Abstract
Clinical proteomics has emerged as an important new discipline, promising the discovery of biomarkers that will be useful for early diagnosis and prognosis of disease. While clinical proteomic methods vary widely, a common characteristic is the need for (i) extraction of proteins from extremely heterogeneous fluids (i.e. serum, whole blood, etc.) and (ii) extensive biochemical processing prior to analysis. Here, we report a new digital microfluidics (DMF) based method integrating several processing steps used in clinical proteomics. This includes protein extraction, resolubilization, reduction, alkylation and enzymatic digestion. Digital microfluidics is a microscale fluid-handling technique in which nanoliter-microliter sized droplets are manipulated on an open surface. Droplets are positioned on top of an array of electrodes that are coated by a dielectric layer - when an electrical potential is applied to the droplet, charges accumulate on either side of the dielectric. The charges serve as electrostatic handles that can be used to control droplet position, and by biasing a sequence of electrodes in series, droplets can be made to dispense, move, merge, mix, and split on the surface. Therefore, DMF is a natural fit for carrying rapid, sequential, multistep, miniaturized automated biochemical assays. This represents a significant advance over conventional methods (relying on manual pipetting or robots), and has the potential to be a useful new tool in clinical proteomics.
Mais J. Jebrail, Vivienne N. Luk, and Steve C. C. Shih contributed equally to this work.
Sergio L. S. Freire's current address is at the University of the Sciences in Philadelphia located at 600 South 43rd Street, Philadelphia, PA 19104.
Protocol
Part 1: Device Fabrication
Clean glass substrates in Piranha solution (3:1 conc. sulfuric acid:30% hydrogen peroxide). Leave the substrates in Piranha solution for 10 min with frequent agitation.
After rinsing in deionized (DI) water and drying the substrates with N2 gas, place the substrates inside the electron beam chamber for chromium deposition (thickness of 250 nm).
To dehydrate the chromium-coated substrate, rinse in isopropanol and then bake on a hot plate for 5 min at 115°C.
Dry the substrates and prime with hexamethyldisilazane (HMDS) by spin-coating (30 s, 3000 rpm). Spin-coat again (using identical parameters) with Shipley S1811 photoresist.
Pre-bake the substrate on a hot-plate (100°C, 2 min), then pattern the photoresist by exposure to ultraviolet (UV) irradiation for 5 s through a photomask.
Develop the UV-exposed substrates in Shipley MF 321 developer for 3 min and wash in DI water. Post-bake on a hot-plate at 100°C for 1 min.
Etch the exposed chromium by immersing in chromium etchant for 30 s. Rinse in DI water, then immerse in AZ300T stripper for 10 min to remove the remaining photoresist. Rinse in DI water and dry with N2 gas.
Deposit 2-5 μm Parylene-C (an insulating polymer) by chemical vapor deposition onto a substrate bearing patterned chromium. Deposit 50 nm of Teflon-AF (to make the surface hydrophobic) by spin-coating a solution (1% wt/wt in Fluorinert FC-40) at 2000 rpm for 60 s. Post-bake on a hot-plate (160 °C, 10 min).
To form the top plate, coat an un-patterned indium tin oxide (ITO) glass substrates 50 nm Teflon-AF, as above.
Post-bake both substrates on a hot-plate (160 °C, 10 min).
Part 2: Device Set-up and Automation
In digital microfluidic devices operated in "two-plate" configuration,1 droplets are positioned between a bottom substrate (with patterned electrodes) and a top substrate (with one contiguous, transparent electrode, typically formed from ITO).
To set the device up, remove polymer coatings from the contact pads of a bottom substrate by gentle scraping with a scalpel. Couple the exposed pads of the bottom substrate with 40-pin connectors (Figure 1a).
Power-on a computer running LabView (National Instruments, Austin, TX), a home-built control box (featuring an array of high-voltage relays that are controlled by signals from a National Instruments DaqPad), a function generator, and an amplifier. The computer/control box facilitates user control over application of 100 VRMS/18 kHz signals to the device via the 40-pin connectors.
Assemble the device by positioning two pieces of double-sided tape (140 μm total thickness) on the edges of the bottom substrate and complete with unpatterned ITO slides (top plate).
To initialize the control system, run the LabView calibration program (written in-house) to calibrate the feedback control.
Load the sample and reagents into the appropriate reservoirs (see section 3, below) and enclose them under the top substrate (Teflon-coated side facing down). Attach the ground connector to the top substrate.
Load the actuation code in LabView (written in-house) and execute the program to initiate droplet actuation.
Part 3: Sample and Reagent Preparation
Prepare working buffer (WB): 100 mM TrisHCl (pH 7.8) and 0.08% Pluronic F127 w/v.
Dissolve protein(s) to be analyzed in WB and pipette sample into reservoir 1 (R-1) on the device, as shown in Figure 1b.
Prepare precipitant, 20% Trichloroacetic acid (TCA) in DI water, and pipette into R-2.
Prepare wash buffer, 70/30 Chloroform/acetonitrile (ACN) and pipette into R-4.
Prepare resolubilizing buffer, 100 mM Ammonium Bicarbonate (pH 8.0) and 0.08% Pluronic F127 w/v, and pipette into R-5.
Dissolve reductant, tris(2-carboxyethyl)phosphine (TCEP), at 10 mM in WB and pipette into R-6.
Dissolve alkylating agent, iodoacetamide (IAM), at 12 mM in WB and pipette into R-7.
Dissolve trypsin in WB at a concentration equal to1/5 of the concentration of the total protein sample (see 3.1, above) and pipette into R-8.
Part 4: Digital Microfluidic Sample Processing
The following procedure is implemented automatically by means of LabView code written in-house. The volume of droplets dispensed from reservoirs is defined by the dimensions of the electrodes (in this case, dispensed droplets are ~600nL).
Dispense a droplet of protein-containing sample from R-1 and a droplet of precipitant from R-2 (Figure 2, Frame 1). Merge the two droplets and allow the combined droplet to incubate for 5 min, resulting in the precipitation of proteins onto the surface (Figure 2, Frames 2-3). Actuate the supernatant away from the precipitated protein to the waste reservoir, R-3 (Figure 2, Frame 4).
Dispense three droplets of wash buffer from R-4 and drive them across the precipitated protein to the waste reservoir, R-3 (Figure 2, Frame 5).
Allow the precipitate to dry, and dispense a droplet of resolubilizing buffer from R-5 to the protein. Allow the buffer to incubate for 20 min until the precipitate has dissolved (Figure 2, Frame 6).
Dispense a droplet of reductant from R-6 and merge it with the sample droplet. Mix the combined droplet by actuating it across 6 electrodes in a circular pattern. Allow the droplet to incubate for 1 h at room temperature in a humidified chamber (Figure 3, Frames 1-2).
Dispense a droplet of alkylating agent from R-7 and merge it with the sample droplet, followed by mixing (as in 4.5). Allow the droplet to incubate for 15 min at room temperature in a humidified chamber, protected from light (Figure 3, Frames 2-3).
Dispense a droplet of trypsin from R-8, and merge it with the sample droplet, followed by mixing (as in 4.5). Allow the droplet to incubate for 3 h at 37°C in a humidified chamber on a hot plate (Figure 3, Frames 3-4).
Part 5: Post-Processing Sample Preparation
Remove top substrate and quench digestion reaction by adding 0.6 mL of 2.5% trifluoroacetic acid in water.
Purify quenched reaction product using C18 ZipTips (Millipore, Billerica, MA) according to manufacturer’s instructions.
Dilute purified sample in DI water to a final volume of 100 mL.
Part 6: Mass Spectrometry
Evaluate the processed sample on a nano-HPLC mated to a tandem mass spectrometer. The HPLC system used here is from Eksigent Technologies and is equipped with a trap column (3 mm dia. C18 beads, 150 μm ID x 2.5 cm) and a reversed-phase separation column (3 μm dia. C18 beads, 75 μm ID x 15 cm). The mass spectrometer used here is an LTQ from Thermo-Fisher Scientific.
Load a 2 μL sample onto the trap column at 5μL/min in a mobile phase comprising 5% acetonitrile in water. Separate the sample on the reversed phase column at 0.5μL/min using a linear gradient from 20% acetonitrile to 80% acetonitrile over 25 min. Continue running at 80% acetonitrile for another 10 min.
Analyze the bands eluting off the column by tandem mass spectrometry. In the work reported here, the eluent is sampled into the mass spectrometer via a nanoelectrospray emitter (20 μm ID tapered to 10 μm ID) from NewObjective.
Identify the proteins in the sample using a database search program such as SEQUEST. In this work, we used the version of SEQUEST incorporated in Bioworks v3.01 (Thermo-Fisher Scientific), and identified proteins had probability scores of less than 1.0E-03 (equivalent to 99.9% confidence interval), and sequence coverages of at least 30% (Figure 4).
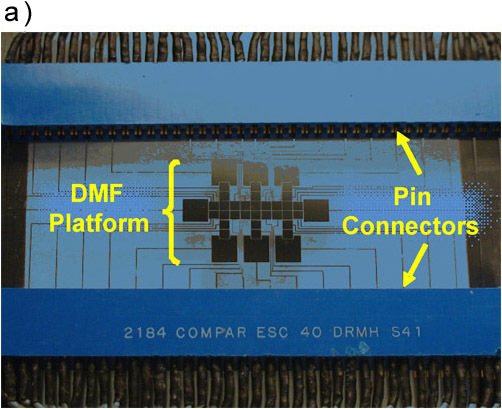
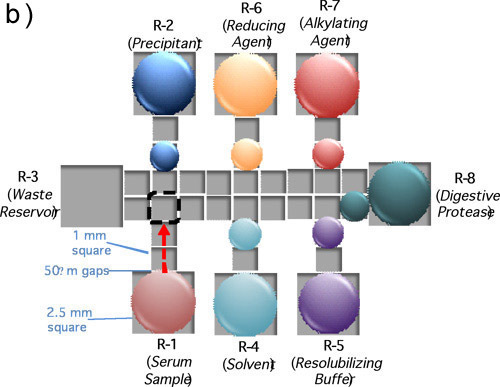 Figure 1. (a) A picture a DMF device mated to 40-pin connectors for automated droplet actuation. (b) A schematic of a device depicting the positioning of sample and reagents required for a proteomic workup.
Figure 1. (a) A picture a DMF device mated to 40-pin connectors for automated droplet actuation. (b) A schematic of a device depicting the positioning of sample and reagents required for a proteomic workup.
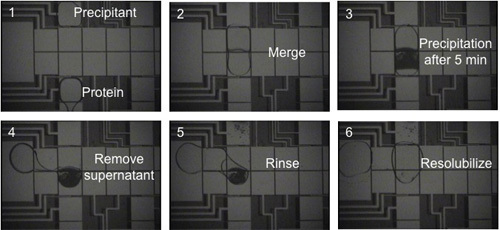 Figure 2. Frames from a movie depicting the automated extraction and purification of BSA in 20% TCA (precipitant) and 70/30% chloroform/acetonitrile (rinse solution). In frame 6, the precipitated protein is redissolved in a droplet of 100 mM ammonium bicarbonate.
Figure 2. Frames from a movie depicting the automated extraction and purification of BSA in 20% TCA (precipitant) and 70/30% chloroform/acetonitrile (rinse solution). In frame 6, the precipitated protein is redissolved in a droplet of 100 mM ammonium bicarbonate.
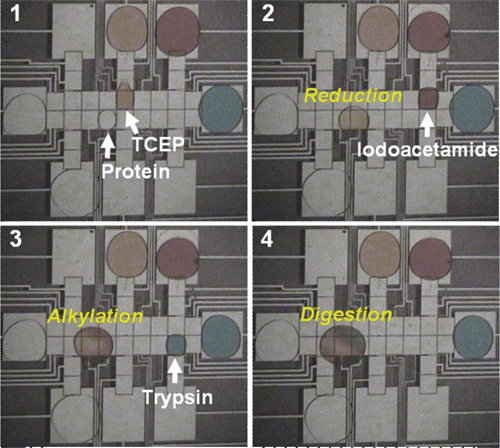 Figure 3. Frames from a movie illustrating sequential reduction, alkylation, and digestion of a droplet of resolubilized protein. In this figure, the reagents are colored with dyes for clarity; in practice, the reagents are not colored.
Figure 3. Frames from a movie illustrating sequential reduction, alkylation, and digestion of a droplet of resolubilized protein. In this figure, the reagents are colored with dyes for clarity; in practice, the reagents are not colored.
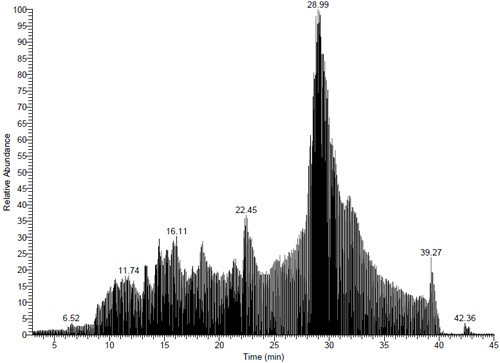 Figure 4. MS chromatogram of a sample of bovine serum albumin processed by digital microfluidics. 25 distinct peptides were identified (99.9% confidence interval) corresponding a sequence coverage of 44%.
Figure 4. MS chromatogram of a sample of bovine serum albumin processed by digital microfluidics. 25 distinct peptides were identified (99.9% confidence interval) corresponding a sequence coverage of 44%.
Discussion
The lack of standardized sample handling and processing in proteomics is a major limitation for the field. In addition, conventional macroscale sample handling involves multiple containers and solution transfers, which can lead to sample loss and contamination. A potential solution to these problems is to form integrated systems for sample processing relying on digital microfluidics1 (DMF). In previous work, DMF was shown to be useful for efficient removal of unwanted contaminants in heterogeneous protein-containing solutions.2 Likewise, DMF was shown to be compatible with integration of multistep solution-phase processing (reduction, alkylation and digestion) on an integrated device.3 Here, we have demonstrated a fully integrated system with automated droplet control for protein extraction by precipitation followed by solution-phase processing. We speculate that if methods such as these are widely adopted, the human error inherent in proteomic sample processing can be largely eliminated, resulting in analyses with better reproducibility. In short, we propose that DMF has the potential for being useful for a broad cross-section of applications, as the conditions can be precisely duplicated in any laboratory in the world.
Acknowledgments
We thank the Natural Sciences and Engineering Research Council (NSERC) and the Canadian Cancer Society for financial support. SCCS thanks NSERC and VNL thanks the Ontario Graduate Scholarship (OGS) program for graduate fellowships. ARW thanks the CRC for a Canada Research Chair.
References
- Abdelgawad M, Wheeler AR. The Digital Revolution: A New Paradigm for Microfluidics. Adv. Mat. 2009;21:920–925. [Google Scholar]
- Jebrail M, Wheeler AR. A Digital Microfluidic Method for Protein Extraction by Precipitation. Anal. Chem. 2009;81:330–335. doi: 10.1021/ac8021554. [DOI] [PubMed] [Google Scholar]
- Luk VN, Wheeler AR. A Digital Microfluidic Approach to Proteomic Sample Processing. Anal. Chem. 2009;81:4524–4530. doi: 10.1021/ac900522a. [DOI] [PubMed] [Google Scholar]


