ABSTRACT
Mechanisms for electron transfer within microbial aggregates derived from an upflow anaerobic sludge blanket reactor converting brewery waste to methane were investigated in order to better understand the function of methanogenic consortia. The aggregates were electrically conductive, with conductivities 3-fold higher than the conductivities previously reported for dual-species aggregates of Geobacter species in which the two species appeared to exchange electrons via interspecies electron transfer. The temperature dependence response of the aggregate conductance was characteristic of the organic metallic-like conductance previously described for the conductive pili of Geobacter sulfurreducens and was inconsistent with electron conduction through minerals. Studies in which aggregates were incubated with high concentrations of potential electron donors demonstrated that the aggregates had no significant capacity for conversion of hydrogen to methane. The aggregates converted formate to methane but at rates too low to account for the rates at which that the aggregates syntrophically metabolized ethanol, an important component of the reactor influent. Geobacter species comprised 25% of 16S rRNA gene sequences recovered from the aggregates, suggesting that Geobacter species may have contributed to some but probably not all of the aggregate conductivity. Microorganisms most closely related to the acetate-utilizing Methanosaeta concilii accounted for more than 90% of the sequences that could be assigned to methane producers, consistent with the poor capacity for hydrogen and formate utilization. These results demonstrate for the first time that methanogenic wastewater aggregates can be electrically conductive and suggest that direct interspecies electron transfer could be an important mechanism for electron exchange in some methanogenic systems.
IMPORTANCE
The conversion of waste organic matter to methane is an important bioenergy strategy, and a similar microbial metabolism of complex organic matter in anaerobic soils and sediments plays an important role in the global carbon cycle. Studies with laboratory cultures have demonstrated that hydrogen or formate can serve as an electron shuttle between the microorganisms degrading organic compounds and methanogens. However, the importance of hydrogen and formate as intermediates in the conversion of organic matter to methane in natural communities is less clear. The possibility that microorganisms within some natural methanogenic aggregates may directly exchange electrons, rather than producing hydrogen or formate as an intermediary electron carrier, is a significant paradigm shift with implications for the modeling and design of anaerobic wastewater reactors and for understanding how methanogenic communities will respond to environmental perturbations.
Introduction
Effective interspecies electron exchange is essential in methanogenic ecosystems (1–3). In order for complex organic matter to be converted to methane and carbon dioxide, the microbial community that metabolizes multicarbon organic compounds other than acetate requires an electron sink. Methanogenic microorganisms can function as this sink, consuming electrons in the reduction of carbon dioxide to methane. Understanding the mechanisms of this electron exchange is key to modeling and/or manipulating methane production in natural methanogenic environments, such as anaerobic soils and sediments, the gastrointestinal systems of diverse animals, and anaerobic wastewater treatment systems.
The first mechanism described for electron exchange in methanogenic systems was interspecies hydrogen transfer, in which microorganisms that require an electron sink reduce protons to produce hydrogen and the methanogens utilize hydrogen as an electron donor (1, 2, 4, 5). Formate may also act as an electron carrier (1, 5–7).
A potential alternative is direct electron transfer between cells. Connections between cells via electrically conductive filaments were suggested as a mechanism for microbes to exchange electrons (8–10). However, only one specific example of direct electron transfer to a methanogen was proposed (9), and subsequent studies demonstrated that the filament connecting the fermentative microorganism and the methanogen was a flagellum, which is not expected to be conductive (2, 11). Therefore, direct electron transfer to the methanogen was unlikely (2).
However, studies in which two Geobacter species were cocultured under conditions that required interspecies electron exchange indicated that direct electron transfer is feasible (12). Adaptation of Geobacter metallireducens and Geobacter sulfurreducens to cooperatively metabolize ethanol resulted in the formation of large (1 to 2 mm in diameter) aggregates. Analysis of a mutation selected over the course of the laboratory evolution of the coculture, as well as studies in which specific mutations were introduced into G. sulfurreducens, suggested that the cells within the aggregates were directly exchanging electrons via a mechanism that involved the multiheme c-type cytochrome OmcS (12). Previous studies (13) demonstrated that OmcS aligns along the conductive pili of G. sulfurreducens, and a similar localization was observed in the aggregates, suggesting that pili were also involved in the electron exchange. Furthermore, the aggregates were electrically conductive, demonstrating that electrons that G. metallireducens released into the extracellular matrix might readily flow to G. sulfurreducens (12). Studies of the capacity for hydrogen and formate metabolism suggested that they were not important electron carriers between the two Geobacter species.
The aggregates that the Geobacter species formed had a morphology similar to that of the aggregates that are found in upflow methanogenic wastewater digesters (14). Methanogenic digester aggregates are comprised of a diverse community of hydrolytic-fermentative bacteria, hydrogen-producing acetogenic bacteria, and methanogens, which cooperate to degrade complex organic compounds to methane and carbon dioxide (1, 2, 15, 16). One factor favoring aggregation is that it prevents cells from being washed out of the system (1, 14, 17–19). Furthermore, aggregation could enhance the exchange of hydrogen and formate between syntrophic partners (20, 21).
An additional potential benefit of aggregation is that it may make it feasible for cells to directly exchange electrons (12, 22). Here we report on conductive properties, community composition, and metabolism of wastewater aggregates consistent with direct exchange of electrons within the aggregates.
RESULTS AND DISCUSSION
General characteristics of the aggregates.
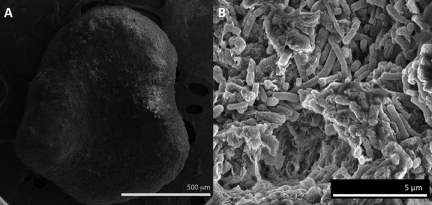
The aggregates obtained from the industrial-scale reactor and the aggregates propagated in the laboratory reactor had similar appearances. They were black and roughly spherical (diameter, 0.5 to 2 mm), comprised primarily of tightly packed rods (Fig. 1).
FIG 1 .
Scanning electron micrographs of an entire aggregate (A) and higher magnification of the aggregate surface (B).
Aggregate conductance.
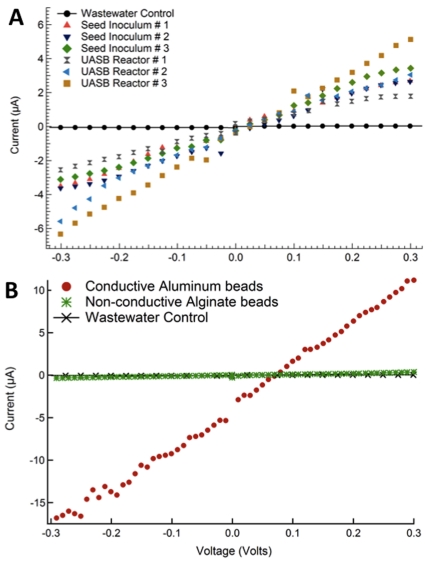
The possibility that the aggregates might be electrically conductive, in a manner similar to that described for the aggregates formed by a coculture of Geobacter species (12), was investigated. Aggregates spanning a nonconductive gap between two gold electrodes exhibited a linear current-voltage response when a DC voltage was applied, consistent with ohmic conductivity (Fig. 2A). The conductivity of the aggregates from the industrial-scale reactor (6.1 ± 0.3 µS/cm [mean ± standard deviation]; n = 3) and that of the aggregates propagated in the laboratory reactor (7.2 ± 3.0 µS/cm) were similar. They were more conductive than the previously described conductive aggregates comprised of a coculture of two Geobacter species (1.4 ± 0.3 µS/cm). Conductive aluminum beads treated in a similar manner had a high conductivity (11.4 ± 0.1 µS/cm), whereas artificial wastewater alone (Fig. 2A) or porous alginate beads in wastewater (Fig. 2B) did not exhibit an ohmic response.
FIG 2 .
Current-voltage response of reactor aggregates (A) and aluminum and alginate control beads serving as positive and negative controls, respectively (B), in artificial wastewater.
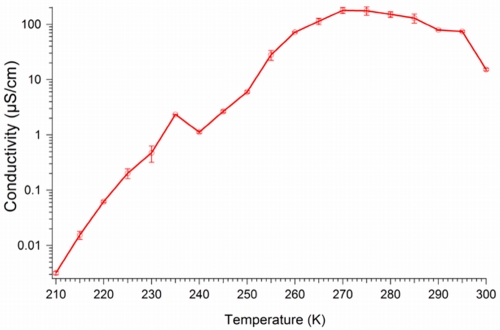
In order to further evaluate the nature of the conductance, aggregate conductance was measured as a function of temperature (Fig. 3). Upon cooling from room temperature, aggregate conductivity increased exponentially by more than an order of magnitude. An exponential increase in conductivity upon cooling is a characteristic signature of organic metals, such as polyaniline, whereas inorganic metals and minerals exhibit only linear increases in conductivity (23). Therefore, even though the aggregates contain inorganic iron that could conceivably contribute to their conductivity, the temperature dependence response rules this out as a major source of conductivity and suggests an organic metal and hence a biological response. This temperature response of the aggregates is similar to that of conductive biofilms and purified pilus preparations of Geobacter species (24). The continued exponential decrease in conductivity with further cooling (Fig. 3) can be attributed to intrinsic disorder present in the aggregate samples (23, 24).
FIG 3 .
Temperature dependence of aggregate conductivity measured using a four-probe setup.
Experimental evaluation of electron sources for methane production.
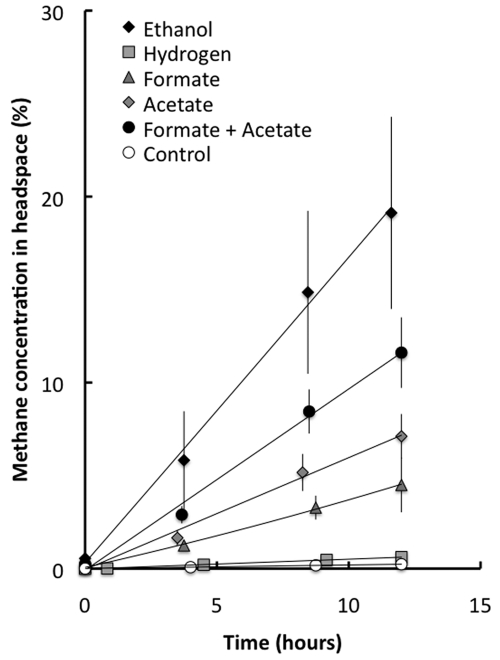
To further evaluate possible mechanisms for electron exchange within the aggregates, the potential for methane production from ethanol and potential intermediates of ethanol metabolism were evaluated (Fig. 4). High concentrations of potential substrates were added in order to estimate the maximum rates at which the compounds might be metabolized. Methane production rates from ethanol exceeded those from other substrates (Fig. 4). There was substantial production of methane from acetate, consistent with the necessity of the community to metabolize the acetate expected to be produced from the ethanol and propionate in the reactor feed and the fact that acetate was directly provided in the reactor feed. The addition of hydrogen did not stimulate methane production, indicating that the aggregates were not adapted to convert hydrogen to methane.
FIG 4 .
Methane production from the aggregates with different potential electron donors. Controls received no electron donor additions. Results are the means and standard deviations of data for six replicates for each treatment.
The aggregates had some potential to convert formate to methane. However, the combined maximum capacity for the aggregates to convert acetate and formate to methane was less than the capacity for conversion of ethanol to methane (Fig. 4). This was the case even though the concentrations of acetate and formate provided to the aggregates were much higher than those the methanogens were likely to experience during syntrophic metabolism of the added ethanol. Thus, if formate was the dominant electron shuttle for ethanol metabolism, combined rates of acetate and formate conversion to methane would be expected to be higher than the rates of methane production from added ethanol. The finding that the combined rates of methane production from added formate and acetate were, in fact, lower that from ethanol suggested that a mechanism other than interspecies hydrogen and formate transfer was required to account for the rapid metabolism of ethanol observed in the aggregates.
Microbial community composition.
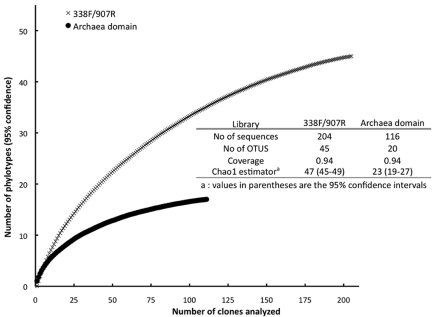
The 16S rRNA gene sequences in the aggregates were evaluated in order to gain further insight into aggregate metabolism. Rarefaction curves for the 16S rRNA gene libraries constructed with universal primers or primers specific to the archaea reached an asymptote, suggesting adequate coverage for assessing the dominant members of the microbial community (Fig. 5). The microbial community of the aggregates propagated in the laboratory reactor (Table 1) was similar to that of the aggregates directly harvested from the industrial-scale reactor (data not shown). The microbial community was phylogenetically diverse, as expected from previous studies (25–27). Bacteria accounted for 79% of the 16S rRNA gene sequences recovered and included members of the Proteobacteria (33.9%), the Chloroflexi (15.8%), the Firmicutes (6.4%), and the Synergistetes (4.4%) (Table 1).
FIG 5 .
Rarefaction curves demonstrating high diversity coverage values for the 16S rRNA gene sequences recovered with the two primer sets employed.
TABLE 1 .
Sequence abundances in 338F/907R and archaeal clone libraries with closest representative in GenBanka
| Library | Phylum | Closest relativeb | No. of clones | Relative abundance (%) |
|---|---|---|---|---|
| 338F/907R primers | Actinobacteria | Bacterium Ellin 6510 (92) | 2 | 1.0 |
| Bacteroidetes | Prolixibacter bellariivorans (89) | 9 | 4.4 | |
| Chloroflexi | Longilinea arvoryzae (90%) | 16 | 7.8 | |
| Anaerobic bacterium MO-CFX2 (89) | 2 | 1.0 | ||
| Bacterium JN18_A7_F* (96) | 13 | 6.4 | ||
| Chloroflexi bacterium BL-DC-9 (86) | 2 | 1.0 | ||
| Deltaproteobacteria | Geobacter daltoniic (97) | 50 | 24.5 | |
| Syntrophobacter sulfatireducens (95) | 11 | 5.4 | ||
| Syntrophorhabdus aromaticivorans (94) | 3 | 1.5 | ||
| Syntrophobacter fumaroxidans MPOB (92) | 3 | 1.5 | ||
| Syntrophus aciditrophicus (95) | 2 | 1.0 | ||
| Euryarchaeota | Methanobacterium petrolearium (97) | 3 | 1.5 | |
| Methanosaeta concilii (96) | 40 | 19.6 | ||
| Firmicutes | Clostridiales sp. SM4/1 (94) | 2 | 1.0 | |
| Oryza sativa Indica (97) | 1 | 0.5 | ||
| Syntrophomonas sp. TB-6 (97) | 10 | 4.9 | ||
| Spirochaetes | Spirochaeta sp. isolate TM-3 (93) | 5 | 2.5 | |
| Spirochaetes bacterium SA-10 (93) | 3 | 1.5 | ||
| Spirochaeta sp. isolate TM-3 (93) | 1 | 0.5 | ||
| Synergistetes | Synergistetes bacterium 7WAY-8-7 (98) | 1 | 0.5 | |
| Aminomonas paucivorans (89) | 8 | 3.9 | ||
| Thermotogae | Thermotogales bacterium MesG1Ag4.2.16S.B (98) | 1 | 0.5 | |
| Unclassified bacteria | 16 | 7.8 | ||
| Arch21F/Arch958R primers | Euryarchaeota | Methanosaeta concilii (99) | 102 | 87.8 |
| Methanobacterium petrolearium (99) | 7 | 6.1 | ||
| Crenarchaeota | Unclassified Desulfurococcales | 7 | 6.1 |
The aggregates from the UASB reactor were used for analyses. Representative sequences comprising more than 10% of the sequences of their respective library are highlighted in grey.
Percentages of nucleotide similarity are shown in parentheses.
Formerly strain Geobacter FRC-32.
The most abundant sequence recovered from the aggregates belonged to Geobacter species most closely related to Geobacter daltonii (Table 1). The finding that Geobacter sequences accounted for 25% of the sequences recovered is consistent with the finding that, in the absence of reactor disturbance, 20 to 30% of the 16S rRNA gene sequences recovered from aggregates treating brewery wastewater were in the family Desulfuromonadales, which includes Geobacter species (28). Although the most intensively studied physiological characteristic of Geobacter species is their capacity for extracellular electron transfer to metals and electrodes (22, 29, 30), they are also capable of syntrophic growth (12, 31–33). Syntrophic oxidation of ethanol and organic acids is their most likely role in brewery digester aggregates.
Also abundant were sequences most closely related to Methanosaeta concilii (Table 1). The importance of this organism was further confirmed with the archaeal domain-specific primer set (Table 1). This is consistent with the known role of Methanosaeta species in initiating aggregate formation in digesters and their usual role as the primary methanogens converting acetate to methane in these systems (34, 35).
Sequences that could be assigned to methanogens capable of utilizing hydrogen or formate were much less abundant than those of the acetate utilizers (Table 1), consistent with the low potential of the aggregates to convert these substrates to methane. The only 16S rRNA gene sequences recovered that were closely related to hydrogen/formate-utilizing methanogens were those related to Methanobacterium petrolearium, a hydrogen-utilizing methanogen (36). These sequences accounted for only 1.5% of the sequences recovered with the primers designed to recover sequences from all microorganisms. The studies with both primer sets indicated that Methanobacterium sequences were less than 10% as abundant as Methanosaeta sequences. These results further suggest that that neither hydrogen nor formate was an important electron shuttle for methane production.
Implications.
These results suggest that direct electron transfer could be an important mechanism for electron exchange within methanogenic wastewater treatment aggregates. The aggregates were even more conductive than the previously described aggregates of G. metallireducens and G. sulfurreducens, which multiple lines of evidence suggested exchanged electrons via direct cell-to-cell electron transfer (12). Geobacter species were present in high abundance in the aggregates and could be expected to facilitate long-range electron transfer via their conductive pili (8, 10, 37). However, the higher conductivity of the methanogenic aggregates suggests that aggregate constituents other than Geobacter species could have been contributing to aggregate conductivity.
Methane production via direct interspecies electron transfer, rather than via interspecies transfer of hydrogen or formate, would require that methanogens have the capacity to directly accept electrons as an electron donor for methane production. The apparent ability of a Methanobacterium-like isolate (38) and a strain of Methanococcus maripaludis (39) to accept electrons directly from metallic iron suggests that this is possible. Therefore, it seems likely that the Methanobacterium species in the methanogenic aggregates could have the capacity for directly accepting electrons from the conductive aggregate matrix. It has also been proposed that the archaea within anaerobic methane-oxidizing aggregates, which are related to Methanosaeta (40, 41), are also capable direct accepting electrons (1, 42) because physiological and modeling studies have suggested that interspecies hydrogen and formate transfer is unlikely within those aggregates (5, 43–47). Although Methanosaeta has been considered to comprise exclusively acetate-utilizing methanogens based on physiological studies, the genome sequences of Methanosaeta species contain genes encoding the enzymes required for the carbon dioxide reduction pathway (41). Therefore, it cannot be ruled out that the highly abundant Methanosaeta species might also have the ability to directly accept electrons within aggregates.
The formation of large, visible aggregates may be only the most extreme instance of cells forming electrical contacts. The possibility of cell-to-cell electron transfer in smaller methanogenic associations and in aggregates carrying out other types of synthrophic processes warrants further investigation.
MATERIALS AND METHODS
Operation of UASB reactor.
Microbial aggregates were obtained from an upflow anaerobic sludge blanket (USAB) treating brewery waste at an Anheuser-Busch facility in Baldwinsville, NY. The industrial UASB reactor had an average chemical oxygen demand of 5.5 g liter−1 and an average hydraulic retention time of 12 h. Aggregates were transported under anaerobic conditions at room temperature before propagation in the laboratory. Aggregates for DNA extraction were stored at −80°C prior to processing.
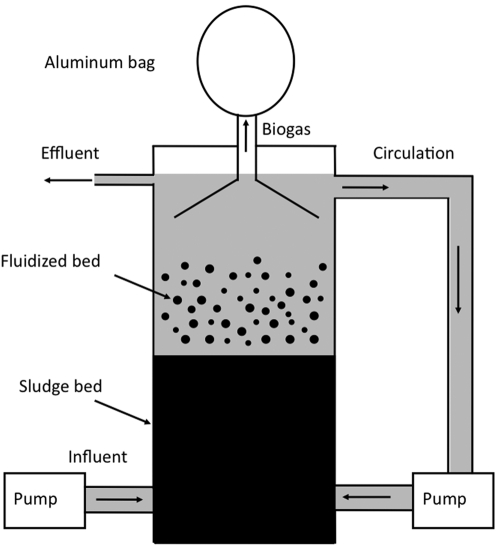
The aggregates were propagated in the laboratory in a mesophilic (37°C) laboratory-scale (0.9 liters) UASB reactor (Fig. 6). The reactor was fed an artificial brewery wastewater, modified from the previously described synthetic brewery wastewater (48). No attempt was made to remove dissolved oxygen from the artificial wastewater prior to its introduction into the reactor, which simulates industrial conditions. The artificial wastewater composition (per liter) was as follows: ethanol, 2.5 ml; sodium propionate, 0.72 g; sodium acetate trihydrate, 1.8 g; urea, 0.23 g; KH2PO4, 0.11 g; K2HPO4, 0.17 g; Na2SO4, 0.05 g; MgCl2 ⋅ 6H2O, 0.1 g; CaCl2 ⋅ 2H2O, 0.05 g; trace element solution, 10 ml; vitamin solution, 10 ml. The composition of the trace element solution (in grams per liter) was as follows: MnSO4 ⋅ H2O, 0.5; FeSO4 ⋅ 7H2O, 0.1; NiCl2 ⋅ 6H2O, 0.04; CoCl2 ⋅ 6H2O, 0.048; ZnCl2, 0.13; CuSO4 ⋅ 5H2O, 0.01; AlK(SO4)2 ⋅ 12H2O, 0.01; H3BO3, 0.01; Na2MoO4 ⋅ 2H2O, 0.025. The composition of the vitamin solution (in grams per liter) was as follows: biotin, 0.002; pantothenic acid, 0.005; B-12, 0.0001; p-aminobenzoic acid, 0.005; thioctic acid (alpha lipoic), 0.005; nicotinic acid, 0.005; thiamine, 0.005; riboflavin, 0.005; pyridoxine HCl, 0.01; folic acid, 0.002. The main carbon sources of the artificial wastewater (ethanol, propionate, and acetate) created an approximate chemical oxygen demand of 5.8 g liter−1. The hydraulic retention time was 1 day.
FIG 6 .
Diagram of the laboratory-scale UASB reactor.
The reactor functioned under steady-state conditions throughout the course of the study. Volatile fatty acids were maintained at low levels (0.18 ± 0.13 mM [mean ± standard deviation]), and chemical oxygen demand removal was 96.7% ± 1.1%. Biogas production rates were 2,074 ± 341 ml liter−1 day−1, with a methane composition of 78.9% ± 4.3%.
Scanning electron microscopy.
Scanning electron micrographs were taken of whole aggregates fixed with 2.5% glutaraldehyde for 4 h at 4°C, dehydrated and dried with hexamethyldisilazane (Sigma Aldrich) as previously described (49), and micrographs were taken as previously described (12).
Aggregate conductance.
Two-probe electrical conductance measurements were performed with two gold electrodes separated by a 50-µm nonconductive gap, as previously described (12). Aluminum and alginate beads and wastewater aggregates were suspended in artificial brewery wastewater before being placed on the gold electrodes, such that materials were spanning the nonconductive gap. Voltage was applied across the gap using a Keithley 2400 source meter. Voltage was scanned from −0.3 V to +0.3 V in steps of 0.025 V and then from +0.3 V to −0.3 V. For each measurement, current was measured 10 s after setting the voltage to allow the exponential decay of the transient ionic current in the gap and to measure steady-state electronic current (50). The LabVIEW data acquisition system (National Instruments, Texas) was used to collect the data. Igor Pro (WaveMetrics Inc., Oregon) was used for data fitting and analysis.
Four-probe electrical conductance measurements were performed as previously described for studies on microbial biofilms and pili (24). Four gold electrodes were separated by 50-µm nonconductive gaps. Electrodes were fabricated using standard photolithography processing. The total electrode size was 1.25 cm by 1 cm. Aggregates were gently crushed to form a homogeneous layer and were placed on the four electrodes. The gentle crushing allowed aggregates to form a confluent film on all four electrodes and kept them from moving apart after drying. A source meter (Keithley 2400) was used to apply a fixed current between the outer of the four electrodes and to measure the potential drop between the two inner electrodes (51), by measuring the voltage for each current every second over a period of 100 s, after reaching the steady state. An additional high-impedance voltmeter (Keithley 2000) was used to record the output voltage of the current source to calculate conductance (Lange and Mirsky [51]).
The temperature dependence of conductivity was measured as previously described (24) with a physical property measurement system (PPMS-6000; Quantum Design, California). The temperature was varied from room temperature (300 K) down to 210 K in steps of 5 K, and four-probe conductance was measured at each temperature. All temperature dependence experiments were performed in vacuum.
Metabolism of ethanol and potential intermediates.
In order to evaluate the relative potential of the aggregates to metabolize ethanol and potential intermediates in ethanol metabolism to methane, 1-ml aliquots of aggregates taken from the lower portion of the laboratory reactor at steady state were added to anaerobic pressure tubes under N2-CO2 (80:20) containing 10 ml of artificial brewery wastewater with the ethanol, propionate, and acetate omitted. NH4Cl (29 mM) was used in place of urea as a nitrogen source. The pH of the medium was adjusted to 7 after autoclaving with 1 M KOH. Ethanol (20 mM), acetate (20 mM), formate (20 mM), or hydrogen (0.7 atm as part of an H2-CO2 [80:20] mix) were added to six replicate tubes for each treatment under strict anaerobic conditions. Methane production at 37°C was monitored by gas chromatography as described below.
Community composition.
Aggregates were frozen at −80°C and ground to a fine powder under liquid nitrogen with a mortar and pestle before processing with Bio 101 FastDNA soil kits (MP Biomedicals, Ohio) according to the manufacturer’s instructions. DNA was quantified with a NanoDrop spectrophotometer (Thermo, Fisher Scientific, Delaware).
Portions of 16S rRNA gene sequences were amplified with the universal primer set 338F (5′ ACTCCTACGGGAGGCAGC 3′) (52) and 907R (5′ CCGTCAATTCCTTTRAGTTT 3′) (53, 54) or the archaeal domain-specific primer set Arch21F (5′ TTCCGGTTGATCCYGCCGGA 3′) and Arch958R (5′ TTCCGGTTGATCCYGCCGGA 3′) (55). DNA was amplified with a minicycler PTC 200 (MJ Research) starting at 95°C for 5 min, followed by 35 cycles consisting of denaturation (45 s at 95°C), annealing (45 s at 50°C for 338F/907R and 55°C for Arch21F/958R), and extension (1 min at 72°C) and a final extension at 72°C for 10 min. PCR products were purified with a gel extraction kit (Qiagen), and clone libraries were constructed with a Topo p2.1 cloning kit (Invitrogen, California) according to the manufacturer’s instructions. Plasmid inserts from each clone library were sequenced with the M13F and M13R primer pair at the University of Massachusetts Sequencing Facility.
The 16S rRNA sequences were trimmed and assembled with the software package Geneious, version 5.3 pro, and were compared to sequences deposited in the GenBank DNA database using the BLAST algorithm (56). 16S rRNA gene sequences were assigned to phylogenetically consistent (>97% similarity) higher-order bacterial taxonomy groups based on a naive Bayesian rRNA classifier using the RDP Classifier software program (57). Alignments were achieved with the ClustalX v. 1.83 program (58) and manually proofread and corrected when necessary with ProSeq v. 2.9 software (59) before performing rarefaction analyses. A distance matrix based on the alignment was constructed using DNAdist from the PHYLIP software program, version 3.6, with default parameters (60, 61). These pairwise distances served as input to the DOTUR software program (distance-based operational taxonomic unit [OTU] and richness), version 1.53 (62), for clustering the sequences into OTUs of 97% defined sequence similarity. These clusters served to generate rarefaction curves and to make calculations of Chao1 (63) estimator and diversity coverage. To estimate the diversity coverage and determine if the number of clones from each library was sufficient, homologous coverage of each DNA library was calculated using the equation CX = 1 − (NX/n), where NX is the number of unique sequences (singletons) and n is the total number of sequences (64).
Analytical methods.
Chemical oxygen demand was determined using Hach’s method 8000 (Hach DR/890 colorimeter procedures manual). In brief, samples were digested with potassium dichromate for 2 h at 150°C to form green chromic ion (Cr3+), which was quantified with a calibrated preprogrammed colorimeter (Hach DR-890). Volatile fatty acids were measured with a high-performance liquid chromatograph, as previously described (65). The ethanol concentration was measured with a gas chromatograph equipped with a headspace sampler and a flame ionization detector (Clarus 600; PerkinElmer Inc., California). Biogas volume was measured by the water displacement method (66). The methane content was determined using gas chromatography with a flame ionization detector (67).
Nucleotide sequence accession numbers.
The sequences determined in this work have been submitted to the EMBL database and assigned accession no. FR823513 to FR823761 for the 16S rRNA bacterial library and FR823762 to FR823877 for the 16S rRNA archaeal library.
ACKNOWLEDGMENTS
We thank T. Cummings, Anheuser-Busch, for providing seed aggregates and M. Tuominen, K. Rachtan, and S. Choi for technical assistance.
This research was supported by the Office of Science (BER), U.S. Department of Energy, award no. DE-SC000448, and the NSF Center for Hierarchical Manufacturing.
Footnotes
Citation Morita M, et al. 2011. Potential for direct interspecies electron transfer in methanogenic wastewater digester aggregates. mBio 2(4):e00159-11. doi:10.1128/mBio.00159-11.
REFERENCES
- 1. Stams AJM, Plugge CM. 2009. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat. Rev. Microbiol. 7:568–577 [DOI] [PubMed] [Google Scholar]
- 2. McInerney MJ, Sieber JR, Gunsalus RP. 2009. Syntrophy in anaerobic global carbon cycles. Curr. Opin. Biotechnol. 20:623–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McInerney MJ, et al. 2008. Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolism. Ann. N. Y. Acad. Sci. 1125:58–72 [DOI] [PubMed] [Google Scholar]
- 4. Bryant M, Wolin E, Wolin M, Wolfe R. 1967. Methanobacillus omelianskii, a symbiotic association of two species of bacteria. Arch. Microbiol. 59:20–31 [DOI] [PubMed] [Google Scholar]
- 5. Stams AJM, et al. 2006. Exocellular electron transfer in anaerobic microbial communities. Environ. Microbiol. 8:371–382 [DOI] [PubMed] [Google Scholar]
- 6. Boone DR, Johnson RL, Liu Y. 1989. Diffusion of the interspecies electron carriers H2 and formate in methanogenic ecosystems and its implications in the measurement of Km for H2 or formate uptake. Appl. Environ. Microbiol. 55:1735–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thiele JH, Zeikus JG. 1988. Control of interspecies electron flow during anaerobic digestion: significance of formate transfer versus hydrogen transfer during syntrophic methanogenesis in flocs. Appl. Environ. Microbiol. 54:20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reguera G, et al. 2005. Extracellular electron transfer via microbial nanowires. Nature 435:1098–1101 [DOI] [PubMed] [Google Scholar]
- 9. Gorby YA, et al. 2006. Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc. Natl. Acad. Sci. U. S. A. 103:11358–11363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reguera G, et al. 2006. Biofilm and nanowire production leads to increased current in Geobacter sulfurreducens fuel cells. Appl. Environ. Microbiol. 72:7345–7348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shimoyama T, Kato S, Ishii S, Watanabe K. 2009. Flagellum mediates symbiosis. Science 323:1574 [DOI] [PubMed] [Google Scholar]
- 12. Summers ZM, et al. 2010. Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science 330:1413–1415 [DOI] [PubMed] [Google Scholar]
- 13. Leang C, Qian X, Mester T, Lovley DR. 2010. Alignment of the c-type cytochrome OmcS along pili of Geobacter sulfurreducens. Appl. Environ. Microbiol. 76:4080–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schmidt JE, Ahring BK. 1996. Granular sludge formation in upflow anaerobic sludge blanket (UASB) reactors. Biotechnol. Bioeng. 49:229–246 [DOI] [PubMed] [Google Scholar]
- 15. Sasaki K, et al. 2011. Bioelectrochemical system accelerates microbial growth and degradation of filter paper. Appl. Microbiol. Biotechnol. 89:449–455 [DOI] [PubMed] [Google Scholar]
- 16. Weiland P. 2010. Biogas production: current state and perspectives. Appl. Environ. Microbiol. 85:849–860 [DOI] [PubMed] [Google Scholar]
- 17. Liu Y, Xu HL, Yang SF, Tay JH. 2003. Mechanisms and models for anaerobic granulation in upflow anaerobic sludge blanket reactor. Water Res. 37:661–673 [DOI] [PubMed] [Google Scholar]
- 18. Grotenhuis JT, et al. 1991. Bacteriological composition and structure of granular sludge adapted to different substrates. Appl. Environ. Microbiol. 57:1942–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lettinga G, Van Velsen A, Hobma S, De Zeeuw W, Klapwijk A. 1980. Use of the upflow sludge blanket (USB) reactor concept for biological wastewater treatment, especially for anaerobic treatment. Biotechnol. Bioeng. 22:699–734 [Google Scholar]
- 20. Schmidt JE, Ahring BK. 1995. Interspecies electron transfer during propionate and butyrate degradation in mesophilic, granular sludge. Appl. Environ. Microbiol. 61:2765–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ozturk SS, Palsson BO, Thiele JH. 1989. Control of interspecies electron transfer flow during anaerobic digestion: dynamic diffusion reaction models for hydrogen gas transfer in microbial flocs. Biotechnol. Bioeng. 33:745–757 [DOI] [PubMed] [Google Scholar]
- 22. Lovley DR. 2011. Powering microbes with electricity: direct electron transfer from electrodes to microbes. Environ. Microb. Rep. 3:27–35 [DOI] [PubMed] [Google Scholar]
- 23. Heeger AJ, Sariciftci NS, Namdas EB. Semiconducting and metallic polymers. Oxford University Press, New York, NY. 2010 [Google Scholar]
- 24. Malvankar NS, et al. 2011. Tunable metallic-like conductivity in microbial nanowire networks. Nat. Nanotechnol. doi: 10.1038/nnano.2011.119 [DOI] [PubMed] [Google Scholar]
- 25. Narihiro T, et al. 2008. Comparative analysis of bacterial and archaeal communities in methanogenic sludge granules from upflow anaerobic sludge blanket reactors treating various food-processing, high-strength organic wastewaters. Microbes Environ. 24:88–96 [DOI] [PubMed] [Google Scholar]
- 26. Díaz EE, Stams AJM, Amils R, Sanz JL. 2006. Phenotypic properties and microbial diversity of methanogenic granules from a full-scale upflow anaerobic sludge bed reactor treating brewery wastewater. Appl. Environ. Microbiol. 72:4942–4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu WT, Chan OC, Fang HH. 2002. Characterization of microbial community in granular sludge treating brewery wastewater. Water Res. 36:1767–1775 [DOI] [PubMed] [Google Scholar]
- 28. Werner JJ, et al. 2011. Bacterial community structures are unique and resilient in full-scale bioenergy systems. Proc. Natl. Acad. Sci. U. S. A. 108:4158–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lovley DR. 2006. Bug juice: harvesting electricity with microorganisms. Nat. Rev. Microbiol. 4:497–508 [DOI] [PubMed] [Google Scholar]
- 30. Lovley DR, Holmes DE, Nevin KP. 2004. Dissimilatory Fe(III) and Mn(IV) reduction. Adv. Microb. Physiol. 49:219–286 [DOI] [PubMed] [Google Scholar]
- 31. Kaden J, Galushko AS, Schink B. 2002. Cysteine-mediated electron transfer in syntrophic acetate oxidation by cocultures of Geobacter sulfurreducens and Wolinella succinogenes. Arch. Microbiol. 178:53–58 [DOI] [PubMed] [Google Scholar]
- 32. Cord-Ruwisch R, Lovley DR, Schink B. 1998. Growth of Geobacter sulfurreducens with acetate in syntrophic cooperation with hydrogen-oxidizing anaerobic partners. Appl. Environ. Microbiol. 64:2232–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Meckenstock RU. 1999. Fermentative toluene degradation in anaerobic defined syntrophic cocultures. FEMS Microbiol. Lett. 177:67–73 [DOI] [PubMed] [Google Scholar]
- 34. Wiegant WM. 1987. The “spaghetti theory” on anaerobic sludge formation, or the inevitability of granulation . In Lettinga G, Zehnder AJB, Grotenhuis JTC, Hulshoff Pol LW, Granular anaerobic sludge: microbiology and technology. PUDOC, Wageningen, The Netherlands [Google Scholar]
- 35. Huser BA, Wuhrmann K, Zehnder AJB. 1982. Methanothrix soehngenii gen. nov. sp. nov., a new acetotrophic non-hydrogen-oxidizing methane bacterium. Arch. Microbiol. 132:1–9 [DOI] [PubMed] [Google Scholar]
- 36. Mori K, Harayama S. 2011. Methanobacterium petrolearium sp. nov. and Methanobacterium ferruginis sp. nov., mesophilic methanogens isolated from salty environments. Int. J. Syst. Evol. Microbiol. 61:138–143 [DOI] [PubMed] [Google Scholar]
- 37. Nevin KP, et al. 2009. Anode biofilm transcriptomics reveals outer surface components essential for high density current production in Geobacter sulfurreducens fuel cells. PLoS One 4:e5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dinh HT, et al. 2004. Iron corrosion by novel anaerobic microorganisms. Nature 427:829–832 [DOI] [PubMed] [Google Scholar]
- 39. Uchiyama T, Ito K, Mori K, Tsurumaru H, Harayama S. 2010. Iron-corroding methanogen isolated from a crude-oil storage tank. Appl. Environ. Microbiol. 76:1783–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Meyerdierks A, et al. 2010. Metagenome and mRNA expression analyses of anaerobic methanotrophic archaea of the ANME-1 group. Environ. Microbiol. 12:422–439 [DOI] [PubMed] [Google Scholar]
- 41. Smith KS, Ingram-Smith C. 2007. Methanosaeta, the forgotten methanogen? Trends Microbiol. 15:150–155 [DOI] [PubMed] [Google Scholar]
- 42. Alperin M, Hoehler T. 2010. The ongoing mystery of sea-floor methane. Science 329:288–289 [DOI] [PubMed] [Google Scholar]
- 43. Sørensen KB, Finster K, Ramsing NB. 2001. Thermodynamic and kinetic requirements in anaerobic methane oxidizing consortia exclude hydrogen, acetate, and methanol as possible electron shuttles. Microb. Ecol. 42:1–10 [DOI] [PubMed] [Google Scholar]
- 44. Nauhaus K, Treude T, Boetius A, Krüger M. 2005. Environmental regulation of the anaerobic oxidation of methane: a comparison of ANME-I and ANME-II communities. Environ. Microbiol. 7:98–106 [DOI] [PubMed] [Google Scholar]
- 45. Treude T, et al. 2007. Consumption of methane and CO2 by methanotrophic microbial mats from gas seeps of the anoxic Black Sea. Appl. Environ. Microbiol. 73:2271–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Orcutt B, Meile C. 2008. Constraints on mechanisms and rates of anaerobic oxidation of methane by microbial consortia: process-based modeling of ANME-2 archaea and sulfate reducing bacteria interactions. Biogeosciences 5:1587–1599 [Google Scholar]
- 47. Alperin MJ, Hoehler TM. 2009. Anaerobic methane oxidation by archaea/sulfate-reducing bacteria aggregates. Part 1. Thermodynamic and physical constraints. Am. J. Sci. 309:869–957 [Google Scholar]
- 48. Kovacik WP, Jr, et al. 2010. Microbial dynamics in upflow anaerobic sludge blanket (UASB) bioreactor granules in response to short-term changes in substrate feed. Microbiology 156:2418–2427 [DOI] [PubMed] [Google Scholar]
- 49. Araujo JC, et al. 2003. Comparison of hexamethyldisilazane and critical point drying treatments for SEM analysis of anaerobic biofilms and granular sludge. J. Electron Microsc. 52:429–433 [DOI] [PubMed] [Google Scholar]
- 50. Du K, et al. 2009. Self-assembled electrical contact to nanoparticles using metallic droplets. Small 5:1974–1977 [DOI] [PubMed] [Google Scholar]
- 51. Lange U, Mirsky VM. 2008. Separated analysis of bulk and contact resistance of conducting polymers: comparison of simultaneous two- and four-point measurements with impedance measurements. J. Electroanal. Chem. 622:246–251 [Google Scholar]
- 52. Amann RI, et al. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lane DJ, et al. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. U. S. A. 82:6955–6959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Huws SA, Edwards JE, Kim EJ, Scollan ND. 2007. Specificity and sensitivity of eubacterial primers utilized for molecular profiling of bacteria within complex microbial ecosystems. J. Microbiol. Methods 70:565–569 [DOI] [PubMed] [Google Scholar]
- 55. DeLong EF. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. U. S. A. 89:5685–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Filatov DA. 2002. ProSeq: a software for preparation and evolutionary analysis of DNA sequence data sets. Mol. Ecol. Notes 2:621–624 [Google Scholar]
- 60. Felsenstein J. 1989. PHYLIP---phylogeny inference package (version 3.2). Cladistics 5:163–166 [Google Scholar]
- 61. Felsenstein J. 2005. PHYLIP (phylogeny inference package), version 3.6.
- 62. Schloss PD, Handelsman J. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chao A. 1984. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 11:265–270 [Google Scholar]
- 64. Good IJ. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264 [Google Scholar]
- 65. Nevin KP, et al. 2008. Power output and columbic efficiencies from biofilms of Geobacter sulfurreducens comparable to mixed community microbial fuel cells. Environ. Microbiol. 10:2505–2514 [DOI] [PubMed] [Google Scholar]
- 66. Urabe A, Tominaga T, Nakamura Y, Wakita H. 1985. Chemical compositions of natural gases in Japan. Geochem. J. 19:11–25 [Google Scholar]
- 67. Kashefi K, et al. 2002. Geoglobus ahangari gen. nov., sp. nov., a novel hyperthermophilic archaeon capable of oxidizing organic acids and growing autotrophically on hydrogen with Fe(III) serving as the sole electron acceptor. Int. J. Syst. Evol. Microbiol. 52:719–728 [DOI] [PubMed] [Google Scholar]