Abstract
Fast evolving regions of many metazoan genomes show a bias toward substitutions that change weak (A,T) into strong (G,C) base pairs. Single-nucleotide polymorphisms (SNPs) do not share this pattern, suggesting that it results from biased fixation rather than biased mutation. Supporting this hypothesis, analyses of polymorphism in specific regions of the human genome have identified a positive correlation between weak to strong (W→S) SNPs and derived allele frequency (DAF), suggesting that SNPs become increasingly GC biased over time, especially in regions of high recombination. Using polymorphism data generated by the 1000 Genomes Project from 179 individuals from 4 human populations, we evaluated the extent and distribution of ongoing GC-biased evolution in the human genome. We quantified GC fixation bias by comparing the DAFs of W→S mutations and S→W mutations using a Mann–Whitney U test. Genome-wide, W→S SNPs have significantly higher DAFs than S→W SNPs. This pattern is widespread across the human genome but varies in magnitude along the chromosomes. We found extreme GC-biased evolution in neighborhoods of recombination hot spots, a significant correlation between GC bias and recombination rate, and an inverse correlation between GC bias and chromosome arm length. These findings demonstrate the presence of ongoing fixation bias favoring G and C alleles throughout the human genome and suggest that the bias is caused by a recombination-associated process, such as GC-biased gene conversion.
Keywords: fixation bias, weak to strong, biased gene conversion, polymorphism, 1000 Genomes Project
Introduction
The most divergent regions of the human genome since its last common ancestor with chimpanzee exhibit a bias favoring substitutions from weak (A,T) base pairs to strong (G,C) base pairs (Dreszer et al. 2007). A similar weak-to-strong (W→S) bias in divergent sequences (BDS) is also found in recent fixed substitutions in several metazoan genomes including mammals, fish, insects, and worms, but not in fungi (Capra and Pollard 2011). BDS is most pronounced in very fast-evolving genome sequences about 1 kb long and is generally strongest in regions with high recombination rates. Many functional regions of the human genome, including human accelerated regions (HARs) (Pollard et al. 2006) and protein-coding exons (Berglund et al. 2009; Ratnakumar et al. 2010), show evidence of W→S bias, underscoring that its causes could be playing a significant role in functional divergence between closely related species.
The evolutionary processes driving BDS are likely related to the large-scale variation in GC content across mammalian genomes—the so-called isochore structure (Bernardi et al. 1985; Eyre-Walker and Hurst 2001; Romiguier et al. 2010). The origins and evolution of the isochores have received considerable attention. Previous studies suggested two possible causes for the isochores and BDS: variation in mutation patterns across genomes and biased fixation of W→S alleles (Eyre-Walker and Hurst 2001). Analyses of patterns of substitution and polymorphism in specific loci in the human genome found evidence of GC-biased allele frequency distributions and little evidence for GC mutation biases (Eyre-Walker 1999; Duret et al. 2002; Lercher and Hurst 2002; Lercher et al. 2002; Webster et al. 2003; Webster and Smith 2004; Spencer et al. 2006). Thus, GC-rich isochores (Duret et al. 2006) and W→S BDS (Dreszer et al. 2007; Capra and Pollard 2011) are unlikely to have been produced by variation in mutation patterns. Fixation bias could result from local selection on GC content (Eyre-Walker and Hurst 2001; Kudla et al. 2006) or nonadaptive processes, such as GC-biased gene conversion (gBGC; Duret and Galtier 2009a). The strong correlation between recombination rates and GC content (Meunier and Duret 2004; Webster et al. 2005; Khelifi et al. 2006; Duret and Arndt 2008), as well as BDS (Dreszer et al. 2007; Capra and Pollard 2011), in the human genome argues for the relevance of gBGC to these evolutionary patterns.
Several recent studies using high-resolution polymorphism data from specific regions of the human genome have supported the hypothesis of ongoing GC-biased evolution. Katzman et al. (2010) performed high-throughput sequencing of neighborhoods around HARs from 22 sets of human chromosomes from 11 Yoruban individuals to determine derived allele frequencies (DAFs) for all single-nucleotide polymorphisms (SNPs) in these regions. The resulting comparison of the DAF spectra of W→S and S→W polymorphisms revealed a significant shift toward high allele frequencies for W→S SNPs around a subset of the HARs. Spencer et al. (2006) identified a local association between recombination hot spots and GC-increasing mutations on human chromosome 20. A very recent study of the effect of meiotic recombination on disease-related mutations found higher frequencies for GC alleles in several likely functional classes of polymorphism identified by the HapMap project (Necşulea et al. 2011).
The recent completion of the pilot phase of the 1000 Genomes (1000G) Project (1000 Genomes Project Consortium 2010) enables the analysis of human polymorphism at an unprecedented scale and resolution. The low-coverage pilot phase data from this project include approximately 15 million SNPs from a total of 179 samples from 4 different HapMap populations (The International Hapmap Consortium 2007): Yorubans from Ibadan, Nigeria (YRI), individuals of European origin in Utah (CEU), Han Chinese from Beijing (CHB), and Japanese from Tokyo (JPT). The 1000G low-coverage data captures in a relatively ascertainment-free manner nearly all (∼95%) of the common polymorphism in the sampled populations in the regions of the human genome that are accessible with current technology. These data give a snapshot of sequence evolution over a much shorter time period than the millions of years that separate sister species, providing the opportunity to capture bias inducing processes in action.
We use the 1000G data to investigate signatures of ongoing W→S bias genome-wide and to test previous hypotheses about its causes. By comparing the DAF spectra of W→S and S→W changes, we find strong evidence for ongoing GC fixation bias across the human genome. The fixation bias is widespread, occurs on a local scale (∼1 kb), and is significantly increased in regions with high recombination rate. Our results shed new light on the scale of and mechanisms responsible for changes in the rate of evolution and GC content in the human genome.
Material and Methods
Data
This report uses the low-coverage pilot data from the 1000G project released in July 2010. These comprise SNP calls for the 22 autosomes in three HapMap population panels: YRI (59 individuals), CEU (60 individuals), and CHB+JPT (60 individuals). The VCF format files contain for each SNP: the position (in NCBI36/hg18 reference coordinates), the count of each allele, and an indication of the ancestral allele. The latter is derived from the Enredo–Pecan–Ortheus (EPO) pipeline (Paten, Herrero, Beal, et al. 2008; Paten, Herrero, Fitzgerald 2008), which determines the common ancestor of human and chimpanzee at a locus by considering alignments of the human, chimpanzee, orangutan, and rhesus macaque genomes. This report eliminated from analysis any positions with more than two alleles among the reference, ancestral, or sample alleles or where the ancestral allele was not determined by the EPO pipeline. Lowercase values of the predicted EPO ancestral allele, which result from various cases without complete evidence in all species, were considered in the main analysis but are excluded in the “Ancestor Match” control (supplementary table S3, Supplementary Material online).
Recombination hot spots and cold spots were taken from the supporting information for Myers et al. (2005), and recombination rates were downloaded from the HapMap Project (The International Hapmap Consortium 2007). The positions of the published centers of the hot spots and cold spots were converted from NCBI34/hg16 coordinates to NCBI36/hg18 coordinates using the liftOver tool of the UCSC Genome Browser (Kent et al. 2002). Two sets of sex-specific recombination rate maps were used. The first set is the pedigree-based data of Kong et al. (2002) as downloaded at the 1 Mb scale from the UCSC Genome Browser's recombRate track. The second set of sex-specific maps with higher resolution was taken from Kong et al. (2010).
The location of the PRDM9 motif (CCTCCCTNNCCAC) sites and associated control motif (CTTCCCTNNCCAC) sites was determined by running the findMotif tool against the human reference genome (hg18) taken from the UCSC Genome Browser (Kent et al. 2002).
BDS of the human genome since divergence from chimpanzee was calculated using alignments of human, chimpanzee, and rhesus macaque as described in Capra and Pollard (2011).
Analysis
Comparison of DAF Spectra.
For a given region or pooled set of regions, the W→S and S→W positions were separately extracted from the set of SNPs and the two spectra of DAFs were constructed from the allele counts in the VCF files. We performed a Mann–Whitney U (MWU) test for a difference between the W→S and S→W DAF spectra using the command “wilcox.test (paired=FALSE, alternative=two.sided)” in the R language (R Development Core Team 2009). For each test, the normalized U statistic “U-norm” was calculated by dividing the U statistic by its maximum possible value in the test, the product of the number of SNPs in the two categories (Bamber 1975). As noted in the Results, this can be interpreted as the probability that a random W→S SNP is segregating at a higher DAF than a random S→W SNP plus one half the probability that they are equal. Since the possible U values for each test are normally distributed with a mean and variance that can be directly computed, confidence intervals for the U-norm estimates were calculated with reference to this normal distribution. As a separate measure of the difference between DAF spectra, the difference in the mean values of the DAFs for W→S and S→W SNPs was calculated for each test.
The MWU test was performed on the set of SNPs from the entire genome, entire chromosomes, entire chromosome arms, and genome-wide sets of nonoverlapping windows of sizes 40 kb, 1 Mb, and 4 Mb. Windows that did not have SNPs in both categories were filtered out before the MWU test. Such regions include portions of the genome that have not been fully sequenced and assembled, regions that were not accessible to the technology used in the 1000G project, and regions in which the EPO pipeline was unable to determine the ancestral allele. At the 40 kb scale, 91% of the 71,703 windows passed these filters.
For the analysis of hot spots, cold spots, and PRDM9 motifs, SNPs were pooled from the neighborhoods of the center position of each feature. For example, for the 200 bp analysis, all SNPs within 100 bp on either side of the 25,644 hot spot centers found in the autosomes were aggregated to perform the MWU test.
Correlation of W→S DAF Skew and Genomic Features.
To quantify the similarity of the spatial distribution of different signals, such as W→S DAF skew, recombination rate, and BDS across the genome, we calculated Spearman's rank correlation coefficient on the statistics measured over nonoverlapping windows of 40 kb and 1 Mb.
Results
Preliminaries
We analyzed autosomal SNPs from all populations in the 1000G low-coverage data set. We focus on results based on the 59 YRI individuals, which had the greatest diversity and yielded results representative of those from the other populations (table 1). Population-specific differences are also described. For each SNP, we determined the ancestral and derived alleles using several outgroup species and the EPO pipeline (Paten, Herrero, Beal, et al. 2008; Paten, Herrero, Fitzgerald, et al. 2008). We then classified the allele on each of the chromosomes in a population (118 for YRI: two from each of the 59 individuals sequenced) as ancestral or derived. Using these counts, we constructed DAF spectra for different sets of SNPs.
Table 1.
Ongoing W→S Fixation Bias in HapMap Populations
| Population | Samples | SNPs | W→S DAF skew |
| YRI | 59 | 8.5M | 0.558 |
| CEU | 60 | 6.1M | 0.552 |
| CHB+JPT | 60 | 4.8M | 0.553 |
NOTE.—W→S DAF skew is quantified by the U-norm statistic.
We tested for an ongoing fixation bias genome-wide and in specific genomic regions by comparing the DAF spectra of W→S and S→W SNPs using the MWU test. Since this is a two-sided test, we further quantify the strength and direction (W→S versus S→W) of bias by calculating a statistic we call “U-norm.” U-norm is the U value from the MWU test divided by the maximum possible U value for the test. U-norm is an estimate of P(Y > X) + 0.5P (Y = X), where X is the DAF of a random S→W SNP and Y is the DAF of a random W→S SNP (Bamber 1975). When U-norm = 0.5, there is no shift in the DAFs. U-norm > 0.5 indicates higher frequencies for the W→S alleles compared to the S→W alleles and U-norm < 0.5 indicates the opposite. Hence, the value of U-norm helps to distinguish W→S fixation bias (U-norm > 0.5), in which strong alleles are rising to higher frequencies in the population, from S→W fixation bias (U-norm < 0.5). We repeated our analyses using an alternative statistic, the difference in average value of the DAF spectra (Necşulea et al. 2011) and obtained similar results (e.g., supplementary table S4, Supplementary Material online).
W→S Changes in All Populations Have Higher DAFs Genome-Wide
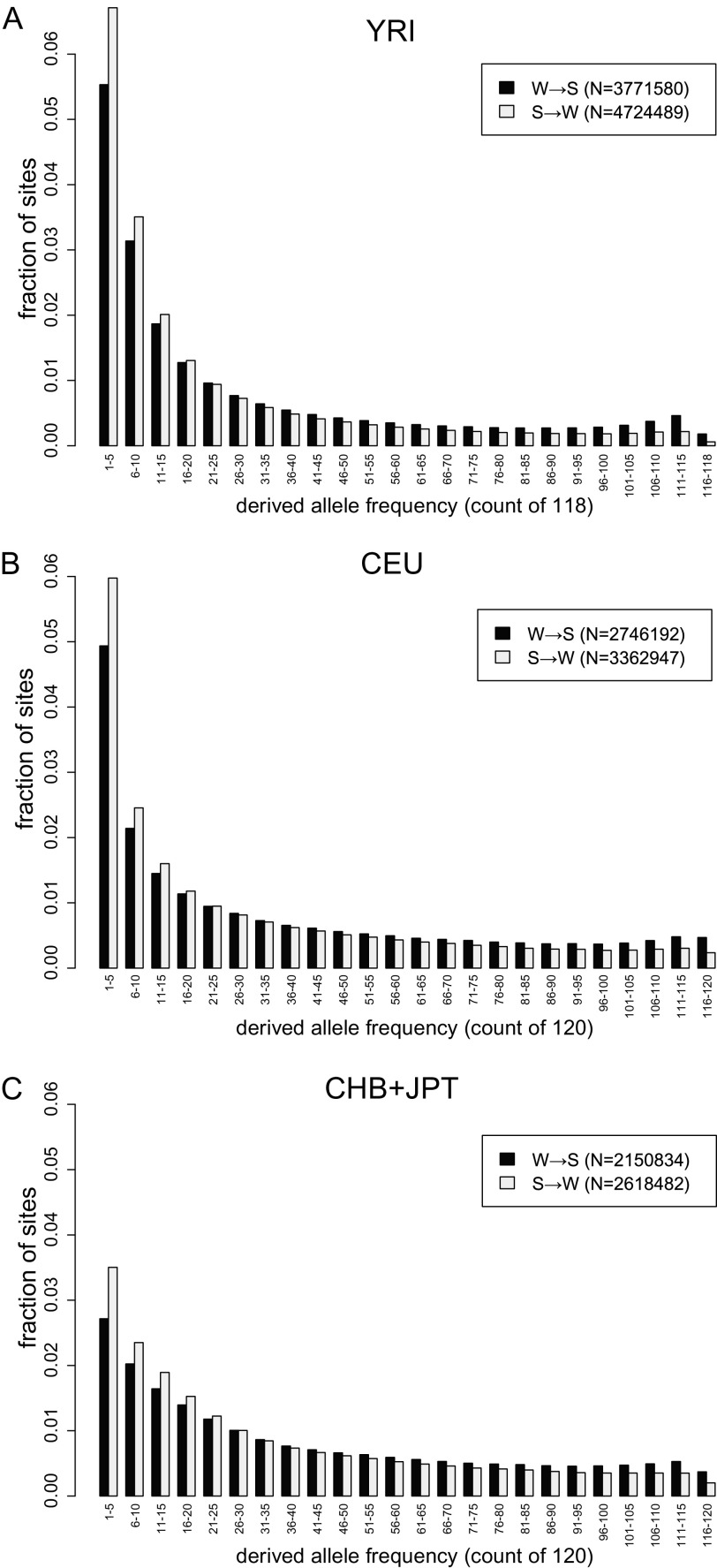
Figure 1A compares the DAFs of W→S and S→W autosomal SNPs in the YRI population. The W→S SNPs have significantly higher DAFs than the S→W changes (U-norm = 0.558; P ≈ 0). In other words, a randomly selected W→S allele is significantly more likely to be observed at high frequency in the population than an S→W allele. We henceforth refer to this pattern as “W→S DAF skew” or “W→S bias.” This result indicates that a fixation bias favoring GC alleles has been active in the recent evolution of the YRI population.
FIG. 1.—
Genome-wide, W→S SNPs have significantly higher DAFs than S→W SNPs. The DAF spectra of W→S SNPs (dark bars) have significantly (P ≈ 0) higher DAF than S→W SNPs (light bars) in the (A) YRI population, (B) CEU population, and (C) CHB+JPT population. Counts are binned for display purposes only. The values in the legends indicate the genome-wide count of SNPs of each category used in the test.
Although we primarily focus on the YRI panel, with its larger set of SNPs, we also repeated our analyses on two additional populations: 60 individuals in the CEU panel and 60 individuals from the combined CHB and JPT panels (table 1). In both these populations, W→S SNPs are also significantly (MWU tests; P ≈ 0) more likely to have higher DAFs than S→W SNPs (fig. 1B and C). SNPs in the latter two data sets are generally segregating at higher frequencies than those in the YRI panel, with mean DAF for CEU SNPs approximately midway between those for YRI and CHB+JPT (supplementary table S4, Supplementary Material online). However, since the differences between populations are nearly the same for both W→S and S→W SNPs, the net effect is less than a 1% difference among populations in the strength of the W→S DAF skew as quantified by U-norm (YRI: 0.558; CEU: 0.552; CHB+JPT 0.553).
W→S DAF Skew Is Widespread across the Genome and Shows Local Variation
The significant difference between the W→S and S→W genome-wide spectra demonstrates a global W→S fixation bias in the genome, but it does not determine its scale or spatial distribution. To investigate these questions, we compared W→S and S→W DAFs within nonoverlapping windows of various sizes across the genome. Starting at a rather large scale—4 Mb windows—we analyzed 699 windows with high-quality data (see Materials and Methods). Of these, a striking 690 windows (98%) show significant W→S DAF skew (MWU test; P < 0.05) in the YRI population, whereas none show S→W skew at P < 0.05. This result clearly demonstrates that there are only a handful of 4 Mb regions of the genome that do not show evidence of W→S fixation bias.
Next, we took advantage of the high density of SNPs in the 1000G data to localize the W→S DAF skew at an even higher resolution. At the scale of 40 kb, 65,510 of the 71,703 windows (91%) in the YRI data set passed our filters for performing the MWU test (see Materials and Methods). Of these 40 kb windows, 14,697 (22.4%) show W→S DAF skew at the P < 0.05 level. On the other hand, only 183 windows (0.3%) show significant S→W DAF skew. This is nearly 10-fold more W→S DAF skew than would be expected in the absence of bias from our two-sided test for significance. The enrichment for W→S DAF skew (and the depletion of S→W DAF skew) in these tests is even higher at more stringent significance levels (supplementary fig. S1, Supplementary Material online). It is also present across a range of smaller window sizes evaluated on a single chromosome (supplementary table S1, Supplementary Material online). For subsequent analyses, 40 kb windows were used because they generally provide enough polymorphism to give the MWU test sufficient power to detect W→S DAF skew and are computationally tractable.
Despite strong evidence that W→S DAF skew is nearly ubiquitous in the human genome, analyses of local fixation bias (40 kb windows and smaller) revealed variation in the magnitude of this bias across the chromosomes. Figure 2 shows the strength of the W→S DAF skew across chromosome 2 as quantified by U-norm. An increase in W→S DAF skew near the telomeres was observed in the majority of chromosomes (supplementary fig. S2, Supplementary Material online).
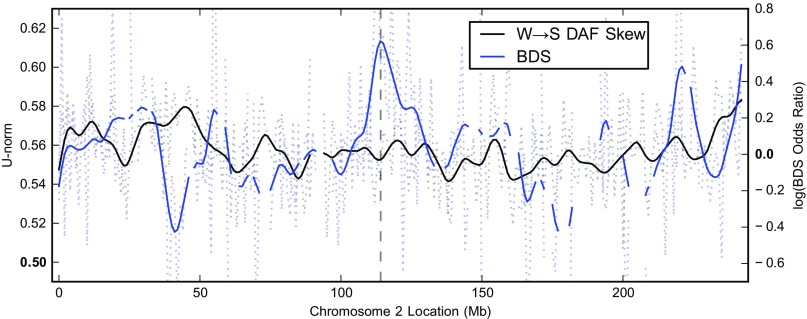
FIG. 2.—
The strength of the W→S fixation bias varies across the genome. W→S DAF skew (black) varies across chromosome 2 and is moderately correlated (ρ = 0.26 smoothed, P ≤ 4.4 × 10-5) with BDS (blue). W→S DAF skew is quantified by calculating U–norm on nonoverlapping 1 Mb windows using the YRI SNP data. This value reflects the probability that a random W→S SNP is segregating at a higher frequency than a random S→W SNP in the same window and would be 0.5 in the absence of fixation bias. The BDS score (described in Capra and Pollard 2011) is calculated over nonoverlapping 1 Mb blocks; gaps in the curve indicate blocks that did not have a sufficient number of substitutions after filtering for the calculation. This score is the logarithm of an odds ratio, so values above 0 indicate a preference for W→S changes in divergent regions. Dotted lines give the raw values, and solid lines give the same data smoothed by convolution with a Hanning window of 12 Mb. The dashed vertical line highlights the location of the chromosome fusion event on the human lineage in 2q13–2q14.1 (hg18: 114077148–114077163). There is a peak of BDS, but not a peak of W→S DAF skew around the fusion.
Taken together, these results argue that the forces driving the fixation bias operate at a local scale, vary across the genome, and are widespread. In the next sections, we examine these patterns of variation in the W→S DAF skew to investigate possible causes.
The Strength of W→S DAF Skew Varies Inversely with Chromosome Arm Length
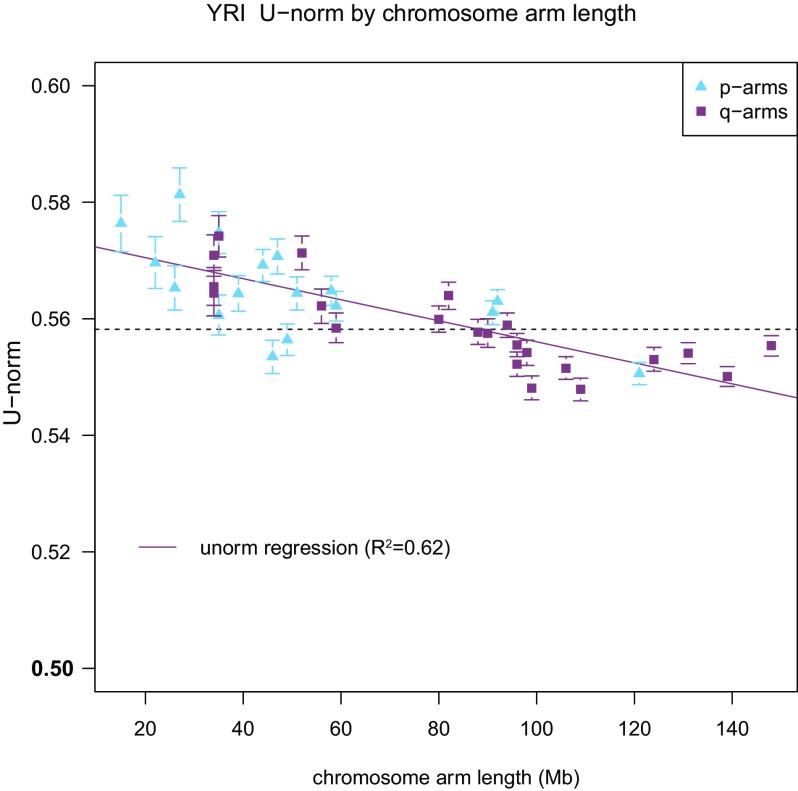
As a first step in investigating the genome-wide distribution of W→S DAF skew, we examined each chromosome arm individually. Several signatures of GC-biased evolution between species, such as current and stationary GC content and BDS, have been found to be inversely correlated with chromosome length (Fullerton et al. 2001; International Chicken Genome Sequencing Consort 2004; Dreszer et al. 2007). To test for this pattern in W→S DAF skew, we calculated U-norm between all W→S and S→W SNPs on each chromosome arm. The magnitude of the W→S DAF skew varies between arms with shorter arms showing stronger bias (fig. 3). Chromosome arm length explains a large amount of the variance in W→S DAF skew between arms (R2 = 0.62). A similar pattern of W→S DAF skew was observed in DAFs over entire chromosomes (R2 = 0.50).
FIG. 3.—
The strength of the W→S fixation bias is inversely correlated with chromosome arm length. For each chromosome p-arm (light blue triangles) or q-arm (purple squares), W→S DAF skew is plotted versus length. Error bars indicate 95% confidence intervals, and the dashed line indicates the genome-wide value for U-norm in the YRI data. The least-squares regression line is plotted with a solid line. Chromosome arm length explains 62% of the variance in W→S DAF skew among chromosomes arms.
Recombination rate is thought to be elevated on shorter chromosome arms compared to longer arms due to the proposed requirement of one chiasma per arm per meiosis (Kaback et al. 1992, 1999; Coop and Przeworski 2007); however, there is still debate about the causes and extent of this pattern (Turney et al. 2004; Fledel-Alon et al. 2009). Correlations with chromosome length have been used to support the theory that a recombination-driven process, such as gBGC, influences patterns of BDS (Duret and Arndt 2008; Duret and Galtier 2009a).
W→S DAF Skew Is Elevated in Neighborhoods of Recombination Hot Spots
Because BDS, which is based on fixed substitutions between sister metazoan species, is correlated with recombination rates in several species, we predicted that W→S DAF skew would also be enriched near current recombination hot spots and would be correlated with recombination rate across the genome. To test the first of these predictions, we investigated W→S DAF skew in neighborhoods around human recombination hot spots and cold spots (regions with no evidence of recombination), identified from patterns of linkage disequilibrium in HapMap polymorphism data (Myers et al. 2005). We computed U-norm on the DAFs of W→S and S→W SNPs in windows of increasing size (from 200 bp to 8 kb) around the center of each recombination hot spot and cold spot.
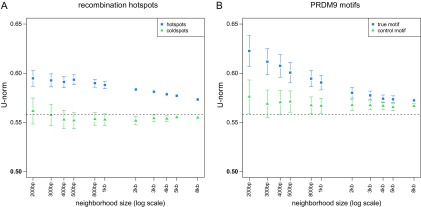
Regions around recombination hot spots show significant W→S DAF skew (fig. 4A). This bias is strongest in small windows (0.595 ± 0.008 for 200 bp compared with 0.573 ± 0.001 for 8 kb). For each window size considered (even windows as large as 8 kb), the W→S DAF skew near recombination hot spots is significantly greater than the genome-wide level (0.558) and the level found in windows of corresponding size around recombination cold spots. SNPs near the cold spots do not show significantly less W→S DAF skew than the genome-wide background level. The increase in W→S DAF skew with proximity to a hot spot argues that the process producing it is likely local, operating on the scale of hundreds of base pairs to a few thousand base pairs. This is in agreement with current estimates of the scale of the conversion tracts in gBGC events (Duret and Galtier 2009a).
FIG. 4.—
W→S DAF skew is stronger in the neighborhood of recombination hot spots and PRDM9-binding motifs. The strength of W→S DAF skew increases with decreasing distance from (blue squares) (A) the center of recombination hot spots and (B) predicted PRDM9-binding motifs (true motif). Control points (green triangles) are (A) recombination cold spots and (B) an altered version of the PRDM9 motif (“control motif”, see Materials and Methods). Each data point is based on the spectra of all YRI SNPs contained in neighborhoods of the indicated size centered on features of a given type. Bars represent 95% confidence intervals. The dashed line indicates the genome-wide level of W→S DAF skew. The increase in bias up to a few hundred base pairs from the centers suggests that the process producing the bias has a very local effect. The stronger W→S DAF skew around PRDM9 motifs than hot spots suggests that hot spots containing this motif could be “hotter” than other hotspots.
W→S DAF Skew Is Elevated around PRDM9-Binding Sites
The human protein PRDM9 has recently been identified as a major determinant of recombination hot spots in human (Baudat et al. 2010; Berg et al. 2010; Myers et al. 2010; Parvanov et al. 2010). This histone methyltransferase contains a highly variable array of DNA-binding C2-H2 zinc finger domains, and the particular DNA sequence specificity of these domains is thought to partially determine sites of recombination. Myers et al. (2008) identified a 13 bp motif (CCTCCCTNNCCAC) that is predicted to be bound by PRDM9 and involved in defining around 40% of all known meiotic hot spots.
We identified ∼7,000 potential PRDM9-binding sites by scanning for the motif genome-wide and then carried out neighborhood-based analyses around these sites. As for recombination hot spots, SNPs near PRDM9-binding motifs show significant W→S DAF skew and are more biased than a set of control sites defined by a very similar, but nonrecombinogenic motif (CTTCCCTNNCCAC) used by the 1000G project (1000 Genomes Project Consortium 2010) (fig. 4B). The overall strength of the W→S DAF skew around PRDM9 motifs is greater than that around predicted hot spots for most neighborhood sizes, although this difference is only significant for 200 bp windows (where hot spot U-norm = 0.595, PRDM9 motif U-norm = 0.623). It is possible that hot spots containing a full copy of the motif are “hotter” than the average hot spot, many of which do not contain the full motif. The W→S DAF skew near control non-PRDM9 sites is slightly elevated over the background (fig. 4B); this could indicate that some of these control motifs actually influence recombination or did so in recent evolution before gaining disabling mutations.
Recombination Rate Is Spatially Correlated with W→S DAF Skew across the Genome
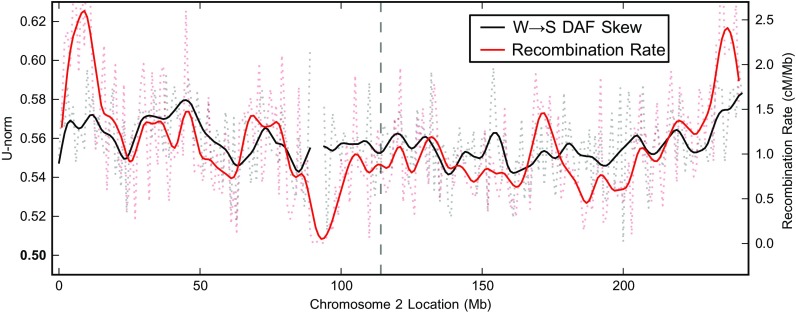
To further explore the relationship of W→S DAF skew and recombination, we correlated the W→S DAF skew in windows across the genome with the estimated recombination rate for each window for the YRI population from HapMap (The International Hapmap Consortium 2007). At a 40 kb window size, the raw Spearman correlation across the entire genome is 0.20 (P ≈ 0). When larger 1 Mb windows are considered, the correlation becomes much stronger (ρ = 0.53; P ≈ 0). Figure 5 illustrates this strong spatial correlation between bias and recombination rate across chromosome 2 (ρ = 0.43; P ≈ 0). The dramatic increase in correlation with increasing window size is consistent with models of the evolution of recombination hot spots in which there is a large amount of local variation between individuals and over time, but this variation is mainly contained within larger-scale regions with fairly constant recombination rates (Myers et al. 2005).
FIG. 5.—
W→S fixation bias and recombination rate are significantly correlated across the genome. The W→S DAF skew (black) (as described in fig. 2) and YRI population–based recombination rate (red) are significantly correlated (ρ = 0.62 smoothed and ρ = 0.43 unsmoothed; both P ≈ 0) across chromosome 2. Raw W→S DAF skew and recombination rate data in 1 Mb windows are plotted with dotted lines, and the data smoothed using a Hanning window of size 12 Mb are shown with solid lines. The dashed vertical line indicates the location of the fusion of two ancestral chromosomes on the human lineage.
The W→S DAF skews in the CEU and CHB+JPT populations are also significantly correlated with HapMap recombination rates (table 2). This holds for both the recombination map specific to the population considered and the combined map based on all polymorphism data. A population-specific map is not available for CHB+JPT. For both the YRI and CEU populations, the correlation is slightly greater with the combined map than the population specific maps (table 2). This result is somewhat surprising but may reflect differences in the hot spot distribution in the ancestral population (in which many high frequency alleles first appeared) compared with modern day populations (see Discussion).
Table 2.
Correlation between W → S Fixation Bias and Recombination across Populations
| Type of Bias | HapMap |
deCODE 2002 |
deCODE 2010 |
|||
| Population Specific | Combined | Male | Female | Male | Female | |
| W→S DAF skew | ||||||
| YRI | 0.528 | 0.535 | 0.371 | 0.330 | 0.403 | 0.471 |
| CEU | 0.442 | 0.454 | 0.318 | 0.275 | 0.351 | 0.396 |
| CHB+JPT | NA | 0.314 | 0.258 | 0.182 | 0.259 | 0.261 |
| BDS | ||||||
| All | NA | 0.189 | 0.179 | 0.0787 | 0.166 | 0.146 |
NOTE.—NA, not applicable. The Spearman rank correlation (ρ) of W→S DAF skew in each population with HapMap population-based (The International Hapmap Consortium 2007) and deCODE sex-specific, pedigree-based (Kong et al. 2002, 2010) recombination maps at the 1 Mb scale is shown. Correlations are all significant but vary in strength across HapMap populations and recombination maps (see Discussion). The correlation of BDS Capra and Pollard (2011) with each recombination map is consistently lower than the correlation of W→S DAF skew and recombination rate. BDS also shows a different sex-specific pattern. These results are similar when considering 40 kb windows (supplementary table S2, Supplementary Material online).
Correlations between W→S DAF Skew and Recombination May Be Less Different between the Sexes than Previously Thought
In species with a pedigree-based recombination map, it is possible to estimate sex-specific recombination rates. Previous sex-specific analyses found stronger correlations between W→S substitution biases and male recombination rate than female recombination rate in several mammalian species (Capra and Pollard 2011), including humans (Webster et al. 2005; Dreszer et al. 2007). To explore the possibility of a similar sex-specific association with W→S DAF skew, we considered two pedigree-based, sex-specific recombination maps from deCODE (Kong et al. 2002, 2010). The first map was estimated from microsatellite markers in 146 Icelandic families; it has a resolution of ∼1 Mb and was used in many previous studies. The second map is based on 15,257 parent–offspring pairs from the Icelandic population and has higher resolution (∼10 kb).
Using both Icelandic recombination maps, W→S DAF skew in all three populations is significantly correlated with recombination rates in males and females (table 2). However, the magnitude of these correlations is smaller than those observed with sex-averaged, population-based HapMap maps. Considering all the maps and populations together yielded a surprising result. The YRI population is more strongly correlated with every recombination map than any other population. This is true even when the other population is more closely related to the source of the map; for example, we might expect the CEU population to have a stronger correlation with the Icelandic maps. But for both male and female rates, it has a smaller correlation than does YRI (table 2).
W→S DAF skew shows less differentiation in its correlation with recombination between the sexes than does BDS (Webster et al. 2005; Dreszer et al. 2007). And although W→S DAF skew is slightly more correlated with male recombination rate than female rate using the initial Kong et al. (2002) maps, the opposite is true using the higher resolution Kong et al. (2010) maps with windows of 1 Mb or 40 kb (table 2; supplementary table S2, Supplementary Material online). These findings suggest that W→S biases in human may not be consistently male-driven as previously hypothesized (Webster et al. 2005; Dreszer et al. 2007).
Comparison of the Spatial Distribution of W→S DAF Skew across Populations
To compare patterns of W→S DAF skew between the three populations, we calculated pairwise correlations of U-norm in 40 kb and 1 Mb windows across the genome. The spatial distribution of the bias is significantly correlated in each pair of populations (Spearman rank correlation; P ≈ 0 for all): ρ = 0.36 for YRI vs. CEU, ρ = 0.29 for YRI vs. CHB+JPT, and ρ = 0.38 for CEU vs. CHB+JPT. As expected the two more closely related populations (CEU and CHB+JPT) show a stronger correlation than either does with the YRI; however, this is no longer true at the 1 Mb scale: ρ = 0.67 for YRI vs. CEU, ρ = 0.54 for YRI vs. CHB+JPT, and ρ = 0.60 for CEU vs. CHB+JPT. Although all these correlations are significant, the lack of stronger correlation at these scales may reflect variation in the location of recombination hot spots between the populations.
Comparison of Spatial Distribution of W→S DAF Skew and BDS
Overall, the spatial distribution of BDS (since divergence with chimpanzee) and W→S DAF skew across the human genome are qualitatively similar. The telomeres show peaks of both types of bias, and each has a significant correlation with recombination rate. To quantify this similarity, we directly compared patterns of W→S DAF skew (U-norm statistic) to those of BDS (log of an odds ratio) computed in 1 Mb windows across the human genome by Capra and Pollard (2011). We found a weak, but significant, correlation between the two measurements (ρ = 0.18; P ≈ 0). The correlation of BDS with current recombination rates (from HapMap) is of similar magnitude (ρ = 0.19; P ≈ 0).
There is, however, one dramatic difference in the patterns of BDS and W→S DAF skew in the human genome. Dreszer et al. (2007) observed a significant peak of BDS in the middle of chromosome 2 (fig. 2). This pattern is consistent with elevated BDS near telomeres (possibly driven by high recombination rates in those regions) because human chromosome 2 resulted from a fusion of two separate chromosomes in the ancestor of human and chimpanzee (Hillier et al. 2005). In contrast, there is not a significant peak of W→S DAF skew in the middle of chromosome 2 (fig. 2). The recombination rate in the region of the fusion is not currently elevated and much of the polymorphism studied in this paper likely occurred after the fusion (estimated to have occurred ∼0.75 Ma by Dreszer et al. (2007)). Hence, the lack of W→S DAF skew near the fusion provides further support to the model in which peaks of W→S DAF skew and BDS are driven by a recombination-associated process that varies in intensity along the chromosomes and over time.
W→S DAF Skew Is Not the Result of CpG Dinucleotide Hypermutability
Context-dependent differences in mutation rate (e.g., as caused by CpG site hypermutability) can lead to misidentification of the ancestral state of an SNP when using parsimony (Hernandez, Williamson, and Bustamante 2007), potentially generating false signatures of a fixation bias (Hernandez, Williamson, Zhu, and Bustamante 2007). Our analysis uses the ancestral state predicted by the 1000G project to infer the derived allele. These predictions were made using Ortheus (Paten, Herrero, Fitzgerald, et al. 2008), a probabilistic alignment-based method. Ortheus considers alignments of human, chimpanzee, orangutan, and rhesus macaque, but it does not explicitly model context-dependent substitution rates. Thus, we expect it to be less subject to the ancestral misidentification bias than parsimony. However, since its sensitivity to CpG hypermutability has not been addressed directly, we performed three control analyses based on either correcting for CpG effects on DAF spectra or removing potential CpG sites from our calculations (see supplementary results S1.1, Supplementary Material online). These analyses consistently show that CpG effects contributed little or no signal to the patterns of W→S DAF skew we observed in the human genome (supplementary fig. S3, Supplementary Material online) and their correlation with recombination rates (supplementary table S3, Supplementary Material online).
Discussion
We used polymorphism data from the 1000G low-coverage pilot project to investigate evidence for ongoing evolutionary biases throughout the human genome. The scale and coverage of the 1000G data allowed us to search recent evolutionary history for signatures of possible causes of these biases at an unprecedented scale and resolution. We found that polymorphic sites in human populations are more likely to have higher DAFs if they convert ancestral A or T nucleotides to G or C nucleotides than vice versa. Our results establish that this W→S DAF skew is widespread across the human genome and increases in strength near recombination hot spots.
W→S DAF Skew Demonstrates an Ongoing GC-Fixation Bias
By considering current variation in the human population, we are able to catch polymorphic sites “in the act” of becoming fixed. The consistent and significant shift of W→S SNPs toward higher frequencies reflects the action of a process that favors strong alleles over weak. An ongoing fixation bias has been proposed previously as a possible cause of the observed BDS (Dreszer et al. 2007; Berglund et al. 2009), wherein fixed changes along the human lineage exhibit W→S bias at certain loci. Several studies have established the presence of a GC fixation bias from analysis of DAFs in specific genomic elements (Lercher and Hurst 2002; Duret et al. 2002; Webster et al. 2003; Webster and Smith 2004; Spencer et al. 2006; Katzman et al. 2010). Our genome-wide results across three populations are consistent with these previous findings and demonstrate the presence of a GC fixation bias throughout the human genome.
gBGC Is Likely a Cause of the Fixation Bias
gBGC is a nonselective, recombination-driven process that produces an evolutionary bias for GC alleles. gBGC results from the DNA repair machinery's handling of mismatches in short (∼1 kb) heteroduplex DNA regions that form near recombination-initiating double-strand breaks. When there is heterozygosity, the alleles from one chromosome are copied to the other, with a bias for conversion of A or T alleles to G or C alleles (Strathern et al. 1995; Marais 2003; Meunier and Duret 2004; Duret and Galtier 2009a). Over time, gBGC is proposed to hasten the fixation of W→S mutations independent of their fitness effect (Galtier and Duret 2007; Duret and Galtier 2009b). The previously observed correlations between recombination rate and GC content (Meunier and Duret 2004; Webster et al. 2005; Duret and Arndt 2008), BDS (Dreszer et al. 2007; Capra and Pollard 2011), and GC-biased polymorphism (Spencer et al. 2006) make gBGC a prime candidate for the cause of GC fixation bias in metazoan genomes.
Our analysis points to gBGC as a likely source of W→S DAF skew. As suggested by the correlation of recombination rate with GC content and BDS, we find a striking association of the W→S DAF skew with three different characterizations of recombination. First, the magnitude of W→S DAF skew increases with decreasing chromosome arm length (R2 = 62%). Because recombination rate per nucleotide per meiosis is thought to correlate with chromosome length (Coop and Przeworski 2007; Fledel-Alon et al. 2009), this ties high rates of recombination to the W→S DAF skew on the chromosome level. Second, our analysis of neighborhoods immediately surrounding recombination hot spots connects W→S DAF skew with recombination on a much more local scale. We find significantly more bias a few thousand base pairs around recombination hot spots than in the genome-wide background. The scale of this effect is consistent with the conclusion of the 1000G project that the extent of recombination hot spots is smaller than previously thought, perhaps only ∼2 kb (1000 Genomes Project Consortium 2010). Finally, the spatial distribution of W→S DAF skew across the genome is correlated with estimates of recombination rate at several scales.
These results argue for gBGC, or some other recombination-associated process, as a cause of W→S DAF skew, but mechanistically testing this hypothesis will require further modeling and experiments.
Why Do Correlations between W→S DAF Skew and Recombination Vary across Data Sets?
Although we consistently observe strong and significant correlations between W→S DAF skew and recombination rates across all 1000G populations and a variety of human recombination maps, the magnitude of these correlations differ. These differences may shed some light on the processes that generated current patterns of W→S DAF skew in the human genome and our ability to detect these processes.
First, W→S DAF skew in all populations is more highly correlated with HapMap linkage-based estimates of recombination than with pedigree-based estimates from the Icelandic population (Kong et al. 2010). This could result from the fact that HapMap recombination data are from the same populations as the W→S DAF skew data. It may also suggest that patterns of W→S DAF skew are driven by a combination of current and recently extinct hot spots, with the latter being detected by linkage but not pedigree estimators. The stronger correlation of the HapMap-combined recombination map versus the population-specific maps for YRI and CEU offers further support for the idea that W→S DAF skew largely reflects differences in the frequencies of older high frequency alleles, many of which likely arose prior to differentiation of the four populations for which we have recombination maps and could have been produced by currently extinct hot spots.
Second, W→S DAF skew in the YRI population is more strongly correlated with recombination compared with the other populations. This result holds across length scales and even when comparing YRI W→S DAF skew to a recombination map from another HapMap population or the Icelandic population. A number of factors could contribute to this pattern. The YRI genome has not experienced the out-of-Africa event and therefore is more diverse, contains a greater number of old polymorphisms, and could potentially have a more stable and/or ancestral distribution of recombination hot spots compared with CEU and CHB+JPT genomes. These variables could lead to more accurate W→S DAF skew estimates and greater correlation between W→S DAF skew and recombination in YRI. Such effects would be particularly strong if current patterns of W→S DAF skew reflect the combination of mutation and recombination processes over hundreds of thousands of years, including biases driven by extinct ancestral hot spots.
Finally, we were surprised to observe a stronger correlation of W→S DAF skew with male recombination rate in the older sex-specific maps and the opposite pattern— stronger correlation with female recombination rate—in the new maps. BDS is more strongly correlated with male recombination rate in both maps; however, the difference between the correlation of BDS with male and female rates is dramatically lower for the newer maps (table 2). This lack of agreement between comparisons using different data sets suggests that there may not be a sex difference in the strength of association between W→S fixation bias and recombination. Rather, sex-specific recombination maps themselves may differ dramatically across studies, even within the same Icelandic population. Indeed, estimated recombination rates are no more correlated between the two Icelandic sex-specific maps than they are between either map and the YRI map (Kong et al. 2010). These differences warrant further exploration.
If we consider only the new, higher resolution sex-specific map, then the correlations of W→S DAF skew and BDS with male and female recombination rates do not agree. Although we believe that W→S DAF skew is likely involved in the creation of BDS, these two phenomena are not analogous. BDS represents historical biases over the entire branch from human to the human–chimp ancestor, whereas W→S DAF skew considers only recent biases detectable in human polymorphism data. Because the distribution of recombination events across the genome evolves rapidly, it is possible that the stronger correlation between W→S DAF skew and female recombination rate represents a recent change in recombination dynamics. For example, a higher fraction of female recombination events might now result in gBGC, whereas in the past the opposite might have been true. Thus, it is possible that over the entire human branch, male recombination rates are a better predictor of gBGC rates. In addition, there are a number of other factors that are likely to influence the strength and genomic distribution of BDS but are unlikely to affect W→S DAF skew. For example, the production of BDS requires variation in recombination rate over time or across the genome (Capra and Pollard 2011), whereas this is not necessary for the production W→S DAF skew. Until we have a better mechanistic understanding of meiotic recombination and its evolution, as well as the observed variation between recombination maps, these discussions will remain speculative.
What Explains W→S DAF Skew Outside of Recombination Hot Spots?
We observe considerable W→S DAF skew outside of recombination hot spots. There are several possible explanations for this observation that are consistent with a recombination-associated processes, such as gBGC, as the main source of W→S DAF skew.
First, our knowledge of recombination hot spots in each population is not complete. Recombination patterns are highly variable between individuals, even within the same population (Coop et al. 2008; Berg et al. 2010; Baudat et al. 2010; Kong et al. 2010). Current maps of recombination do not fully capture the dynamics of its evolution. In addition, many recombination events do not result in crossover, and current linkage disequilibrium–based methods for detecting recombination from population data are not able to detect these events. Non-crossover events are thought to have different distributions across the genome than crossovers; Holloway et al. (2006) found this in sperm-typing studies at individual hot spots. Since gBGC still occurs without crossover (Duret and Galtier 2009a), this would lead to W→S DAF skew outside the current hot spot map. Supporting this interpretation, Gay et al. (2007) found ∼1.5× more gene conversion than expected from crossovers alone in a region of human chromosome 1. It is possible that integrating W→S DAF skew into models of the evolution of recombination could help identify previously unrecognized hot spots and refine maps of recombination.
Another possibility is that gBGC is not the sole cause of the W→S DAF skew. Natural selection favoring GC alleles has been proposed as a possible source of the isochore structure of the genome (Eyre-Walker and Hurst 2001). Many theories about potential benefits of increased GC content—from greater thermal stability of DNA (Bernardi et al. 1985) to positive effects on gene expression (Kudla et al. 2006)—have been suggested. Distinguishing gBGC from natural selection is complicated by the fact that, on the population level, the action of gBGC resembles selection for GC alleles (Nagylaki 1983). Our observation of widespread W→S DAF skew across the genome argues against selection as a main source of bias since the vast majority of SNPs considered are in noncoding sequence and are expected to be fitness neutral (Kimura 1983). The significant correlation of W→S DAF skew with recombination rate strongly supports the gBGC hypothesis, but it is not necessarily inconsistent with selection on GC content. The efficiency of selection is thought to increase in regions of high recombination due to a reduction in Hill–Robertson interference (Hill and Robertson 1966), and Berglund et al. (2009) proposed that this could also lead to a signal that varies with recombination. However, Duret and Arndt (2008) argue that Hill–Robertson effects are not strong enough to produce a significant correlation with recombination rate. Overall, our results argue most strongly for gBGC as a cause of the fixation bias, but they do not definitively exclude the possibility of selection impacting the W→S DAF skew at specific loci.
Why Do Some Recombination Hot Spots Lack Bias?
A small number of recombination hot spots are not biased. When considering 40 kb windows centered on hot spots, 745 (2.9%) have significantly lower W→S DAF skew than the genome-wide background, and 34 have U-norm significantly less than 0.5. As noted above, hot spots change location frequently within populations. These unbiased hot spots may be very young and thus have not been active for a sufficient amount of time to push W→S alleles to higher frequency. It is possible that, with the proper modeling framework, comparison of DAF spectra could provide a way to “age” hot spots.
We did not directly consider the effects of selection, because as described above, it is generally accepted that the vast majority of mutations are fitness neutral. However, on a local scale, variation in selective pressure could influence our ability to detect W→S DAF skew if it is caused by gBGC.
Missing Data Are Likely to Reinforce Evidence for GC Bias
The 1000G project provides the most unbiased set of human SNPs available to date. However, the low-coverage pilot project did not have sufficient power to find all rare variants. For example, it is estimated that only ∼25% of SNPs occurring in a single chromosome (singletons) were identified (1000 Genomes Project Consortium 2010). We observed more S→W than W→S SNPs at low frequencies. Furthermore, the shift of W→S SNPs to higher DAFs that we observed throughout the genome suggests that over time W→S SNPs move into the higher frequency bins of the spectrum more readily than do S→W SNPs. Thus, we would expect the “missing data” in the low-frequency bins (such as singletons) to contain more S→W than W→S SNPs. If such data were not missing, it would magnify the strength of the W→S DAF skew by putting comparatively more weight in the low-frequency S→W bins than W→S bins. Therefore, we conclude that our results likely underestimate the strength of ongoing GC bias.
Conclusions
The 1000G low-coverage pilot project enabled us to perform a deep, genome-wide analysis that demonstrates ongoing GC-biased evolution in the human genome. In contrast to the limited scope of previous, more localized studies, we can conclude that this phenomenon is widespread across the genome. By showing that W→S DAF skew is significantly elevated in close neighborhoods of recombination hot spots, we add support to the hypothesis that (adaptively neutral) gBGC is its main driving force. These findings have important implications for population genetics modeling in general and for methods that use the shape of DAF spectra to draw conclusions about natural selection in particular. Previous studies of fixed substitutions between species argued that gBGC can produce false positives in common tests for positive selection between species (Berglund et al. 2009; Ratnakumar et al. 2010). Our results suggest that this evolutionarily neutral force is currently active throughout the genome in human populations, so models of sequence evolution and tests for selection based on polymorphism data must also take the resulting biases into account.
Supplementary Material
Supplementary results, figures S1–S3, and tables S1–S4 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our-journals/gbe/).
Acknowledgments
We thank Needhi Bhalla for discussions of rates of recombination during meiosis, Benedict Paten for clarification of aspects of the EPO pipeline, Ed Green for discussions of variation detectable along the human lineage, Dennis Kostka for discussions of the effects of gBGC, and Ryan Hernandez for supplying the software to correct DAF spectra for context-dependent mutation bias. S.K. was supported by the California Institute for Quantitative Biosciences. D.H. was supported by the Howard Hughes Medical Institute. K.S.P. and J.A.C. were supported by the National Institute of General Medical Sciences (grant #GM82901).
References
- 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamber D. The area above the ordinal dominance graph and the area below the receiver operating characteristic graph. J Math Psychol. 1975;12(4):387–415. [Google Scholar]
- Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327(5967):836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg IL, et al. Prdm9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet. 2010;42(10):859–863. doi: 10.1038/ng.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund J, Pollard KS, Webster MT. Hotspots of biased nucleotide substitutions in human genes. PLoS Biol. 2009;7:e26. doi: 10.1371/journal.pbio.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi G, et al. The mosaic genome of warm-blooded vertebrates. Science. 1985;228(4702):953–958. doi: 10.1126/science.4001930. [DOI] [PubMed] [Google Scholar]
- Capra JA, Pollard KS. Substitution patterns are GC-biased in divergent sequences across the metazoans. Revision. Genome Biol Evol. 2011 doi: 10.1093/gbe/evr051. http://gbe.oxfordjournals.org/content/3/516. Advance Access published June 13, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Przeworski M. An evolutionary view of human recombination. Nat Rev Genet. 2007;8(1):23–34. doi: 10.1038/nrg1947. [DOI] [PubMed] [Google Scholar]
- Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science. 2008;319:1395–1398. doi: 10.1126/science.1151851. [DOI] [PubMed] [Google Scholar]
- Dreszer T, Wall G, Haussler D, Pollard K. Biased clustered substitutions in the human genome: the footprints of male-driven biased gene conversion. Genome Res. 2007;17:1420–1430. doi: 10.1101/gr.6395807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Arndt P. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008;4:e1000071. doi: 10.1371/journal.pgen.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Eyre-Walker A, Galtier N. A new perspective on isochore evolution. Gene. 2006;385:71–74. doi: 10.1016/j.gene.2006.04.030. [DOI] [PubMed] [Google Scholar]
- Duret L, Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genome Hum G. 2009a;10(1):285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- Duret L, Galtier N. Comment on “human-specific gain of function in a developmental enhancer”. Science. 2009b;323:714. doi: 10.1126/science.1165848. author reply 714. [DOI] [PubMed] [Google Scholar]
- Duret L, Semon M, Piganeau G, Mouchiroud D, Galtier N. Vanishing gc-rich isochores in mammalian genomes. Genetics. 2002;162(4):1837–1847. doi: 10.1093/genetics/162.4.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A. Evidence of selection on silent site base composition in mammals: potential implications for the evolution of isochores and junk DNA. Genetics. 1999;152(2):675–683. doi: 10.1093/genetics/152.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Hurst LD. The evolution of isochores. Nat Rev Genet. 2001;2(7):549–555. doi: 10.1038/35080577. [DOI] [PubMed] [Google Scholar]
- Fledel-Alon A, et al. Broad-scale recombination patterns underlying proper disjunction in humans. PLoS Genet. 2009;5(9):e1000658. doi: 10.1371/journal.pgen.1000658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton SM, Bernardo Carvalho A, Clark AG. Local rates of recombination are positively correlated with GC content in the human genome. Mol Biol Evol. 2001;18(6):1139–1142. doi: 10.1093/oxfordjournals.molbev.a003886. [DOI] [PubMed] [Google Scholar]
- Galtier N, Duret L. Adaptation or biased gene conversion? Extending the null hypothesis of molecular evolution. Trends Genet. 2007;23:273–277. doi: 10.1016/j.tig.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Gay J, Myers S, McVean G. Estimating meiotic gene conversion rates from population genetic data. Genetics. 2007;177(2):881–894. doi: 10.1534/genetics.107.078907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez RD, Williamson SH, Bustamante CD. Context dependence, ancestral misidentification, and spurious signatures of natural selection. Mol Biol Evol. 2007;24(8):1792–1800. doi: 10.1093/molbev/msm108. [DOI] [PubMed] [Google Scholar]
- Hernandez RD, Williamson SH, Zhu L, Bustamante CD. Context-dependent mutation rates may cause spurious signatures of a fixation bias favoring higher GC-content in humans. Mol Biol Evol. 2007;24(10):2196–2202. doi: 10.1093/molbev/msm149. [DOI] [PubMed] [Google Scholar]
- Hill W, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966;8:269–294. [PubMed] [Google Scholar]
- Hillier LW, et al. April. Generation and annotation of the DNA sequences of human chromosomes 2 and 4. Nature. 2005;434(7034):724–731. doi: 10.1038/nature03466. [DOI] [PubMed] [Google Scholar]
- Holloway K, Lawson VE, Jeffreys AJ. Allelic recombination and de novo deletions in sperm in the human β-globin gene region. Hum Mol Genetics. 2006;15(7):1099–1111. doi: 10.1093/hmg/ddl025. [DOI] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consort. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432(7018):695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Kaback D, Guacci V, Barber D, Mahon J. Chromosome size-dependent control of meiotic recombination. Science. 1992;256(5054):228–232. doi: 10.1126/science.1566070. [DOI] [PubMed] [Google Scholar]
- Kaback DB, Barber D, Mahon J, Lamb J, You J. Chromosome size-dependent control of meiotic reciprocal recombination in Saccharomyces cerevisiae: The role of crossover interference. Genetics. 1999;152(4):1475–1486. doi: 10.1093/genetics/152.4.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman S, Kern AD, Pollard KS, Salama SR, Haussler D. GC-biased evolution near human accelerated regions. PLoS Genet. 2010;6:e1000960. doi: 10.1371/journal.pgen.1000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khelifi A, Meunier J, Duret L, Mouchiroud D. GC content evolution of the human and mouse genomes: insights from the study of processed pseudogenes in regions of different recombination rates. J Mol Evol. 2006;62:745–752. doi: 10.1007/s00239-005-0186-0. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- Kong A, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- Kong A, et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467(7319):1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- Kudla G, Lipinski L, Caffin F, Helwak A, Zylicz M. High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol. 2006;4(6):e180. doi: 10.1371/journal.pbio.0040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Hurst LD. Can mutation or fixation biases explain the allele frequency distribution of human single nucleotide polymorphisms (SNPs)? Gene. 2002;300:53–58. doi: 10.1016/s0378-1119(02)01040-5. [DOI] [PubMed] [Google Scholar]
- Lercher MJ, Smith NGC, Eyre-Walker A, Hurst LD. The evolution of isochores: evidence from SNP frequency distributions. Genetics. 2002;162(4):1805–1810. doi: 10.1093/genetics/162.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais G. Jun. Biased gene conversion: implications for genome and sex evolution. Trends Genet. 2003;19:330–338. doi: 10.1016/S0168-9525(03)00116-1. [DOI] [PubMed] [Google Scholar]
- Meunier J, Duret L. Recombination drives the evolution of GC-content in the human genome. Mol Biol Evol. 2004;21:984–990. doi: 10.1093/molbev/msh070. [DOI] [PubMed] [Google Scholar]
- Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- Myers S, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327(5967):876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008;40(9):1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- Nagylaki T. Evolution of a finite population under gene conversion. Proc Natl Acad Sci U S A. 1983;80:6278–6281. doi: 10.1073/pnas.80.20.6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necşulea A, et al. Meiotic recombination favors the spreading of deleterious mutations in human populations. Hum Mutat. 2011;32(2):198–206. doi: 10.1002/humu.21407. [DOI] [PubMed] [Google Scholar]
- Parvanov ED, Petkov PM, Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327(5967):835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paten B, Herrero J, Beal K, Fitzgerald S, Birney E. Enredo and Pecan: genome-wide mammalian consistency-based multiple alignment with paralogs. Genome Res. 2008;18:1814–1828. doi: 10.1101/gr.076554.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paten B, et al. Genome-wide nucleotide-level mammalian ancestor reconstruction. Genome Res. 2008;18:1829–1843. doi: 10.1101/gr.076521.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard K, et al. Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006;2:e168. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2009. [Google Scholar]
- Ratnakumar A, et al. Detecting positive selection within genomes: the problem of biased gene conversion. Philos Trans R Soc B Biol Sci. 2010;365(1552):2571–2580. doi: 10.1098/rstb.2010.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romiguier J, Ranwez V, Douzery EJP, Galtier N. Contrasting GC-content dynamics across 33 mammalian genomes: Relationship with life-history traits and chromosome sizes. Genome Res. 2010;20(8):1001–1009. doi: 10.1101/gr.104372.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer CC, et al. The influence of recombination on human genetic diversity. PLoS Genet. 2006;2:e148. doi: 10.1371/journal.pgen.0020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern J, Shafer B, McGill C. DNA synthesis errors associated with double-strand-break repair. Genetics. 1995;140:965–972. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Hapmap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turney D, de los Santos T, Hollingsworth NM. Does chromosome size affect map distance and genetic interference in budding yeast? Genetics. 2004;168(4):2421–2424. doi: 10.1534/genetics.104.033555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MT, Smith NGC. Fixation biases affecting human SNPs. Trends Genet. 2004;20:122–126. doi: 10.1016/j.tig.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Webster MT, Smith NGC, Ellegren H. Compositional evolution of noncoding DNA in the human and chimpanzee genomes. Mol Biol Evol. 2003;20(2):278–286. doi: 10.1093/molbev/msg037. [DOI] [PubMed] [Google Scholar]
- Webster MT, Smith NGC, Hultin-Rosenberg L, Arndt PF, Ellegren H. Male-driven biased gene conversion governs the evolution of base composition in human alu repeats. Mol Biol Evol. 2005;22(6):1468–1474. doi: 10.1093/molbev/msi136. [DOI] [PubMed] [Google Scholar]