB, given the dogma that UVA absorption of DNA is weak, and owing to the methodological ease in utilizing UVC and UVB for experimental purposes (4). In recent years, however, the recognition of a potential role of UVA in the genesis of human melanoma has shifted the focus of photocarcinogenesis research from UVC/UVB to UVA (8, 9). Notwithstanding the surge in investigating UVA carcinogenicity, the genotoxic effects of UVA, specifically as they relate to DNA damage and mutagenesis, have been less than straightforward (10–12). The uncertainties in profiling UVA-induced DNA damage and mutations have arisen from the irreproducible and/or nonconclusive results of various in vitro and/or in vivo studies (reviewed in refs. 13, 14). For the most part, the divergent findings reported by various research groups could be ascribed to differences in experimental model systems (e.g., animals or cell types) and/or variations in experimental settings, e.g., UV irradiation source, UV dose, and treatment protocols used in different studies (13, 14). Of these, the use of UV irradiation sources with varying emission spectra and fluence rates seems to have caused considerable discrepancies in the results obtained from different studies. For example, UVA sources emitting contaminating wavelengths, especially in the UVB range, produce distorted genotoxic effects that are not specific for UVA (15, 16). Or, low-fluence-rate UVA sources can only be used for experimental purposes if there is prolonged exposure of samples in suboptimal conditions (i.e., long irradiation of naked DNA or cells in buffer solution), which is not necessarily physiologically relevant (17–27).
In the present study, we have investigated the DNA-damaging and mutagenic effects of solar UV using a tunable laser system (λ tuning range 260–350 nm), while accounting for the above-mentioned variables that may confound the results of any given experiment. More specifically, we have irradiated the genomic DNA of transgenic Big Blue mouse fibroblasts with a constant dose of UV at 13 different, yet precisely defined, serial wavelengths, inclusive of UVC, UVB, and UVA under uniform experimental conditions. Subsequently, we have determined the formation of two typical UV-induced photodimers, cis-syn-cyclobutane pyrimidine dimers (CPDs) and pyrimidine (6–4) pyrimidone photoproducts [(6–4)PPs], and two photooxidation-derived lesions, 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxo-dG) and 2,6-diamino-4-hydroxy-5-formamidopyrimidine (Fapy) (ref. 28 and Fig. 1) in the irradiated samples, using a combination of enzymatic digestion assays coupled with gel electrophoresis, which take advantage of specialized DNA repair enzymes that recognize and cleave specific types of lesions (29), and immunodot blot assays that utilize highly specific antibodies, which are raised against specific types of UV-induced lesions (30). We have further performed DNA footprinting (31) to locate the nucleotide positions in which UV-induced DNA lesions are formed in a mutational target gene (cII; ref. 32) in irradiated samples. Finally, we have sought correlation between the locations of UV-induced DNA lesions in the cII transgene of irradiated samples and the frequency distribution and codon position of sunlight-induced cII mutations in mouse fibroblast cells irradiated with simulated sunlight (SSL).
Figure 1.
Chemical structures of the investigated UV-induced DNA lesions. A) CPDs. B) (6–4)PPs. C) 8-Oxo-dG. D) Fapy.
MATERIALS AND METHODS
Cell culture
Primary mouse embryonic fibroblasts, prepared from 13.5-d-old embryos of Big Blue mice, were grown in Dulbecco's modified Eagle's medium (Irvine Scientific, Santa Ana, CA, USA) supplemented with 10% fetal bovine serum. When reaching near confluence, all cultures were harvested by trypsinization and subjected to genomic DNA isolation using a standard phenol and chloroform extraction and ethanol precipitation protocol (33). The DNA was dissolved in TE buffer (10 mM Tris-HCl and 1 mM EDTA, pH 7.5), and kept at −80°C until further analysis. Prior to UV irradiation, all DNA samples were reconstituted in phosphate buffered saline (PBS) to aliquots of 8 μg DNA in 100 μl PBS.
UV irradiation
Rather than a continuous lamp, the UV source we used here to irradiate the DNA samples was a quasi-continuous amplified Ti:sapphire-based laser system (Mira 900/RegA 9000; Coherent, Inc., Santa Clara, CA, USA). The laser system produces ∼60-fs pulses at 800 nm with a pulse repetition rate of 250 kHz. The output was frequency doubled to pump an optical parametric amplifier (OPA; OPA 9400; Coherent) capable of tuning from 500 to 700 nm. The appropriately tuned OPA output was then frequency doubled with a type I 0.25-mm-thick β-barium borate (BBO) crystal to produce the final desired UV wavelength. Care was taken to employ an appropriately thick BBO crystal to decrease the unconverted bandwidth in order to make the laser radiation as monochromatic as possible. The spectral profile was measured at each tuned wavelength with a spectrometer (USB2000; Ocean Optics, Inc., Dunedin, FL, USA) to monitor center wavelength as well as bandwidth. Typical bandwidths recorded were 2–4 nm. The tunability, spectral purity, clean near-gaussian beam profile, and near-monochromatic emission of this laser system make it an ideal light source for such experiments, where the determination of accurate wavelength dependence of radiation damage is sought. The use of a pulsed laser system, however, requires that careful consideration be given to avoid possible multiphoton effects. By employing a near-centimeter spot size in the sample, the peak intensity of each pulse is at least 4 orders of magnitude below the threshold to induce appreciable 2-photon ionization of the solvent (34), and about an order of magnitude below any significant photoionization of the DNA (35), processes that could lead to oxidative DNA damage (36, 37). These comparisons suggest that absorption of UV light in this experiment is in the linear regime, as with sunlight or lamp irradiation.
To irradiate DNA samples at each specified wavelength, a new sample was exposed at an irradiation dose of 0.5 J/cm2, by adjusting the average irradiance (W/cm2) for the determined spot size (Table 1), while keeping the exposure time approximately constant at ∼320 s to eliminate any possible variations that might be attributed to the length of time the sample was on the laser table and not on ice. The 0.5-J/cm2 dose was chosen based on our preliminary experiments, in which we could verify that this dose is above the damage threshold for wavelengths as long as 309 nm (i.e., λ≤309 nm). Despite the generation of damage at 0.5 J/cm2, there exists no observable effect on the absorption spectrum due to radiation exposure at doses as high as 8.6 J/cm2, as judged by the UV-visible (UV-vis) spectrum established by a single-monochromator spectrophotometer (Varian Cary 50; Varian Inc., Lake Forest, CA, USA; see Fig. 2). Because the objective of our study was to determine the correlation between UV-induced DNA damage and mutations, we chose this dose of UV irradiation after verifying that it could elicit a mutagenic response in our experimental model system. In our preliminary studies, we confirmed that irradiation of transgenic mouse embryonic fibroblasts with SSL for the same duration of time and under the same experimental conditions as described here, was significantly mutagenic, as reflected by the elevation of background cII mutant frequency from 3.45 ± 0.95 to 15.03 ± 3.54; P = 0.0079 (median ± sd; Wilcoxon rank-sum test). Of note, we installed an atmospheric attenuation filter (Air Mass 1.0; Newport, Stratford, CT, USA) in this SSL to correct the output emission to match the solar spectrum at ground level when the sun is directly overhead.
Table 1.
Irradiation conditions for mouse genomic DNA at a series of UV wavelengths
| Sample | Actual λ (nm)a | FWHM (nm) | Spot size (cm2) | Power (mW) | Irradiation time (s) | Dose (J/cm2)b | Attenuation correctionc |
|---|---|---|---|---|---|---|---|
| 1 | 349 | 4.1 | 0.319 | 0.50 | 319 | 0.5 | 1.00 |
| 3 | 339 | 3.6 | 0.245 | 0.38 | 323 | 0.5 | 1.00 |
| 2 | 330 | 2.8 | 0.330 | 0.52 | 317 | 0.5 | 1.00 |
| 4 | 321 | 3.1 | 0.289 | 0.45 | 322 | 0.5 | 1.00 |
| 5 | 315 | 3.2 | 0.327 | 0.51 | 320 | 0.5 | 1.00 |
| 6 | 310 | 2.9 | 0.330 | 0.51 | 323 | 0.5 | 1.00 |
| 7 | 305 | 2.8 | 0.297 | 0.46 | 323 | 0.5 | 1.00 |
| 8 | 300 | 2.8 | 0.317 | 0.50 | 317 | 0.5 | 1.00 |
| 10 | 296 | 2.8 | 0.277 | 0.43 | 322 | 0.5 | 0.99 |
| 9 | 290 | 2.6 | 0.250 | 0.39 | 320 | 0.5 | 0.98 |
| 11 | 282 | 2.2 | 0.289 | 0.45 | 322 | 0.5 | 0.97 |
| 12 | 270 | 2 | 0.314 | 0.49 | 320 | 0.5 | 0.96 |
| 13 | 261 | 2.1 | 0.364 | 0.57 | 320 | 0.5 | 0.95 |
| Control | NA | NA | NA | NA | 319 | NA | NA |
NA, not applicable.
Center wavelength (±0.5 nm) and bandwidth about center were measured with spectrometer. Near-constant average dose is achieved by adjusting the laser power (±0.01 mW) after determining irradiation spot size (±0.005 cm2). In this way, irradiation time is kept almost constant.
Calculated dose based on irradiance in laser spot and exposure time.
Correction factor for average attenuation along 1-mm exposure path.
Figure 2.
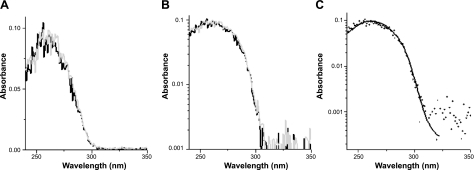
UV-vis spectra of mouse genomic DNA before and after laser irradiation. Laser exposure of genomic DNA was performed at a dose of 8.6 J/cm2 at wavelength 309 nm, as described in Materials and Methods. A, B) UV absorption of DNA before (black trace) and after (gray trace) laser exposure on linear (A) and logarithmic scales (B). A steep rolloff in the absorbance of DNA between wavelengths 290 and 305 nm is clearly visible in both panels. C) Higher signal-to-noise ratio for a reference absorption spectrum from unirradiated sheared herring sperm DNA recorded at higher concentration (black trace) and overlaid and scaled to the mouse genomic DNA absorption spectrum (small dots). The spectra are virtually identical.
The frozen sample pellets were thawed using only body heat immediately prior to laser exposure. After thawing, the sample tube was gently tapped to ensure homogeneity of the solution, which was then placed in a 1- × 1-cm quartz cuvette, positioned on the laser table. The laser radiant flux (W) was measured using a NIST-traceable calibrated power meter (Lasermate/Q; Coherent). We expanded the beam from 2 to 3 mm out of the laser to a 0.5- to 0.6-cm full-width at half-maximum (FWHM) waist, which allowed for the most homogeneous illumination of the sample solution spread out at the bottom of the cuvette while using virtually all of the fluence (necessary because of the relatively low average power of the laser). The expanded beam was directed downward through the square opening of the cuvette directly onto the surface of the DNA sample, at which point the beam width was ∼0.5–0.6 cm at FWHM. The spot area (cm2) was then calculated assuming a gaussian beam profile. After exposure, each sample was immediately removed from the cuvette and placed in a clean tube on ice. Once the sample had been removed, the cuvette was rigorously cleaned with both dilute HCl and NaOH, followed by multiple doubly distilled H2O rinses to ensure no cross-contamination between the individual samples. Finally, a control sample was placed in the cuvette and allowed to sit on the laser table for an equivalent amount of time as other samples, but without laser exposure.
Regarding the uniformity of exposure at different regions of the cuvette, both transverse (x, y) to the direction the laser propagates and along the laser path length (z), we note that the choice of experimental geometry was dictated by experimental constraints. With the need to achieve the required dose within a reasonably short exposure time, fully expanding the beam so that the cuvette lay only in the central “flat” portion of the gaussian x, y spatial mode, and not using the entire fluence, was unrealistic. Furthermore, in the ideal experiment, the path length or DNA concentration would be so low that the laser irradiance would not decrease along the sample path length in z. Of course, this is unrealistic if macroscopic amounts of damaged DNA need to be assayed. A compromise is achieved where a small attenuation of the laser along the path occurs, but the variation of irradiance of DNA molecules at different depths in the cuvette is relatively small. As shown in Table 1, the greatest attenuation is at shortest wavelengths. For our experimental configuration, at 261 nm, the worst-case wavelength, attenuation at the bottom face is only 10%; for all other wavelengths the attenuation is much smaller. These are calculated using a pathlength of 1 mm, which is the top-to-bottom thickness at the center of the sample volume (the region irradiated). So, this introduces a correction factor to get the average exposure along the whole path of 5% at 261 nm and a much smaller correction for other wavelengths (Table 1).
UV-vis spectroscopy of genomic DNA
We performed standard UV-vis spectroscopy (Varian Cary 50 UV-visible single-monochromator spectrophotometer) on the genomic DNA of Big Blue mouse fibroblasts before and after laser irradiation (see above). For comparison purposes, we also performed parallel analysis on concentrated unirradiated sheared herring sperm DNA (Sigma-Aldrich Inc., St. Louis, MO, USA).
Lesion-specific cleavage assays with T4 endonuclease V (T4 Endo V) and formamidopyrimidine DNA glycosylase (Fpg)
To determine the formation of a characteristic UV-induced photodimer (i.e., CPD), and two representative photooxidation-derived lesions (i.e., 8-oxo-dG and Fapy) in irradiated DNA samples, we have used enzymatic digestion assays coupled with gel electrophoresis, as described earlier (28). Methodologically, the assays are based on the recognition and cleavage of specific types of DNA damage by specialized DNA repair enzymes, followed by alkaline gel electrophoresis (29). We used T4 Endo V and Fpg enzymes for the detection of CPD (38) and oxidized (or ring-opened) purines, i.e., 8-oxo-dG and Fapy, combined (39, 40), respectively. Briefly, DNA was digested with an excess amount of T4 Endo V (Epicenter, Madison, WI, USA) or Fpg (Trevigen, Gaithersburg, MD, USA) enzymes in buffer solutions supplied by the respective manufacturers. After ethanol precipitation, the digests were loaded onto a 1.5% alkaline agarose gel and run at 3.0 V/cm for 4 h, with constant recirculation of running buffer. The electrophoretic profiles were determined using standard ethidium bromide staining. Scanning was performed using Bio-Rad imaging equipment with Quantity One image analysis software (Bio-Rad Laboratories, Life Science Group, Hercules, CA, USA). For quantification of results, we performed the average DNA break frequency analysis as described by Drouin et al. (41). To determine the average break frequency from the DNA weight distribution (i.e., ethidium bromide fluorescence intensity) along an electrophoresis gel, one needs to assert the molecular weight of the peak fraction. When an infinitely long polymer is partially cleaved (e.g., enzymatically digested DNA), so that each bond is broken with the same probability P (42), the weight distribution of the resulting molecules quickly approaches a certain distribution, “Kuhn's approximation to the Montroll and Simha equation” (43). This distribution is similar to a Poisson distribution in that one number P or its reciprocal Mn determines the entire normalized distribution. Thus, P is the probability of a bond being broken, Mn is the number-average molecular weight, which equals half of the weight-average molecular weight Mw (42). When such a randomly cleaved distribution is fractionated along an electrophoresis gel, the mobility of each fragment is proportional to the log of the molecular weight throughout the middle of the mobility range (44). Therefore, the molecular weight corresponding to mobility of the mass distribution's peak is approximately equal to Mw, and Mn is the average break frequency in a population of gel-fractionated molecules (41).
Immunodot blot assays with α-CPD and α-(6–4)PP antibodies
To assess the formation of two predominant UV-induced photodimers [i.e., CPD and (6–4)PP; ref. 31] in irradiated DNA samples, we have used immunodot blot assays, which utilize specific antibodies raised against CPD and (6–4)PP, respectively (30). The assays were performed as described earlier, with some modifications (30). Briefly, heat-denatured DNA (0.5 μg) was dot-blotted onto a nitrocellulose membrane using the Convertible Filtration Manifold System (Life Technologies, Gaithersburg, MD, USA). The membrane was laid over an absorbent paper presoaked with 0.4 N NaOH for 20 min at room temperature. Subsequently, the membrane was blocked by incubating in PBS plus 0.2% Tween 20 (PBS-T) containing 5% nonfat milk (NFM) overnight at 4°C. After multiple washes with PBS-T, the membrane was incubated with Anti-Thymine Dimer mAb (clone KTM53; specific for CPD) and Anti-(6–4) Photoproducts mAb [clone KTM50; specific for (6–4)PP] (Kamiya Biomedical Co., Seattle, WA, USA) for a duration of 2 h at room temperature [dilution 1:2000 in PBS-T plus NFM for both α-CPD and α-(6–4)PP antibodies]. The membrane was washed thoroughly with PBS-T and further incubated with an anti-mouse horseradish peroxidase-conjugated immunoglobulin (Promega, Madison, WI, USA) for 1 h at room temperature (dilution 1:10,000 in PBS-T plus NFM). To reveal peroxidase activity, the membrane was stained with the Enhanced Chemiluminescence Detection System (Amersham Biosciences UK Ltd., Little Chalfont, UK) according to the manufacturer's instructions. The stained membrane was exposed to X-ray film, and the relative intensity of luminescence was determined using the Bio-Rad imaging equipment and Quantity One image analysis software.
To quantitate the immunodot blot assay results and express the relative intensity of luminescence as the number of CPDs or (6–4)PPs per megabase of DNA, we prepared standard controls with known quantities of each photodimer and subsequently established calibration curves for luminescence signals specific for the respective photodimers using our published protocol (30). The standard controls were made from the genomic DNA of mouse embryonic fibroblasts irradiated with increasing doses of UVC (i.e., 1, 10, 25, 50, 100, 150, 200, and 250 J/m2) emitted from a germicidal lamp, and of UVB (i.e., 200, 600, 1200, and 2400 J/m2) generated by three Philips TL 20 W/12R fluorescent tubes filtered through a cellulose acetate sheet (Philips, Eindhoven, The Netherlands). The standard controls for CPD determination were subjected to T4 Endo V digestion, and then run on a 1.5% alkaline/agarose electrophoresis gel with various DNA-length ladder markers (New England Biolabs Inc., Ipswich, MA, USA). The standard controls for (6–4)PP determination were subjected to two consecutive treatments with CPD photolyase reactivation and digestion with UV-damage endonuclease (UVDE; Trevigen), and subsequently run on an electrophoresis gel as described above. The UVDE is known to cleave both CPDs and (6–4)PPs (45–47). Therefore, a pretreatment with CPD photolyase reactivation, which rids DNA of all CPDs, followed by digestion with UVDE can specifically identify the remaining (6–4)PPs in each sample (30).
Terminal transferase-dependent polymerase chain reaction (TD-PCR)
To map the formation of UV-induced DNA lesions in the cII transgene in irradiated DNA samples, we have used the TD-PCR footprinting assay, which enables detection of DNA damage at the level of nucleotide resolution (48). Methodologically, TD-PCR is based on the concept that DNA polymerase cannot synthesize past certain types of lesions, e.g., bulky lesions, such as CPDs and (6–4)PPs (31). Because the TD-PCR assay detects CPDs and (6–4)PPs indiscriminately (31), the lesions detected by this technique are collectively referred to here as UV-induced photodimers. Briefly, genomic DNA was subjected to 9 cycles of primer extension using a custom-made biotinylated primer (cII.P1: 5′-CAACAGCATAAATAACCCCGCTCTTAC-3′; Tm=59.8°C) in a Vent(exo–) DNA polymerase mix reaction (New England Biolabs). The thermocycler settings were as follows: 3 min at 95°C, 5 min at 60°C, and 10 min at 72°C. The extension products were mixed with streptavidin-coupled magnetic beads (Dynal Biotech ASA, Oslo, Norway), and gently rotated for 45 min at room temperature to allow binding of the products to the beads. The beads were washed thoroughly with 1× TE (pH 7.5) in a magnetic particle concentrator (Dynal Biotech), and the bead-bound DNA was resuspended in 0.1× TE, pH 7.5. After denaturation with 0.15 M NaOH, the single-stranded DNA underwent 3′ ribotailing by terminal deoxynucleotidyl transferase (49). The ribotailed products were ligated overnight at 17°C, and afterward were exponentially amplified by PCR using a nested primer (cII.P2: 5′-CCGCTCTTACACATTCCAGCCCTG-3′; Tm=63.2°C) and the LP25 linker primer (48) in an AmpliTaq DNA polymerase mix reaction (Applied Biosystems, Foster City, CA, USA). The thermocycler settings were as follows: 2 min at 95°C; 2 min at 61°C; 3 min at 72°C; 21 cycles of 45 s at 95°C, 2 min at 61°C, and 3 min at 72°C; 45 s at 95°C; 2 min at 61°C; and 10 min at 72°C. The amplified products were labeled using a fluorescence infrared dye-labeled primer (cII.P3: 5′-CCGCTCTTACACATTCCAGCCCTG-3′; Tm=63.2°C; IRDye 700; LI-COR, Lincoln, NE, USA) in a mixture of AmpliTaq DNA polymerase (Applied Biosystems). The thermocycler settings were as follows: 2 min at 95°C; 2 min at 66°C; 3 min at 72°C; 3 cycles of 45 s at 95°C, 2 min at 65°C, and 3 min at 72°C; 1 min at 95°C; 2 min at 60°C; and 10 min at 72°C. Following the labeling reaction, the IRDye 700 fluorescence-labeled fragments were run on polyacrylamide–urea gel electrophoresis, which is part of a computerized Long Read IR 4200 DNA Sequencing system (LI-COR). The system is equipped with an IRDye 700/800-laser (dual) detector and data acquisition software, which enable real time scanning of the sequencing gel throughout the run (50).
RESULTS
UV-absorption spectrum of genomic DNA
To establish the absorbance of DNA in our experimental model system, we performed UV-vis spectroscopy on the genomic DNA obtained from Big Blue mouse fibroblasts before and after laser irradiation. Figure 2A, B shows UV-vis spectra of mouse genomic DNA before and after laser exposure on linear and logarithmic scales, respectively. In both cases, nearly identical absorption spectra of the genomic DNA before and after laser irradiation, as well as a steep rolloff in the absorbance of DNA between wavelengths 290 and 305 nm, were readily noticeable. The small quantities of DNA available from each sample required dilution of all samples for this analysis, resulting in relatively low absorption signals in the cuvette-based spectrophotometer. To obtain a reference absorption spectrum at high signal-to-noise ratio, which could be used for verifying the accuracy of our spectroscopic analysis of mouse genomic DNA, we performed parallel UV-vis spectroscopy on unirradiated concentrated sheared herring sperm DNA. Subsequently, we overlaid the scaled-down absorption spectrum of the reference DNA to that of mouse genomic DNA. We confirmed that the UV-absorption spectra of the mouse genomic DNA and the reference herring sperm DNA are virtually identical (Fig. 2C). As shown in Fig. 2C, for regions where we hit the noise level for the mouse DNA, the absorption continues on to redder wavelengths longer than 320 nm for the herring sperm DNA, albeit at very much weaker molar absorbance. So, based on these observations and those from the published literature (51), it is clear that DNA continues to absorb at longer wavelengths, although with much less efficiency.
DNA damage in the genome
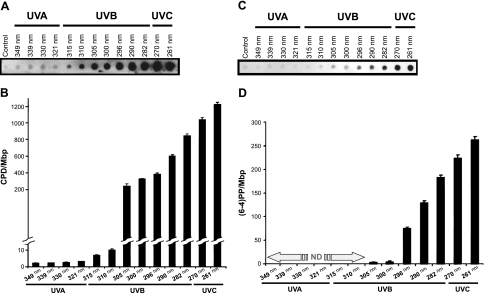
We assessed the formation of two typical UV-induced pyrimidine dimers [i.e., CPDs and (6–4)PPs], and two photooxidation-derived lesions (i.e., 8-oxo-dG and Fapy) in irradiated DNA samples using a combination of enzymatic digestion assays coupled with gel electrophoresis (28), and immunodot blot assays (30). The T4 Endo V digestion and electrophoresis assay, which is commonly used for the detection of CPDs (28, 38), showed a wavelength dependence of CPD formation in the irradiated DNA samples, as demonstrated by progressively increased T4 Endo V-sensitive sites in samples exposed to UVC and part of the UVB range (Fig. 3). The formation of CPDs, which was initially detectable at an irradiation wavelength of 305 nm, intensified as the wavelength of irradiation decreased. However, in samples irradiated at wavelengths ≥ 310 nm, CPD formation was less appreciable (we note that the detection limit constraints of this assay prevented us from quantitative determination of CPD formation in these samples). Conversely, the Fpg digestion and electrophoresis assay, which detects oxidized (or ring-opened) purines (i.e., 8-oxo-dG and Fapy combined; refs. 28, 39, 40) did not show any discernable formation of these lesions in the irradiated DNA samples at any of the 13 tested wavelengths, which encompass UVC, UVB, and UVA (Fig. 4).
Figure 3.
Determination of CPD formation in mouse genomic DNA irradiated with various UV wavelengths. Genomic DNA of mouse embryonic fibroblasts was irradiated with a series of UV wavelengths (λ=261–349 nm) at a constant dose of 0.5 J/cm2 in comparison with control, as described in Materials and Methods. Aliquots of DNA samples (1 μg) were digested with T4 Endo V enzyme, and subsequently subjected to alkaline agarose gel electrophoresis, followed by standard visualization procedure. M, molecular size marker. A) Digestion buffer plus T4 Endo V enzyme was added to the reaction mix. B) Quantification of results from panel A. Values are expressed as the number of T4 Endo V-sensitive sites per megabase of DNA. Average results from 2 independent experiments are shown for UV-irradiated samples. Error bars = sd. C) Digestion buffer only; no enzyme was added to the reaction mix.
Figure 4.
Determination of oxidized (or ring-opened) purine formation in mouse genomic DNA irradiated with various UV wavelengths. Genomic DNA of mouse embryonic fibroblasts was irradiated with a series of UV wavelengths (λ=261–349 nm) at a constant dose of 0.5 J/cm2 in comparison with control, as described in Materials and Methods. Aliquots of DNA samples (1 μg) were digested with Fpg enzyme, and subsequently subjected to alkaline agarose gel electrophoresis, followed by standard visualization procedure. As positive control, genomic DNA treated with methylene blue plus light (Mb+Light), which is known to produce a combination of 8-oxo-dG and Fapy lesions, was run in all analyses. M, molecular size marker. A) Digestion buffer plus Fpg enzyme was added to the reaction mix. B) Digestion buffer only; no enzyme was added to the reaction mix.
In confirmation of the T4 Endo V digestion and electrophoresis assay results, immunodot blot assay with an α-CPD antibody showed a wavelength-dependent formation of CPDs in irradiated DNA samples. As shown in Fig. 5A, B, the formation of CPDs intensified with decreasing wavelengths of irradiation; whereas substantial levels of CPDs were found in samples irradiated with UVC, and UVB wavelengths borderlined with UVA, there were small but detectable levels of these lesions in samples irradiated with longer wavelengths. Furthermore, immunodot blot assay with an α-(6–4)PP antibody revealed a wavelength dependence of (6–4)PP formation in irradiated DNA samples. Although substantial formation of (6–4)PPs was detectable in samples irradiated at 296 nm, which increased as the irradiation wavelength decreased, only small quantities of these lesions were found in samples irradiated at 300–305 nm, with no detectable level of (6–4)PPs in samples irradiated with longer wavelengths (Fig. 5C, D).
Figure 5.
Determination of CPD and (6–4)PP formation in mouse genomic DNA irradiated with various UV wavelengths. Genomic DNA of mouse embryonic fibroblasts was irradiated with a series of UV wavelengths (λ=261–349 nm) at a constant dose of 0.5 J/cm2 in comparison with control, as described in Materials and Methods. Aliquots of DNA samples (0.5 μg) were subjected to immunodotblot assays using Anti-Thymine Dimer mAb (Clone KTM53) and Anti-(6–4) Photoproducts mAb (Clone KTM50; Kamiya Biomedical Co.), which are specific for CPD and (6–4)PP, respectively. A) Results of immunodotblot assay for the detection of CPDs. B) Quantification of results from panel A. Values are expressed as number of CPDs per megabase of DNA. Average results from 2 independent experiments are shown for UV-irradiated samples. Error bars = sd. C) Results of immunodotblot assay for the detection of (6–4)PPs. D) Quantification of results from panel C. Values are expressed as number of (6–4)PPs per megabase of DNA. Average results from 2 independent experiments are shown for UV-irradiated samples. ND, not detectable.
DNA damage in the cII transgene
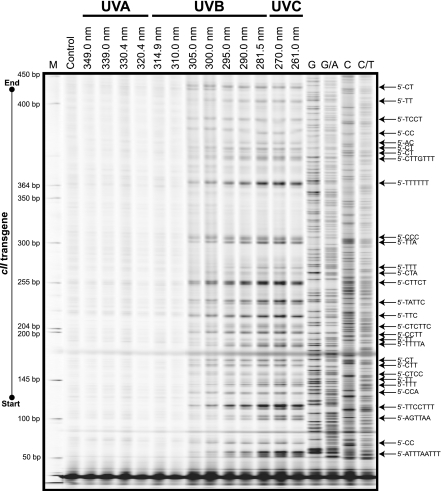
We mapped the formation of UV-induced photodimers (i.e., CPDs and (6–4)PPs combined) in irradiated DNA samples using the TD-PCR footprinting assay (31). As shown in Fig. 6, preferential formation of UV-induced photodimers was observed in the cII transgene of irradiated DNA samples, which was initially detectable at an irradiation wavelength of 305 nm, and intensified as the wavelength of irradiation decreased. The UV-induced photodimers in the cII transgene of irradiated DNA samples formed exclusively in pyrimidine-rich sequences, with the overwhelming majority having di-, tri-, or multipyrimidine sequence contexts. Of these, sequences containing tandem thymines were the sites of most pronounced formation of UV-induced photodimers (Fig. 6). In agreement with the enzymatic digestion and electrophoresis assay and immunodot blot assay results (see above), less than appreciable formation of UV-induced photodimers was observed in the cII transgene of DNA samples irradiated with long UV wavelengths (Fig. 6).
Figure 6.
Mapping of UV-induced DNA damage in the cII transgene in mouse genomic DNA irradiated with various UV wavelengths. TD-PCR footprinting of the full-length cII transgene was done using genomic DNA of mouse embryonic fibroblasts irradiated with a series of UV wavelengths (λ=261–349 nm) at a constant dose of 0.5 J/cm2 in comparison with control, as described in Materials and Methods. In addition to the IRDye 700 Sizing Standard (LI-COR), Maxam and Gilbert chemical reactions, prepared from control genomic DNA and subjected to ligation-mediated PCR (LM-PCR; ref. 50), were run in parallel to all samples (last 4 lanes: G, G/A, C, C/T). This strategy will help locate the exact position of each base along the reference DNA sequence. LM-PCR bands migrate ∼3 bases faster than the corresponding TD-PCR bands due to the addition of 3 riboguanosine triphosphates to all primer extension products in the latter method (31, 48). M, molecular size marker.
cII Mutations in relation to DNA damage
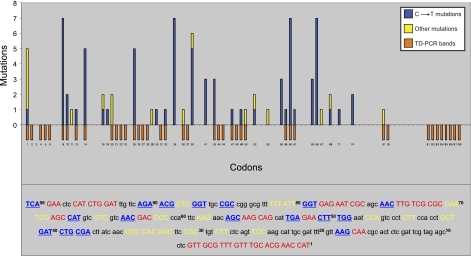
To find the contribution of UV-induced DNA damage to solar mutagenesis, we used our previously established sunlight-induced cII mutation spectrum (28), and sought correlation between the locations of UV-induced photodimers in the cII transgene of mouse genomic DNA irradiated with various UV wavelengths and the frequency distribution and codon position of sunlight-induced cII mutations in mouse cells irradiated with SSL. For comparison purposes, we classified all the induced cII mutations into two categories: category I includes single/tandem C→T transitions, which are known signature mutations of CPDs and (6–4)PPs (13); category II includes all other types of mutation, including base substitutions, i.e., transversions, transitions other than C→T mutations, insertions, or deletions. As shown in Fig. 7, the vast majority (80%) of the induced cII mutations in mouse cells irradiated with SSL consisted of single/tandem C→T transitions within pyrimidine dinucleotides, which implicate CPDs and (6–4)PPs as the driving force behind this type of mutation (13). Of all cII-mutated codons harboring the hallmark single/tandem C→T transitions in SSL-irradiated mouse cells, only half (13 of 26 codons) colocalized with the preferential sites of photodimer formation found in UV-irradiated mouse genomic DNA (Fig. 7). Likewise, of all sunlight-induced cII-mutated codons containing category I and/or category II mutations in SSL-irradiated mouse cells, only half (17 of 34 codons) colocalized with the preferential sites of lesion formation found in UV-irradiated mouse genomic DNA (Fig. 7).
Figure 7.
Correlation of UV-induced DNA damage and mutations. Relationship between TD-PCR footprinting data (Fig. 6) and sunlight-induced mutagenicity data (adapted from ref 28) were sought by plotting the locations of UV-induced DNA lesions in mouse genomic DNA irradiated with a series of UV wavelengths (λ=261–349 nm) against the frequency distribution and codon position of sunlight-induced mutations in mouse embryonic fibroblasts irradiated with SSL (28). Red bars represent positions of UV-induced DNA lesions in the reference cII sequence (numbers below red bars indicate codon positions). Blue and/or yellow bars represent frequency distribution and codon position of sunlight-induced mutations in the reference cII sequence. Blue bars indicate sunlight-induced single/tandem C→T transition mutations at dipyrimidines only; yellow bars indicate all other types of mutation, including base substitutions (i.e., transversions, transitions other than C→T mutations, insertions, or deletions). Detailed DNA footprinting and mutagenicity data in the cII transgene are shown in a color-coded sequence context at bottom. Codons in which UV-induced DNA lesions are formed are shown in red capitals; sunlight-induced mutated codons are shown in yellow capitals (mutations with no accompanying UV-induced lesions) or blue underscored capitals (mutations with accompanying UV-induced lesions). Superscript numbers indicate codon positions in the reference cII sequence.
DISCUSSION
To specifically determine the contribution of various UV wavelengths to solar mutagenesis, we used a tunable laser source of UV light, and investigated the induction of DNA damage, both in the overall genome and at the nucleotide resolution level, in the genomic DNA of transgenic Big Blue mouse fibroblasts irradiated with a series of UV wavelengths, inclusive of UVC (λ<280 nm), UVB (λ=280–320 nm), and UVA (λ>320 nm). Subsequently, we sought correlation between the locations of UV-induced DNA lesions in the cII transgene of irradiated DNA samples and the frequency distribution and codon position of sunlight-induced cII mutations in counterpart mouse cells irradiated with SSL.
Our UV-vis spectroscopy of the genomic DNA showed a sharp cutoff in the ability of DNA to absorb UV at wavelengths 300–305 nm, with a 100-fold decrease in DNA absorption between wavelengths 290 and 305 nm, both before and after laser exposure (Fig. 2). In agreement with previously published literature (51), this observation indicates that the biological consequences of UV irradiation at UVB wavelengths bordering the UVA range and the longer wavelengths may not solely depend on direct UV absorption of DNA. In confirmation, we found relatively small quantities of UV-induced photodimers in DNA samples irradiated at long UV wavelengths, as determined by a combination of T4 Endo V digestion assay coupled with gel electrophoresis (Fig. 3) and immunodot blot assay with α-CPD and α-(6–4)PP antibodies (Fig. 5). We should stress that of all the assays used here for the detection of DNA damage, the immunodot blot assay has the greatest sensitivity and specificity owing to its use of highly sensitive and specific monoclonal antibodies (30).
The photodimeric CPDs and (6–4)PPs are induced primarily through direct UV absorption of DNA, as a result of covalent-bond formation between two adjacent pyrimidines (13, 52). Because the frequency of CPD formation is 3- to 10-fold higher than that of (6–4)PPs after UV irradiation of DNA or cells, and due to repair resistance and high bypass tolerance of CPD, this lesion is considered the principal contributor to solar mutagenesis (13, 52–54). The predominant formation of CPDs relative to (6–4)PPs was confirmed in our immunodot blot analysis of genomic DNA irradiated with various UV wavelengths (Fig. 5). Of significance, we also observed a divergence of CPD and (6–4)PP formation at 296-nm irradiation in mouse genomic DNA. In both cases, however, the intensities of lesion formation were increased as the wavelength of irradiation decreased (Fig. 5). Whereas the unique wavelength-dependent formation of both photodimers deserves special attention, the distinct formation of CPDs as compared to (6–4)PPs at longer UV wavelengths concurs with the predominant formation of CPD in sunlight-irradiated DNA or cells (52, 54, 55). Because the terrestrial sunlight spectrum rolls off sharply at wavelengths ∼300 nm (e.g., ref. 56), the relatively low detection of (6–4)PPs at around this wavelength or the longer wavelengths in irradiated genomic DNA (Fig. 5C, D) suggests that CPDs are the principal lesions accounting for most DNA damage-dependent biological effects of sunlight.
Our TD-PCR footprinting of UV-induced lesions in the cII transgene of irradiated DNA samples confirmed the above-mentioned wavelength-dependent formation of photodimers in the genome overall of irradiated samples (Fig. 6). Although the sensitivity of TD-PCR assay is lower than that of immunodot blot assay (30, 31), the footprinting results revealed a consistent formation of UV-induced photodimers in the cII transgene of irradiated samples, initially detectable in samples irradiated with midrange UVB wavelengths, and intensifying as the wavelength of irradiation decreased. The induced lesions in the cII transgene of irradiated samples were mapped exclusively to pyrimidine-rich sequences, with the overwhelming majority being in di-, tri-, or multipyrimidine sequence contexts, which corroborates the CPD and/or (6–4)PP nature of the detected lesions. In addition, we found that sequences containing tandem thymines were the sites of most pronounced formation of UV-induced photodimers (Fig. 6), which implicates CPDs as the primary lesions formed within these sequences, because CPDs are known to form preferentially at TT dipyrimidines, whereas (6–4)PPs hardly form within this sequence context (13, 57, 58). The relatively low formation of UV-induced dimeric lesions in the cII transgene of DNA samples irradiated with long wavelengths UV (Fig. 6), together with the nondetection of photooxidation-derived lesions in the overall samples (Fig. 4) indicates that UV-induced photosensitization reactions are less likely to have occurred, at substantial levels, in our experimental system. Such photosensitization reactions, especially at long UV wavelengths, are more pronounced in a cellular milieu, where the intracellular chromophores/photosensitizors are abundantly present, as compared to naked genomic DNA (28, 30, 40, 59–61).
Using different irradiation protocols and UV sources as compared to those used in the present study, other researchers have reported an induction of photooxidation-derived DNA lesions in samples irradiated with long UV wavelengths (62–64). For instance, Kuluncsics et al. (62) irradiated drops of plasmid DNA deposited onto plastic and glass Petri dishes for a maximal exposure time of 120 min, which corresponded to a dose of 1008 kJ/m2 of UVA radiation (vs. 5 kJ/m2 delivered through ∼5 min of irradiation in the present study). The researchers have investigated the possible contribution of artifacts generated through iron-catalyzed Fenton reactions to the overall detected Fpg-sensitive sites, as an indicator of oxidative DNA damage, in the irradiated plasmid DNA (e.g., effects of the matrix onto which DNA was deposited during irradiation and/or the composition of dilution buffer were discussed) (62). Likewise, Jiang et al. (63) have used irradiation of plasmid DNA placed in NMR tubes for exposure times of 48–222 min, which corresponded to doses of 1300–6000 kJ/m2 of UV radiation. Although we do not refute the possibility of photooxidation of DNA consequent to high-dose UVA irradiation in certain experimental settings (62–64), we note that it is unlikely to have similar findings while comparing the results of different studies that have employed different experimental procedures. At the same time, we acknowledge that the lack of detection of photooxidized purines in UV-irradiated samples in the present study was shown by an enzymatic digestion assay, which like any other assay is constrained by its detection limits.
Dewar valence photoisomers are an important class of UV-induced lesions that are derived from (6–4)PPs, which are maximally induced in the UVC range and to a lesser extent in the UVB range, that on subsequent irradiation with long UV wavelengths undergo photoisomerization, thereby giving rise to valence isomers (22, 53, 65, 66). In the present study, however, we have used UV irradiation only at a single fixed wavelength for all UV-exposure experiments. So, the above specified 2-step reaction in which the initially produced (6–4)PPs generated at UVB and UVC ranges are converted to their valence photoisomers on subsequent irradiation with long-wavelength UV (53, 65) is not likely to have happened, at substantial levels, in our experiments. We also stress that the antibody used for the detection of (6–4)PPs in our immunodot blot analysis of UV-induced photodimers does not detect Dewar valence photoisomers (67). Nonetheless, as a polymerase-blocking lesion, Dewar valence photoisomers are readily detectable by TD-PCR analysis (31). In our study, however, TD-PCR analysis of UV-irradiated samples did not reveal any new lesion introduced into the DNA, nor did it show a reappearance of the lesions that were detectable at shorter UV wavelengths [i.e., CPDs and (6–4)PPs combined]. Altogether, it is less likely that Dewar valence photoisomers were produced at significant levels in our UV-irradiated samples.
Comparison of lesion formation in the cII transgene of mouse genomic DNA irradiated with various UV wavelengths and sunlight-induced cII mutations in mouse cells irradiated with SSL revealed that only 50% of all cII-mutated codons harboring the characteristic C→T transitions within pyrimidine dinucleotides (13) in SSL-irradiated cells colocalized with the preferential sites of damage formation found in UV-irradiated genomic DNA (Fig. 7). Likewise, of all sunlight-induced cII-mutated codons in SSL-irradiated cells, only half colocalized with the preferential sites of lesion formation found in UV-irradiated genomic DNA (Fig. 7). These findings, together with the observation that UV absorption of DNA accounts for nearly all the induced lesions in irradiated genomic DNA, imply that many of the sunlight-induced mutations found in SSL-irradiated mouse cells might be ascribed to lesions that are not generated through direct UV absorption of DNA. The latter lesions are likely to have arisen, at least partially, from photosensitization reactions, perhaps also in the UVA range (12–14, 68), which would implicate UVA as a key contributor to solar mutagenesis.
However, we acknowledge that many of the UV-induced lesions formed in the cII transgene of irradiated DNA samples, which did not have a corresponding mutated codon in SSL-irradiated cells, could fall into codon positions (e.g., third codon base) that do not provide a selectable phenotype in the cII mutagenesis assay. Alternatively, they are likely to have undergone DNA repair in mouse cells (69), although the induced lesions in a cell-free environment, such as the naked genomic DNA tested here, are not subject to this repair pathway. The lack of mutability of UV-induced lesions in specific genomic loci may also be explained by the fact that a major component of UV mutagenesis is deamination of cytosines within CPDs (13), which can be sequence-dependent (70). Furthermore, other features, such as replicative bypass of DNA damage by various DNA polymerases, whose fidelity and efficiency can be sequence dependent, may varyingly influence the mutagenic potential of UV-induced DNA lesions in different sequence contexts (71). Altogether, the imperfect match between the locations of UV-induced DNA lesion and mutation sites in the cII transgene, which accords with a recent report by Vreeswijk et al. (72), underscores the importance of multiple influential factors in determining the mutagenicity of UV-induced DNA damage. Clearly, lesion formation by direct UV absorption of DNA is only one parameter that explains the mutational signature of genes in sunlight-irradiated cells. However, as far as specific lesion formation by direct DNA absorption of wavelengths from sunlight is concerned, our data suggest that the contribution from (6–4)PPs is relatively minor, whereas the vast majority of DNA damage occurring between 300 and 315 nm, a part of the solar spectrum that reaches the earth's surface (56), can be ascribed to the CPDs.
CONCLUSIONS
The novel approach of our study is the use of a laser system to generate tunable radiation of varying wavelengths that encompass the UV component of sunlight, which enabled us to investigate sunlight-induced DNA damage and mutagenesis in a validated model system. The significance of our study lies in our demonstration of a unique wavelength-dependent formation of photodimeric lesions, based on direct UV absorption of DNA, in UV-irradiated mouse genomic DNA, which can partially explain the induction of mutations in mouse cells irradiated with simulated sunlight. Wavelengths of sunlight that penetrate the atmosphere and can cause direct DNA damage (300–315 nm) produce mostly CPDs but elicit small quantities of (6–4)PPs.
Acknowledgments
This study was supported by grants from the American Cancer Society (RSG-11-083-01-CNE) to A.B., and the National Institute of Environmental Health Sciences (ES06070) to G.P.P.
REFERENCES
- 1. Woodhead A. D., Setlow R. B., Tanaka M. (1999) Environmental factors in nonmelanoma and melanoma skin cancer. J. Epidemiol. 9, S102–S114 [DOI] [PubMed] [Google Scholar]
- 2. Diepgen T. L., Mahler V. (2002) The epidemiology of skin cancer. Br. J. Dermatol. 146(Suppl. 61), 1–6 [DOI] [PubMed] [Google Scholar]
- 3. Setlow R. B. (1974) The wavelengths in sunlight effective in producing skin cancer: a theoretical analysis. Proc. Natl. Acad. Sci. U. S. A. 71, 3363–3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Diffey B. L. (2002) Sources and measurement of ultraviolet radiation. Methods 28, 4–13 [DOI] [PubMed] [Google Scholar]
- 5. Kelfkens G., de Gruijl F. R., van der Leun J. C. (1990) Ozone depletion and increase in annual carcinogenic ultraviolet dose. Photochem. Photobiol. 52, 819–823 [DOI] [PubMed] [Google Scholar]
- 6. De Gruijl F. R. (2002) Photocarcinogenesis: UVA vs. UVB radiation. Skin Pharmacol. Appl. Skin Physiol. 15, 316–320 [DOI] [PubMed] [Google Scholar]
- 7. De Gruijl F. R., Sterenborg H. J., Forbes P. D., Davies R. E., Cole C., Kelfkens G., van Weelden H., Slaper H., van der Leun J. C. (1993) Wavelength dependence of skin cancer induction by ultraviolet irradiation of albino hairless mice. Cancer Res. 53, 53–60 [PubMed] [Google Scholar]
- 8. Garibyan L., Fisher D. E. How sunlight causes melanoma. Curr. Oncol. Rep. 12, 319–326 [DOI] [PubMed] [Google Scholar]
- 9. Von Thaler A. K., Kamenisch Y., Berneburg M. The role of ultraviolet radiation in melanomagenesis. Exp. Dermatol. 19, 81–88 [DOI] [PubMed] [Google Scholar]
- 10. Runger T. M., Kappes U. P. (2008) Mechanisms of mutation formation with long-wave ultraviolet light (UVA). Photodermatol. Photoimmunol. Photomed. 24, 2–10 [DOI] [PubMed] [Google Scholar]
- 11. McMillan T. J., Leatherman E., Ridley A., Shorrocks J., Tobi S. E., Whiteside J. R. (2008) Cellular effects of long wavelength UV light (UVA) in mammalian cells. J. Pharm. Pharmacol. 60, 969–976 [DOI] [PubMed] [Google Scholar]
- 12. Cadet J., Douki T., Ravanat J. L., Di Mascio P. (2009) Sensitized formation of oxidatively generated damage to cellular DNA by UVA radiation. Photochem. Photobiol. Sci. 8, 903–911 [DOI] [PubMed] [Google Scholar]
- 13. Pfeifer G. P., You Y. H., Besaratinia A. (2005) Mutations induced by ultraviolet light. Mutat. Res. 571, 19–31 [DOI] [PubMed] [Google Scholar]
- 14. Cadet J., Sage E., Douki T. (2005) Ultraviolet radiation-mediated damage to cellular DNA. Mutat. Res. 571, 3–17 [DOI] [PubMed] [Google Scholar]
- 15. Woollons A., Kipp C., Young A. R., Petit-Frere C., Arlett C. F., Green M. H., Clingen P. H. (1999) The 0.8% ultraviolet B content of an ultraviolet A sunlamp induces 75% of cyclobutane pyrimidine dimers in human keratinocytes in vitro. Br. J. Dermatol. 140, 1023–1030 [DOI] [PubMed] [Google Scholar]
- 16. Ikehata H., Kudo H., Masuda T., Ono T. (2003) UVA induces C–>T transitions at methyl-CpG-associated dipyrimidine sites in mouse skin epidermis more frequently than UVB. Mutagenesis 18, 511–519 [DOI] [PubMed] [Google Scholar]
- 17. Courdavault S., Baudouin C., Charveron M., Favier A., Cadet J., Douki T. (2004) Larger yield of cyclobutane dimers than 8-oxo-7,8-dihydroguanine in the DNA of UVA-irradiated human skin cells. Mutat. Res. 556, 135–142 [DOI] [PubMed] [Google Scholar]
- 18. Drobetsky E. A., Turcotte J., Chateauneuf A. (1995) A role for ultraviolet A in solar mutagenesis. Proc. Natl. Acad. Sci. U. S. A. 92, 2350–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mouret S., Baudouin C., Charveron M., Favier A., Cadet J., Douki T. (2006) Cyclobutane pyrimidine dimers are predominant DNA lesions in whole human skin exposed to UVA radiation. Proc. Natl. Acad. Sci. U. S. A. 103, 13765–13770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sage E., Lamolet B., Brulay E., Moustacchi E., Chateauneuf A., Drobetsky E. A. (1996) Mutagenic specificity of solar UV light in nucleotide excision repair-deficient rodent cells. Proc. Natl. Acad. Sci. U. S. A. 93, 176–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Palmer C. M., Serafini D. M., Schellhorn H. E. (1997) Near ultraviolet radiation (UVA and UVB) causes a formamidopyrimidine glycosylase-dependent increase in G to T transversions. Photochem. Photobiol. 65, 543–549 [DOI] [PubMed] [Google Scholar]
- 22. Perdiz D., Grof P., Mezzina M., Nikaido O., Moustacchi E., Sage E. (2000) Distribution and repair of bipyrimidine photoproducts in solar UV-irradiated mammalian cells. Possible role of Dewar photoproducts in solar mutagenesis. J. Biol. Chem. 275, 26732–26742 [DOI] [PubMed] [Google Scholar]
- 23. Courdavault S., Baudouin C., Charveron M., Canguilhem B., Favier A., Cadet J., Douki T. (2005) Repair of the three main types of bipyrimidine DNA photoproducts in human keratinocytes exposed to UVB and UVA radiations. DNA Repair 4, 836–844 [DOI] [PubMed] [Google Scholar]
- 24. Mathonnet G., Leger C., Desnoyers J., Drouin R., Therrien J. P., Drobetsky E. A. (2003) UV wavelength-dependent regulation of transcription-coupled nucleotide excision repair in p53-deficient human cells. Proc. Natl. Acad. Sci. U. S. A. 100, 7219–7224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pflaum M., Boiteux S., Epe B. (1994) Visible light generates oxidative DNA base modifications in high excess of strand breaks in mammalian cells. Carcinogenesis 15, 297–300 [DOI] [PubMed] [Google Scholar]
- 26. Kappes U. P., Luo D., Potter M., Schulmeister K., Runger T. M. (2006) Short- and long-wave UV light (UVB and UVA) induce similar mutations in human skin cells. J. Invest. Dermatol. 126, 667–675 [DOI] [PubMed] [Google Scholar]
- 27. Kappes U. P., Runger T. M. (2005) No major role for 7,8-dihydro-8-oxoguanine in ultraviolet light-induced mutagenesis. Radiation Res. 164, 440–445 [DOI] [PubMed] [Google Scholar]
- 28. Besaratinia A., Kim S. I., Pfeifer G. P. (2008) Rapid repair of UVA-induced oxidized purines and persistence of UVB-induced dipyrimidine lesions determine the mutagenicity of sunlight in mouse cells. FASEB J. 22, 2379–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Besaratinia A., Kim S. I., Hainaut P., Pfeifer G. P. (2009) In vitro recapitulating of TP53 mutagenesis in hepatocellular carcinoma associated with dietary aflatoxin B1 exposure. Gastroenterology 137, 1127–1137, 1137, e1121–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Besaratinia A., Synold T. W., Chen H. H., Chang C., Xi B., Riggs A. D., Pfeifer G. P. (2005) DNA lesions induced by UV A1 and B radiation in human cells: comparative analyses in the overall genome and in the p53 tumor suppressor gene. Proc. Natl. Acad. Sci. U. S. A. 102, 10058–10063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Besaratinia A., Pfeifer G. P. (2006) Investigating human cancer etiology by DNA lesion footprinting and mutagenicity analysis. Carcinogenesis 27, 1526–1537 [DOI] [PubMed] [Google Scholar]
- 32. Jakubczak J. L., Merlino G., French J. E., Muller W. J., Paul B., Adhya S., Garges S. (1996) Analysis of genetic instability during mammary tumor progression using a novel selection-based assay for in vivo mutations in a bacteriophage lambda transgene target. Proc. Natl. Acad. Sci. U. S. A. 93, 9073–9078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pfeifer G. P., Chen H. H., Komura J., Riggs A. D. (1999) Chromatin structure analysis by ligation-mediated and terminal transferase-mediated polymerase chain reaction. Methods Enzymol. 304, 548–571 [DOI] [PubMed] [Google Scholar]
- 34. Elles C. G., Jailaubekov A. E., Crowell R. A., Bradforth S. E. (2006) Excitation-energy dependence of the mechanism for two-photon ionization of liquid H(2)O and D(2)O from 8.3 to 12.4 eV. J. Chem. Phys. 125, 44515. [DOI] [PubMed] [Google Scholar]
- 35. Marguet S., Markovitsi D., Talbot F. (2006) One- and two-photon ionization of DNA single and double helices studied by laser flash photolysis at 266 nm. J. Phys. Chem. B 110, 11037–11039 [DOI] [PubMed] [Google Scholar]
- 36. Friedberg E. C., Walker G. C., Siede W., Wood R. D., Schultz R. A., Ellenberger T. (2005) DNA Repair and Mutagenesis, ASM Press, Washington, D. C [Google Scholar]
- 37. Slavicek P., Winter B., Faubel M., Bradforth S. E., Jungwirth P. (2009) Ionization energies of aqueous nucleic acids: photoelectron spectroscopy of pyrimidine nucleosides and ab initio calculations. J. Am. Chem. Soc. 131, 6460–6467 [DOI] [PubMed] [Google Scholar]
- 38. Mitchell D. L., Jen J., Cleaver J. E. (1991) Relative induction of cyclobutane dimers and cytosine photohydrates in DNA irradiated in vitro and in vivo with ultraviolet-C and ultraviolet-B light. Photochem. Photobiol. 54, 741–746 [DOI] [PubMed] [Google Scholar]
- 39. Boiteux S., O'Connor T. R., Laval J. (1987) Formamidopyrimidine-DNA glycosylase of Escherichia coli: cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J. 6, 3177–3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cadet J., Ravanat J. L., Martinez G., Medeiros M., Di Mascio P. (2006) Singlet oxygen oxidation of isolated and cellular DNA: product formation and mechanistic insights. Photochem. Photobiol. 82, 1219–1225 [DOI] [PubMed] [Google Scholar]
- 41. Drouin R., Gao S., Holmquist G. P. (1996) Agarose gel electrophoresis for DNA damage analysis. In Technologies for Detection of DNA Damage and Mutations, Vol. 1 (Pfeifer G. P. ed.). pp. 37–43, Plenum Press, New York [Google Scholar]
- 42. Tanford C. (1961) Physical Chemistry of Macromolecules, Wiley, New York [Google Scholar]
- 43. Hamer D. H., Thomas C. A. J. (1975) The cleavage of Drosophila melanogaster DNA by restriction endonucleases. Chromosoma 49, 243–255 [Google Scholar]
- 44. Willis C. E., Willis D. G., Holmquist G. P. (1988) An equation for DNA electrophoretic mobility in agarose gels. Appl. Theor. Electrophor. 1, 11–18 [PubMed] [Google Scholar]
- 45. Freyer G. A., Davey S., Ferrer J. V., Martin A. M., Beach D., Doetsch P. W. (1995) An alternative eukaryotic DNA excision repair pathway. Mol. Cell. Biol. 15, 4572–4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yajima H., Takao M., Yasuhira S., Zhao J. H., Ishii C., Inoue H., Yasui A. (1995) A eukaryotic gene encoding an endonuclease that specifically repairs DNA damaged by ultraviolet light. EMBO J. 14, 2393–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kaur B., Avery A. M., Doetsch P. W. (1998) Expression, purification, and characterization of ultraviolet DNA endonuclease from Schizosaccharomyces pombe. Biochemistry 37, 11599–11604 [DOI] [PubMed] [Google Scholar]
- 48. Chen H. H., Kontaraki J., Bonifer C., Riggs A. D. (2001) Terminal transferase-dependent PCR (TDPCR) for in vivo UV photofootprinting of vertebrate cells. Sci. STKE 2001, pl1. [DOI] [PubMed] [Google Scholar]
- 49. Komura J., Riggs A. D. (1998) Terminal transferase-dependent PCR: a versatile and sensitive method for in vivo footprinting and detection of DNA adducts. Nucleic Acids Res. 26, 1807–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Besaratinia A., Pfeifer G. P. (2009) DNA-lesion mapping in mammalian cells. Methods 48, 35–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sutherland J. C., Griffin K. P. (1981) Absorption spectrum of DNA for wavelengths greater than 300 nm. Radiation Res. 86, 399–409 [PubMed] [Google Scholar]
- 52. Sage E. (1993) Distribution and repair of photolesions in DNA: genetic consequences and the role of sequence context. Photochem. Photobiol. 57, 163–174 [DOI] [PubMed] [Google Scholar]
- 53. Rosenstein B. S., Mitchell D. L. (1987) Action spectra for the induction of pyrimidine(6–4)pyrimidone photoproducts and cyclobutane pyrimidine dimers in normal human skin fibroblasts. Photochem. Photobiol. 45, 775–780 [DOI] [PubMed] [Google Scholar]
- 54. You Y. H., Lee D. H., Yoon J. H., Nakajima S., Yasui A., Pfeifer G. P. (2001) Cyclobutane pyrimidine dimers are responsible for the vast majority of mutations induced by UVB irradiation in mammalian cells. J. Biol. Chem. 276, 44688–44694 [DOI] [PubMed] [Google Scholar]
- 55. Yoon J. H., Lee C. S., O'Connor T. R., Yasui A., Pfeifer G. P. (2000) The DNA damage spectrum produced by simulated sunlight. J. Mol. Biol. 299, 681–693 [DOI] [PubMed] [Google Scholar]
- 56. Jacovides C. P., Gianourakos G. P., Asimakopoulos D. N., Steven M. D. (1998) Measured spectra of solar ultraviolet irradiances at Athens basin, Greece. Theor. Appl. Climatol. 59, 107–119 [Google Scholar]
- 57. Setlow R. B., Carrier W. L. (1966) Pyrimidine dimers in ultraviolet-irradiated DNA's. J. Mol. Biol. 17, 237–254 [DOI] [PubMed] [Google Scholar]
- 58. Mitchell D. L., Jen J., Cleaver J. E. (1992) Sequence specificity of cyclobutane pyrimidine dimers in DNA treated with solar (ultraviolet B) radiation. Nucleic Acids Res. 20, 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dahle J., Brunborg G., Svendsrud D. H., Stokke T., Kvam E. (2008) Overexpression of human OGG1 in mammalian cells decreases ultraviolet A induced mutagenesis. Cancer Lett. 267, 18–25 [DOI] [PubMed] [Google Scholar]
- 60. Kozmin S., Slezak G., Reynaud-Angelin A., Elie C., de Rycke Y., Boiteux S., Sage E. (2005) UVA radiation is highly mutagenic in cells that are unable to repair 7,8-dihydro-8-oxoguanine in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 102, 13538–13543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Agar N. S., Halliday G. M., Barnetson R. S., Ananthaswamy H. N., Wheeler M., Jones A. M. (2004) The basal layer in human squamous tumors harbors more UVA than UVB fingerprint mutations: a role for UVA in human skin carcinogenesis. Proc. Natl. Acad. Sci. U. S. A. 101, 4954–4959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kuluncsics Z., Perdiz D., Brulay E., Muel B., Sage E. (1999) Wavelength dependence of ultraviolet-induced DNA damage distribution: involvement of direct or indirect mechanisms and possible artefacts. J. Photochem. Photobiol. B. 49, 71–80 [DOI] [PubMed] [Google Scholar]
- 63. Jiang Y., Rabbi M., Kim M., Ke C., Lee W., Clark R. L., Mieczkowski P. A., Marszalek P. E. (2009) UVA generates pyrimidine dimers in DNA directly. Biophys. J. 96, 1151–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang Y., Zhu X., Smith J., Haygood M. T., Gao R. Direct observation and quantitative characterization of singlet oxygen in aqueous solution upon UVA excitation of 6-thioguanines. J. Phys. Chem. B 115, 1889–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mitchell D. L., Rosenstein B. S. (1987) The use of specific radioimmunoassay to determine action spectra for the photolysis of (6–4) photoproducts. Photochem. Photobiol. 45, 781–786 [DOI] [PubMed] [Google Scholar]
- 66. Taylor J. S., Lu H. F., Kotyk J. J. (1990) Quantitative conversion of the (6–4) photoproduct of TpdC to its Dewar valence isomer upon exposure to simulated sunlight. Photochem. Photobiol. 51, 161–167 [DOI] [PubMed] [Google Scholar]
- 67. Mori T., Nakane M., Hattori T., Matsunaga T., Ihara M., Nikaido O. (1991) Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or (6–4)photoproduct from the same mouse immunized with ultraviolet-irradiated DNA. Photochem. Photobiol. 54, 225–232 [DOI] [PubMed] [Google Scholar]
- 68. Tyrrell R. M. (2000) Role for singlet oxygen in biological effects of ultraviolet A radiation. Methods Enzymol. 319, 290–296 [DOI] [PubMed] [Google Scholar]
- 69. Sancar A., Lindsey-Boltz L. A., Unsal-Kacmaz K., Linn S. (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73, 39–85 [DOI] [PubMed] [Google Scholar]
- 70. Tu Y., Dammann R., Pfeifer G. P. (1998) Sequence and time-dependent deamination of cytosine bases in UVB-induced cyclobutane pyrimidine dimers in vivo. J. Mol. Biol. 284, 297–311 [DOI] [PubMed] [Google Scholar]
- 71. McCulloch S. D., Kunkel T. A. (2006) Multiple solutions to inefficient lesion bypass by T7 DNA polymerase. DNA Repair 5, 1373–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Vreeswijk M. P., Meijers C. M., Giphart-Gassler M., Vrieling H., van Zeeland A. A., Mullenders L. H., Loenen W. A. (2009) Site-specific analysis of UV-induced cyclobutane pyrimidine dimers in nucleotide excision repair-proficient and -deficient hamster cells: Lack of correlation with mutational spectra. Mutat. Res. 663, 7–14 [DOI] [PubMed] [Google Scholar]