Comparing different TLR-4-activated macrophages reveals metabolite quantity dependence on COX-2 level and higher PGIS activity relative to other prostanoid syntheses that outcompetes with mPGES-1.
Keywords: lipidomics, prostaglandin, cyclooxygenase, metabolism, coupling, synthase
Abstract
Eicosanoid metabolism differs in profile and quantity between macrophages of different tissue origin and method of elicitation, as well as between primary and immortalized macrophages after activation with inflammatory stimuli. Using a lipidomic approach, we comprehensively analyzed the eicosanoids made by murine RPMs, TGEMs, BMDM, and the macrophage-like cell line RAW after stimulation with the TLR-4-specific agonist KLA. Direct correlation among total COX metabolites, COX side-products (11-HETE, 15-HETE), COX-2 mRNA, and protein at 8 h was found when comparing each cell type. Comprehensive qPCR analysis was used to compare relative transcript levels between the terminal prostanoid synthases themselves as well as between each cell type. Levels of PGE2, PGD2, and TxB2 generally correlated with enzyme transcript expression of PGES, PGDS, and TBXS, providing evidence of comparable enzyme activities. PGIS transcript was expressed only in RPM and TGEM macrophages and at an exceptionally low level, despite high metabolite production compared with other synthases. Presence of PGIS in RPM and TGEM also lowered the production of PGE2 versus PGD2 by approximately tenfold relative to BMDM and RAW cells, which lacked this enzyme. Our results demonstrate that delayed PG production depends on the maximal level of COX-2 expression in different macrophages after TLR-4 stimulation. Also, the same enzymes in each cell largely dictate the profile of eicosanoids produced depending on the ratios of expression between them, with the exception of PGIS, which appears to have much greater synthetic capacity and competes selectively with mPGES-1.
Introduction
Eicosanoids are a class of bioactive lipid signaling molecules derived primarily from AA, as well as other polyunsaturated fatty acids, including EPA and DHA. Through activation of their natural receptors, they mediate numerous physiological processes, including pain, fever, platelet aggregation, vascular constriction, and the promotion and resolution of acute inflammation. Eicosanoids are formed after esterified AA is hydrolyzed from membrane phospholipids at the sn-2 position [1, 2]. AA is then metabolized by a variety of COX [3–5], LOX [3], and CYP enzymes [6]. Further metabolism occurs by a vast number of enzymes downstream of these major arms of the pathway, which leads to hundreds of known lipid species that comprise the eicosanoid pathway. The major steps of eicosanoid biosynthesis, structural identification of the majority of the lipid species, their pharmacological roles, and their medical significance are well known [7].
Macrophages represent one of the important immune effector cells of focus [8] in understanding the pathogenesis of various inflammatory diseases. These mononuclear phagocytes originate from hematopoietic stem cells, circulate through the blood as monocytes, and populate tissues as macrophages in the steady-state and during inflammation [9]. Expression of a wide variety of TLRs and other PRRs allows macrophages to respond efficiently to danger signals by phagocytosis and secretion of inflammatory eicosanoids and cytokines. Additionally, they exhibit vast heterogeneity in terms of phenotype, depending on their tissue of residence and other environmental cues. More specifically, differences in the patterns of COX pathway eicosanoids and total quantities produced among RPM, TGEM, BMDM, and immortalized RAW cells have been observed after various stimuli [10–13]. Several biochemical mechanisms have been proposed to explain the unique PG profiles between different cell types and after different activations, which include spatial compartmentalization of COX-1 and COX-2 that leads to preferential coupling to certain prostanoid synthases [14], induced expression of certain enzymes after activation [15], and differences in substrate affinity and kinetics between prostanoid synthases [16, 17]. However, no direct comparison of these four macrophage phenotypes under controlled condition exists. It ultimately remains unclear how the various COX pathway enzymes are expressed in different cells.
Of these four macrophage types, the RAW is a transformed cell line that is most often used as a model cell, because it exhibits many of the functional characteristics of primary macrophages [18], it eliminates the need for an animal source, and its rapid replication yields many cells efficiently for experiments. RAW cells were isolated from a tumor induced by the Abelson MuLV, which contained the Moloney-MuLV as a helper virus, although other viruses may be present [18, 19]. BMDM represent a homogeneous macrophage population that has matured and differentiated directly from hematopoietic cells in the presence of M-CSF in vitro; thus, it has lost some in vivo context. TGEM are peritoneal macrophages lavaged from the peritoneal cavity several days after an inflammatory assault created by thioglycollate injection (a classic model of sterile, acute inflammation). They infiltrate from the blood to aid in phagocytosis of apoptotic cells and represent an activated macrophage population. RPMs are present during peritoneal homeostasis and are distinct from elicited macrophages. These cells are considered to be sentinels that respond to antigens by secreting chemokines and eicosanoids that initiate acute inflammation. RPM eicosanoid production undoubtedly has a clearer role in the inflammatory response than the other macrophages and exhibits a response common to cells from other tissues, including vascular endothelial cells, ciliary epithelial cells, gingival fibroblasts, and myometrial cells, where primarily, PGI2 and PGE2 are formed after stimulations with LPS, TNF-α, or IL-1 [20]. Only ∼1 million RPM cells are obtainable from a given mouse, and experiments often call for amounts far beyond what is feasible. Thus, some studies opt for one of the other cell types as a representative macrophage somewhat arbitrarily. All of these cells have a distinct phenotype that can be explored for their inherent function and disease relevance, assuming the right question is being asked. Here, it is more interesting to observe an essentially complete lipid mediator fingerprint for each macrophage that will be more widely applicable to different biological topics.
We have developed an LC MS/MS methodology that comprehensively detects nearly all of the known metabolites in the eicosanoid class. This has been applied to understanding eicosanoid production in macrophages after a variety of stimuli using the RAW as a model cell [21]. Recently, this methodology was used to create a predictive kinetic model of eicosanoid flux and signaling in RAW cells stimulated with the TLR-4 specific agonist, KLA [22, 23]. In this study, we have applied a quantitative lipidomic and transcriptomic analysis to RPM, TGEM, BMDM, and RAW macrophage cells stimulated with KLA. From a comprehensive, comparative approach, we have established a near-complete view of the differences in various macrophage phenotypes and propose a seamless mechanism for macrophage eicosanoid production that integrates the different biochemical mechanisms described above.
MATERIALS AND METHODS
Materials
LC-grade solvents were purchased from EMD Biosciences (San Diego, CA, USA). Synergy C18 reverse-phase HPLC column and Strata-X SPE columns were purchased from Phenomenex (Torrance, CA, USA). Eicosanoids were purchased from Cayman Chemicals (Ann Arbor, MI, USA) and Biomol (Plymouth Meeting, PA, USA). KLA [22] was obtained from Avanti Polar Lipids (Alabaster, AL, USA). PBS was purchased from VWR (West Chester, PA, USA). RPMI-1640 medium, FBS, and Quant-iT broad-range DNA assay kit were purchased from Invitrogen (Carlsbad, CA, USA). RT-PCR primers were purchased from PrimerBank (Cambridge, MA, USA). Antibodies for Western blotting were purchased from Cell Signaling Technology (Danvers, MA, USA).
Animals
Male C57bl/6 mice were purchased from Charles River Laboratories (Wilmington, MA, USA). Mice exhibiting skin lesions or visible tumors were excluded from the study. All experiments were carried out according to protocols approved by the Institutional Animal Care Committee of the University of California, San Diego (USA).
RPM harvesting and preparation
Mice were killed via CO2 asphyxiation. Peritoneal exudate cells were obtained by peritoneal lavage with cold Ca2+- and Mg2+-free PBS. Cells were centrifuged and resuspended in RPMI-1640 medium supplemented with 10% heat-inactivated FBS and 100 units/ml penicillin/streptomycin at 37°C in a humidified 5% CO2 atmosphere. Cells were plated in six-well culture plates with 2.0 ml media (1×106 cells/well) and allowed to adhere for 16 h. Nonadherent cells were removed by aspiration, and 1.0 ml fresh media was added. After 1 h, cells were stimulated by adding another 1.0 ml media containing 2× KLA (200 ng/ul), bringing the total volume to 2.0 ml and KLA concentration to 100 ng/ul. Control cells were given 1.0 ml media containing the same amount of PBS as stimulated cells. Media during stimulation contained 10% heat-inactivated FBS.
TGEM harvesting and preparation
Mice were injected with 2.5 mL, 2 months or older autoclaved thioglycollate medium i.p. Four days after injection, mice were killed, and cells were harvested, plated, and stimulated as described above for RPMs.
BMDM harvesting and preparation
BM cells were removed aseptically from mouse femurs and tibiae. The bone ends were cut, and the marrow cavity was flushed out from one end of the bone using a sterile, 22-gauge needle. The BM suspension was agitated carefully, and the resulting suspension was washed twice, resuspended in RPMI-1640 medium, and supplemented with 10% FBS, 100 units/ml penicillin/streptomycin, and 50 ng/ml M-CSF, incubated on nontissue culture-treated Petri dishes at 37°C in a humidified 5% CO2 atmosphere. After 4 days, an additional volume of fresh medium was added, and incubation was continued. After 6 days, medium was aspirated, and cells were removed from dishes with PBS containing 5 mM EDTA using gentle agitation. Cells were washed twice, resuspended in fresh medium, and then plated and stimulated as described above for RPMs.
RAW cell culture and preparation
RAW murine macrophage-like cells were cultured in DMEM, supplemented with 10% FBS and 100 units/ml penicillin/streptomycin. Cells were propagated for at least five passages (no more than 20 passages) before plating and stimulation, as described above for RPMs.
Eicosanoid cell media sample preparation
Media samples were analyzed for extracellular eicosanoid release. After stimulation with KLA, the entire 2.0 ml medium was removed, and each sample was supplemented with 50 μl internal standards (200 pg/μl, EtOH). Samples were centrifuged for 5 min at 3000 rpm to remove cellular debris and were then purified. Eicosanoids were extracted using Strata-X SPE columns, which were washed with 3 ml MeOH and then 3 ml H2O. After applying the sample, the columns were washed with 10% MeOH, and the eicosanoids were then eluted with 1 ml MeOH and stored at –20°C. The eluant was dried under vacuum and redissolved in 100 μl LC solvent A for LC/MS/MS analysis.
Cell quantitation
Eicosanoid levels were normalized to cell DNA. After the extracellular media was removed, the cells were washed twice in cold PBS, then scraped in 250 μl PBS, and stored at –20°C for DNA quantitation using the Quant-iT broad-range DNA assay kit, according to the manufacturer's instructions.
LC and MS of eicosanoids
Eicosanoid analysis was performed by LC-MS/MS, as described previously [24]. Briefly, eicosanoids were separated by a 25-min reverse-phase LC gradient using solvent A and acetonitrile-isopropyl alcohol (50:50, v/v; solvent B). Eicosanoids were analyzed subsequently using a tandem quadrupole MS (ABI 4000 Q-Trap, Applied Biosystems, Foster City, CA, USA) via MRM in negative-ion mode. Eicosanoids were identified in samples by matching their MRM signal and LC retention time with those of a pure standard.
RNA isolation and qRT-PCR
Total RNA was isolated from the macrophage cells using the RNeasy mini kit (Qiagen, Valencia, CA, USA) and DNase 1 (Invitrogen). First-strand cDNA was synthesized using SuperScript 111 and random hexamers (Invitrogen). Samples were run in 20 ul reactions using an ABI 7300 (Applied Biosystems). Samples were incubated at 95°C for 15 min, followed by 40 cycles at 95°C for 10 s, 56°C for 20 s, and 72°C for 30 s. SYBR Green oligonucleotides were used for detection and quantification of a given gene, expressed as relative mRNA level compared with a standard housekeeping gene (GAPDH) using the Δ CT method, as described by the manufacturer (Invitrogen). The GAPDH value used for normalization was an average of the CT values obtained from each cell type as a result of a noticeable difference between the RAW and primary cells.
Western blotting
Cells were washed twice with cold PBS and scraped into 250 μl cold buffer containing 50 mM HEPES, pH 7.4, 150 mM NaCl, 200 mM NaF, 20 mM sodium pyrophosphate, 10% glycerol, 1% Triton X-100, 4 mM sodium orthovanadate, 2 mM PMSF, and 1 mM EDTA and Complete mini protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN, USA). Protein concentrations were determined and normalized using the BCA protein assay kit (Fisher Scientific, Waltham, MA, USA). Total protein (10 μg) was loaded onto 4–12% Bis-Tris SDS polyacrylamide gel (Invitrogen), electrophoresed, and transferred onto a PVDF membrane (BioRad, Hercules, CA, USA). The membrane was blocked with 100% methanol for 5 min, dried for 10 min, and then incubated with the appropriate antibody overnight in 5% (w/v) milk protein in ultrapure water containing 0.1% Tween 20. Membranes were then washed three times in TBS-Tween buffer and incubated with the appropriate secondary antibody (Cell Signaling Technology) for 1 h. All membranes were washed three times before development using the ECL Plus Western blotting detection system (Amersham Biosciences, Piscataway, NJ, USA).
RESULTS
Eicosanoid production in TLR-4-stimulated macrophages
Media from the different macrophages were collected at 0, 8, and 24 h after KLA stimulation to examine the lipid changes during the delayed phase (beyond the first 2 h of stimulation) of COX metabolite production. We used a middle time-point of 8 h, as it was found to be near the maximal rate of eicosanoid production based on time-course studies (data not shown), and 24 h was chosen as an arbitrary endpoint. During the delayed phase, there is a robust induction of COX-2 and mPGES-1 expression and down-regulation of 5-LOX; the immediate phase of eicosanoid production (0–2 h after stimulation) is dependent on the constitutively expressed COX-1 and terminal prostanoid synthases. COX-1 metabolism contributes minimally to the delayed-phase levels, as all cell types produced more than one order of magnitude less COX metabolites by 2 h compared with 8 h or 24 h. Further, a short-term, COX-1-dependent metabolism using ATP stimulation, which mobilizes Ca2+, produces maximal prostanoid levels by 30 min and is more than one order of magnitude lower than maximal, long-term KLA stimulation [21].
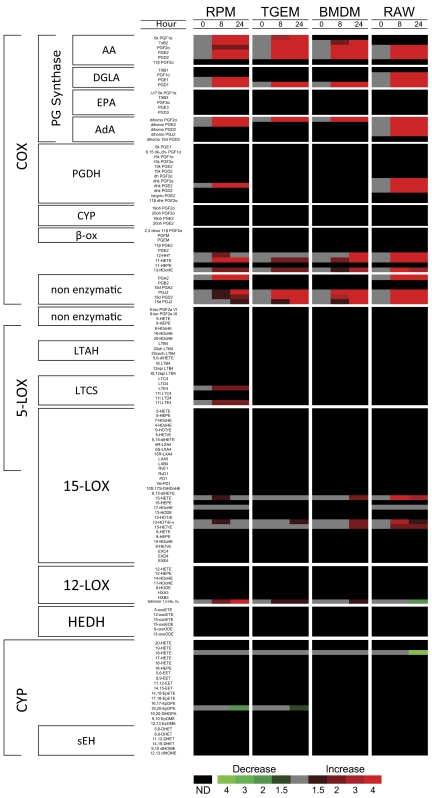
Eicosanoids were analyzed using LC coupled with MS/MS. In total, 26, 19, 17, and 26 distinct molecular species were detected in the extracellular medium of RPM, TGEM, BMDM, and RAW macrophages, respectively. We arranged the lipidomic data in an array format similar to a gene array and clustered the different species based on the enzyme responsible for their production (Fig. 1). In the COX pathway, the eicosanoids are further clustered by fatty acids (AA, EPA, DGLA, AdA); however, this was not done in other pathways for simplicity. As expected, the vast majority of metabolites produced by RPM, TGEM, BMDM, and RAW cells were AA-derived and from the COX pathway, being greatly increased (greater than fourfold) after KLA stimulation versus control. We also observed COX products derived from DGLA and AdA in all of the cell types. Only a few metabolites from 5-LOX, 12-LOX, 15-LOX, and CYP pathways were detected which did not greatly differ between KLA stimulation and control. The minimal production (RPM) or absence (TGEM, BMDM, and RAW) of 5-LOX products is due to down-regulation of the transcript in all of the cell types (data not shown) and the lack of Ca2+ mobilization by KLA, which is required for activity. All of the macrophages ultimately respond to TLR-4 stimulation by greatly up-regulating COX-2 and mPGES-1 and down-regulating 5-LOX (data not shown) from a transcriptomic standpoint.
Figure 1. Lipidomic analysis of RPM, TGEM, BMDM, and RAW macrophages.
Heat map representing fold-change in the extracellular medium levels of 140 eicosanoid species after stimulation with the TLR-4-specific receptor agonist KLA (100 ng/ml) relative to unstimulated PBS control at 0, 8, and 24 h time-points. Increases in metabolite levels are indicated by red, decreases by green, and detectable but unchanged levels by gray. Metabolites below the limit of detection are indicated by black; n = 3 individual biological replicates/time-point/group. PGDH, PG dehydrogenase; β-ox, β-oxidation; LTAH, LTA4 hydrolase; LTCS, LTC4 synthase; HEDH, HETE dehydrogenase; sEH, soluble epoxide hydrolase.
As we quantitate all of the stable arachidonate metabolites, we can also see that the bulk of this fatty acid substrate in each cell is largely acted on by COX (Table 1). Enzymatic and nonenzymatic breakdown products of the various prostanoids are included in this total accounting of COX metabolites. Of all the prostanoids, PGD2 requires particular attention, as it is degraded further into three dehydration products: PGJ2, 15d PGD2, and 15d PGJ2, whereas PGE2 is degraded into analogous molecules at a significantly slower rate. Previously, we have estimated the degradation rates of the major COX metabolites in KLA-stimulated RAW cells, including these PGD2 breakdown products [23]. As seen in Figs. 1 and 2, all of the cell types produced these metabolites in minor quantities. Additionally, RPM cells produced a small amount of the oxidation product 13,14-dihydro-15k PGE2, and RAW cells produced low quantities of this same product, along with 13,14-dihydro-15k PGD2 and 13,14-dihydro-15k PGF2α. These metabolites were minor and not included in our metabolite profile comparison but were included in the total COX metabolite analysis, which will be addressed further. This initial lipidomic screening confirmed that all of the macrophage phenotypes produce mostly COX metabolites in response to TLR-4 activation.
Table 1. Total Macrophage Eicosanoid Production.
| Enzymatic pathway | Time poststimulation | KLA (–/+) | Eicosanoid production (pmol/μg DNA) |
|||
|---|---|---|---|---|---|---|
| Resident | TGEM | BMDM | RAW | |||
| COX | 8 h | – | 9.2 ± 1.6 | 5.4 ± 0.3 | 2.7 ± 0.4 | 2.3 ± 0.7 |
| + | 202.6 ± 12.2 | 31.4 ± 3.2 | 21.9 ± 4.7 | 432.4 ± 69.2 | ||
| 24 h | – | 14.8 ± 1.9 | 5.5 ± 0.4 | 1.9 ± 0.4 | 1.5 ± 0.4 | |
| + | 242.6 ± 45.0 | 40.3 ± 4.4 | 24.3 ± 5.7 | 438.0 ± 105.2 | ||
| 5-LOX | 8 h | – | 1.1 ± 0.3 | 0.0 | 0.0 | 0.0 |
| + | 2.5 ± 0.3 | 0.0 | 0.0 | 0.0 | ||
| 24 h | – | 2.2 ± 0.2 | 0.0 | 0.0 | 0.0 | |
| + | 6.4 ± 0.7 | 0.0 | 0.0 | 0.0 | ||
| 15-LOX | 8 h | – | 0.5 ± 0.0 | 0.2 ± 0.0 | 1.7 ± 0.1 | 0.6 ± 0.1 |
| + | 1.4 ± 0.2 | 0.3 ± 0.0 | 1.8 ± 0.3 | 0.7 ± 0.1 | ||
| 24 h | – | 1.0 ± 0.1 | 0.2 ± 0.0 | 0.9 ± 0.2 | 0.5 ± 0.2 | |
| + | 5.5 ± 0.6 | 0.3 ± 0.0 | 1.6 ± 0.4 | 0.2 ± 0.1 | ||
| 12-LOX | 8 h | – | 1.7 ± 0.2 | 1.0 ± 0.2 | 0.6 ± 0.1 | 1.1 ± 0.1 |
| + | 2.4 ± 0.1 | 1.3 ± 0.1 | 0.7 ± 0.2 | 2.1 ± 0.0 | ||
| 24 h | – | 4.8 ± 0.3 | 1.2 ± 0.1 | 0.6 ± 0.2 | 1.1 ± 0.3 | |
| + | 6.4 ± 0.5 | 2.1 ± 0.1 | 1.2 ± 0.4 | 1.5 ± 0.4 | ||
| CYP | 8 h | – | 1.3 ± 0.1 | 3.9 ± 0.8 | 1.4 ± 0.1 | 0.9 ± 0.1 |
| + | 1.4 ± 0.2 | 4.3 ± 0.5 | 1.2 ± 0.2 | 0.9 ± 0.1 | ||
| 24 h | – | 3.3 ± 0.3 | 9.6 ± 1.2 | 2.2 ± 0.3 | 1.2 ± 0.2 | |
| + | 2.3 ± 0.1 | 8.9 ± 1.3 | 1.5 ± 0.2 | 0.3 ± 0.1 | ||
Quantitative analysis of eicosanoids formed through COX, 5-LOX, 15-LOX, 12-LOX, and CYP pathways, expressed as pmol eicosanoid per μg DNA. The data are from the same experiment as in Fig. 1 and are expressed as mean values ± sem of three biological replicates.
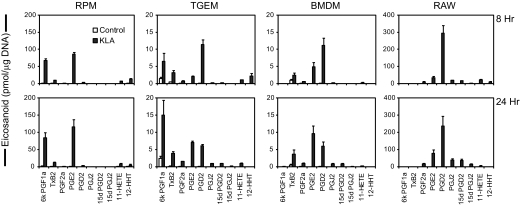
Figure 2. Quantitative eicosanoid production profiles of different TLR-4 agonist-stimulated macrophages.
Different macrophage types incubated in the absence (open bars) and presence (shaded bars) of KLA (100 ng/ml) from the same experiment as Fig. 1. Extracellular medium was removed at 8 h (upper row) and 24 h (lower row) and was analyzed for eicosanoid levels by MS. The data are expressed as mean values ± sem of three biological replicates.
Next, the quantitative profiles of COX metabolites after 8 h and 24 h in the four macrophage types were compared to identify the more discreet differences in this pathway (Fig. 2). Each cell type produced a unique profile of prostanoid species upon TLR-4 stimulation. RPM formed predominantly 6k PGF1a (the stable breakdown product of PGI2) and PGE2 at roughly equal amounts and much lower levels of other COX metabolites; this profile remained essentially the same from 8 h to 24 h. TGEM macrophages produced PGD2, 6k PGF1a, TxB2 (the stable breakdown product of TxA2) + 12-HHT, and PGE2 at a ratio of 4:2:2:1, respectively, at 8 h. After 24 h, the TGEM profile shifted to 6k PGF1a, PGE2, PGD2, and TxB2 + 12-HHT at 4:2:2:1, respectively. BMDM macrophages at 8 h produced predominantly PGD2, PGE2, and TxB2 + 12-HHT at a ratio of 4:2:1, respectively. This profile in BMDM shifted to PGE2, PGD2, and TxB2 +12-HHT at 3:2:1. RAW cells produced predominantly PGD2 and approximately tenfold less PGE2 at 8 h; at 24 h, the RAW level of PGD2 decreased (as a result of increased metabolite breakdown), and PGE2 roughly doubled, resulting in approximately four times more PGD2 than PGE2. In general, the major change from 8 h to 24 h in all of the cell types was a higher PGE2:PGD2 ratio, likely as a result of the induction of mPGES-1 expression (this issue will be addressed further later in the text). All of the cell types produced smaller levels of PGF2a, 11-HETE, and the PGD2 nonenzymatic-breakdown products, PGJ2, 15d PGD2, and 15d PGJ2, relative to the metabolites just mentioned above. RAW cells did not produce detectable levels of 6k PGF1a or TxB2, BMDM cells did not produce detectable levels of 6k PGF1a, and RPM and TGEM cells produced some level of all of the major COX metabolites monitored with our method. It is noteworthy that individual stimulation of the major TLR receptors (TLR-1 to -7, and -9) in RAW [21] leads to the same eicosanoid profile observed in Fig. 2. We found similar basal levels of 5-LOX mRNA between RAW and RPM (∼1.3-fold higher in RPM) and lower levels in TGEM and BMDM, which leads us to expect that the primary macrophage profiles observed in Fig. 2 would be similar upon long-term stimulation with other TLR receptors.
Induced COX-2 expression controls total prostanoid production in different macrophage cell types
Our lipidomic data (Figs. 1 and 2 and Table 1) allowed us to quantify the total COX metabolites made by each cell. The cell order (from highest to lowest) of total COX metabolite production at 8 h was RAW, RPM, TGEM, and BMDM at a roughly 20:10:1.5:1 ratio (Fig. 3A), respectively, which remained the same at 24 h (data not shown).
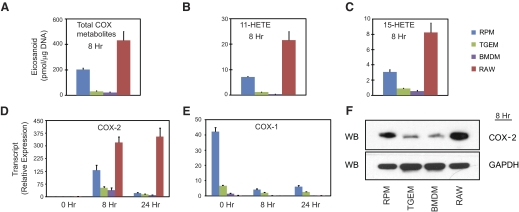
Figure 3. COX-2 activity and expression.
Comparison of 8 h (A) total arachidonate COX-derived eicosanoids, (B) 11-HETE, and (C) 15-HETE from the same experiment as in Fig. 1; relative expression level of (D) COX-2 mRNA and (E) COX-1 mRNA at 0, 8, and 24 h; and (F) 8 h COX-2 protein in RPM, TGEM, BMDM, and RAW macrophages after KLA (100 ng/ml) stimulation. The data are expressed as mean values ± sem of three biological replicates. WB, Western blot.
Next, we compared this profile with the levels of 11-HETE and 15-HETE in each cell. In addition to making PGH2, COX-1 and -2 have LOX activity, which produces small levels of 11-HETE and 15-HETE [25–27]. The cell comparison profiles of 11-HETE and 15-HETE (Fig. 3B and C) correlated directly with the total COX metabolite profile (Fig. 3A), giving an initial indication that the level of COX expression in each cell is the rate-limiting step. Although 15-HETE can also be made by 12/15-LOX, the direct correlation with COX-2 expression and lack of other 12/15-LOX products suggests that this metabolite is indeed COX-derived.
We then compared the expression level of COX-2 mRNA and protein (Fig. 3D and F), which matched directly with the aforementioned profiles of total COX metabolites, 11-HETE, and 15-HETE. Despite drastic differences in total COX product quantities, it was verified that similar proportions of PGH2-derived metabolites, 11-HETE, and 15-HETE were made in each cell type (Table 2) and that these metabolite distributions were consistent with that of the in vitro distribution produced by purified COX protein [26]. In a strikingly clear way, these results show that the level of maximal COX-2 expression in macrophages after TLR-4 stimulation directly controls the level of COX metabolites in the delayed phase.
Table 2. COX-2 and LOX acitivity.
| Product | Arachidonate product (% total COX) |
|||
|---|---|---|---|---|
| RPM | TGEM | BMDM | RAW | |
| PGH2-derived products | 94.6 ± 0.2 | 90.9 ± 1.2 | 92.8 ± 0.6 | 92.7 ± 0.9 |
| 11-HETE | 3.9 ± 0.2 | 6.1 ± 0.7 | 4.8 ± 0.4 | 5.4 ± 0.7 |
| 15-HETE | 1.5 ± 0.1 | 3.0 ± 0.5 | 2.4 ± 0.2 | 1.9 ± 0.2 |
COX-derived arachidonate eicosanoids, formed after 8 h of KLA (100 ng/ml) stimulation, were summed, and the percentages of PGH2-derived and HETE products were calculated in RPM, TGEM, BMDM, and RAW. The data are from the same experiment as Fig. 1 and are expressed as mean values ± sem of three biological replicates.
Terminal prostanoid synthase expression
AA is primarily converted to PGH2 before being isomerized into PGE2, PGD2, PGF2a, PGI2, TxA2, or 12-HHT, depending on the expression of the enzymes responsible for their production. We semiquantitatively compared the transcript expression of the terminal prostanoid synthases (at 0, 8, and 24 h after KLA stimulation) to observe the enzymatic transcript composition in the four macrophage types, which lead to the eicosanoid profiles that we have observed (Fig. 2). PGF2 synthase was left out of our analysis, as PGF2a was a minimal product in all of the cells and can be additionally converted from PGE2 by a 9-ketoreductase.
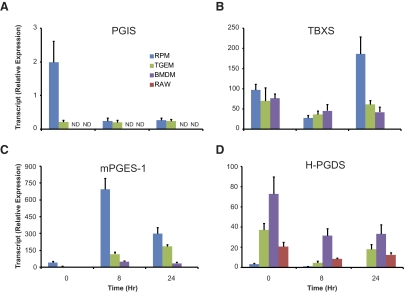
We only detected PGI2 in RPM and TGEM (not in BMDM or RAW), and the same was true for PGIS transcript expression (Fig. 4A). RAW was the only cell type that did not produce detectable levels of TxB2 and expressed a level of TBXS much lower than the other macrophages (Fig. 4B). It is worth noting that RPM, TGEM, and BMDM expressed similar levels of TBXS mRNA, which is consistent with the literature regarding RPM and TGEM [28].
Figure 4. Expression of terminal prostanoid synthases.
Comparison of transcript expression of (A) PGIS, (B) TBXS (TBXAS), (C) mPGES-1, and (D) H-PGDS synthases after 0, 8, and 24 h of KLA (100 ng/ml) stimulation in RPM (blue), TGEM (green), BMDM (purple), and RAW (red) macrophages. The data are expressed as mean values ± sem of three biological replicates.
Of the three enzymes able to isomerize PGH2 to PGE2 (mPGES-1, mPGES-2, and cPGES), mPGES-1 is the primary isoform considered in numerous inflammatory models, where its expression is increased greatly in concert with COX-2. Also, mPGES-1 and COX-2 are both primarily localized in the perinuclear and ER membranes [29], whereas mPGES-2 and cPGES are localized in the Golgi membrane and cytoplasm, respectively [30, 31]. We observed a dramatic increase in mPGES-1 expression from 0 h to 8 h in RPM, TGEM, BMDM, and RAW, although at different levels (Fig. 4C). Several studies have failed to detect mPGES-1 protein in RAW and have speculated this to be the reason for low production of PGE2 relative to PGD2 in this cell type [10, 32]. In contrast, we detected mPGES-1 transcript in RAW but at a considerably lower level than in RPM, TGEM, and BMDM. Similar levels of mPGES-2 and cPGES mRNA were measured between each type of macrophage, which did not change after 8 h or 24 h of KLA stimulation (data not shown).
Two PGDS isoforms produce PGD2, H-PGDS and L-PGDS, although only H-PGDS was detected in our qPCR transcript analysis. There is little to be found regarding the contribution of L-PGDS in macrophage PGD2 production, where it is not constitutively expressed but may be induced by various stimuli. One study has reported that small interfering RNA knockdown of induced L-PGDS significantly lowered the level of PGD2 in LPS-stimulated RAW cells [33], although the effect on other eicosanoids was not examined. This is a possible concern, as we have observed a global COX metabolite decrease after small hairpin RNA knockdown or chemical inhibition of mPGES-1 in the same cells using KLA stimulation in a separate study (unpublished results). We observed the lowest H-PGDS transcript in RPM, and the highest levels were in BMDM and TGEM (Fig. 4D). Although RAW cells produce high levels of mostly PGD2, this does not appear to be a result of an exorbitantly higher level of PGDS relative to other macrophages.
Enzyme competition for PGH2 differs between cells with and without PGIS
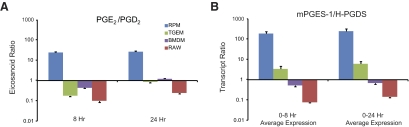
We then focused on the ratios of PGE2/(PGD2+breakdowns) produced in each type of macrophage compared with the ratios of mPGES-1/H-PGDS expressed to see whether there is a general correlation between metabolite and enzyme. The average transcript expression ratios between 0 h and 8 h and between 0 h and 24 h were compared with the eicosanoid ratios at 8 h and 24 h, respectively (Fig. 5). The two extremes in ratios were between RPM, having a very high PGE/PGD metabolite and transcript (>10), and RAW, where the PGE/PGD metabolite and transcript ratios were both very low (∼0.1). TGEM and BMDM had ratios of PGE/PGD closer to one in eicosanoid amount and enzyme transcript expression. This ratio increased from 8 h to 24 h, as mPGES-1 transcript expression remained induced, and the constitutively expressed H-PGDS transcript decreased after stimulation. Of the four macrophage types, BMDM and RAW lack expression of PGIS (mentioned above) and therefore, provide a simpler system to study PGE2 and PGD2 production. These cells produced ratios of PGE2/PGD2 that were nearly equal to the mPGES-1/H-PGDS transcript ratios, which suggests similar activities in the two enzymes and competition, based directly on protein quantity. In the specific case of RAW, low PGE2 versus PGD2 production appears to be caused, not by a complete lack of mPGES-1 but by the relative dominance of H-PGDS expression. In RPM and TGEM macrophages, there is a much lower (approximately one order of magnitude) ratio of PGE2/PGD2 than mPGES-1/H-PGDS transcript compared with RAW and BMDM, which is perhaps a result of specific competition between PGIS and mPGES-1 for the substrate. This is most noticeable in TGEM, where PGE2/PGD2 was <1, and PGES/PGDS was >1.
Figure 5. Correlation between PGE2:PGD2 ratio and respective enzyme transcript expression ratio.
(A) Eicosanoid ratios of PGE2:PGD2 after 8 h and 24 h KLA stimulation, where PGD2, PGJ2, 15d PGD2, and 15d PGJ2 were summed. (B) Transcript expression ratios of mPGES-1:H-PGDS using average expression of 0 h and 8 h time-points (0–8 h) and 0, 8, and 24 h time-points (0–24) in RPM (blue), TGEM (green), BMDM (purple), and RAW (red) macrophages stimulated with KLA (100 ng/ml). The data are expressed as mean values ± sem of three biological replicates.
The correlations between PGE and PGD transcript and metabolite, which we could compare in all of the cell types together, prompted us to determine proportionality relationships among all of the terminal prostanoid synthases and their metabolites (Table 3). Overall, expression among mPGES-1, H-PGDS, and TBXS and the levels of their respective eicosanoid products were proportionate, in clear contrast to PGIS and PGI2. As PGIS mRNA was expressed at a disproportionately low level compared with the other prostanoid synthases relative to metabolite production, this enzyme likely has a much greater synthetic capacity, perhaps as a result of a locational advantage to COX-2. Altogether, we can see several degrees of complexity in prostanoid synthase expression and metabolite production between macrophage phenotypes (Fig. 6).
Table 3. Proportionality of COX Metabolites and Enzyme Transcripts.
| Cell type | Metabolite (% total PGH2) |
Enzyme transcript (% total prostanoid synthase mRNA) |
Metabolite/enzyme transcript (% total PGH2)/(% total prostanoid synthase mRNA) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PGI2 | TxB2 + 12-HHT | PGE2 | PGD2 | PGIS | TBXS | mPGES-1 | H-PGDS | PGI2/PGIS | TxB2 + 12-HHT/TBXS | PGE2/mPGES-1 | PGD2/H-PGDS | |
| RPM | 35.3 | 12.4 | 45.2 | 1.8 | 0.3 | 14.5 | 84.8 | 0.5 | 137.4 | 0.9 | 0.5 | 3.6 |
| TGEM | 22.6 | 19.2 | 7.5 | 40.5 | 0.2 | 38.8 | 45.8 | 15.2 | 147.6 | 0.5 | 0.2 | 2.7 |
| BMDM | N.D. | 11.9 | 24.6 | 55.2 | N.D. | 44.0 | 18.6 | 37.4 | N.D. | 0.3 | 1.3 | 1.5 |
| RAW | N.D. | N.D. | 8.5 | 73.2 | N.D. | N.D. | 7.0 | 93.0 | N.D. | N.D. | 1.2 | 0.8 |
COX-derived prostanoids formed after 8 h of KLA (100 ng/ml) stimulation, represented as percentages of total PGH2 (11-HETE and 15-HETE exluded from total) and percentage of average individual prostanoid synthase transcript expression between 0 h and 8 h relative to total prostanoid synthase mRNA. Metabolite versus enzyme transcript ratios were calculated using metabolite and enzyme transcript percentages. Values expressed are representative of the mean of three biological triplicates.
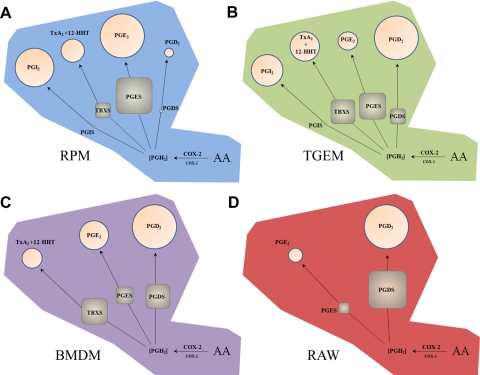
Figure 6. Overview of prostanoid synthase expression and relative activity in different macrophage cell types after long-term TLR-4 activation.
Metabolism of AA in (A) RPM, (B) TGEM, (C) BMDM, and (D) RAW through induced COX-2 leads to a pool of PGH2 that is metabolized by TBXS, PGIS, PGDS (H-PGDS), and induced PGES (mPGES-1). Percentages of metabolites (circles) and enzyme transcripts (squares) in each cell type are represented by area after 8 h KLA stimulation. Metabolites and expressed levels of PGES, PGDS, and TBXS are roughly proportional, although the presence of PGIS appears to selectively draw substrate away from PGES but not PGDS. Low constitutive expression level of PGIS in RPM and TGEM macrophages with disproportionately high levels of PGI2 suggests a significantly higher synthetic rate compared with other prostanoid synthases.
DISCUSSION
In an earlier study, we extensively examined RAW eicosanoid production in response to many different TLR agonists and found that the profile of COX metabolites was dominated by PGD2 with far smaller levels of PGE2 and PGF2a, regardless of which TLR was stimulated [21]. The same PGD2-dominated profile was also seen with several short-term Ca2+ agonists (aside from the production of 5-LOX products). It was noted that RAW and primary cells differed substantially with regard to the profiles of COX metabolites that they produced [21]. Here, we set out to fully characterize these differences (in RPM, TGEM, BMDM, and RAW) and describe explicitly how the RAW cell can accurately serve as a model cell for primary macrophage eicosanoid production in the context of TLR-4 stimulation. Previous studies have compared the temporal changes in enzyme levels but have not fully addressed how they are quantitatively expressed relative to one another. Now, we can more accurately describe the entire eicosanoid pathway and how it operates in different macrophage phenotypes.
Lipidomic analysis of different macrophage phenotypes
Previous eicosanoid analyses of macrophages have typically monitored only a few of the major PGs and LTs made by the COX and 5-LOX pathways, respectively. Metabolites, such as PGE2, PGI2, TxA2, LTA4, LTC4, and others, are considered the more important species for signaling, although monitoring only these metabolites leaves out useful information about the complete metabolism of AA. Here, we have used a lipidomics approach capable of monitoring 140 unique eicosanoid lipid species that cover the entire pathway, including COX, 5-, 12-, and 15-LOX, CYP, and nonenzymatic pathways upon which AA can be acted (Fig. 1). Also included in the 140 metabolites simultaneously monitored are numerous EPA, DHA, and DGLA versions of the various eicosanoids, along with novel DHA and EPA molecules such as protectins and resolvins [34].
It is clear from our screening that different macrophage types indeed produce mostly arachidonate COX products but not metabolites from other pathways such as the anti-inflammatory, proresolving hepoxilins, lipoxins, protectins, and resolvins. We did observe the formation of AdA-derived dihomo-PGs [35] and DGLA-derived 1 series PGs in all four macrophage types. Whether these metabolites elicit a significant biological effect could be of interest in future studies.
Macrophages share a singular mechanism of COX-2 metabolism
We examined the obvious differences in total prostanoid production (Table 1) by looking for a correlation among total COX metabolites, 11-HETE, and 15-HETE (Fig. 3). Biological analyses involving COX metabolism usually ignore 11-HETE and 15-HETE, as both are produced at much lower levels than PGH2 and have yet to show any major bioactivity. Here, we were able to demonstrate their usefulness as a direct readout of COX activity. As many studies measure changes in PGE2 or other bioactive COX products to indicate changes in COX activity, this assumes that no alterations in PGH2 metabolism have occurred. Measuring differences in total COX protein assumes that all of the protein is active, which may not be true, as “suicide inactivation” can occur after many substrate-product turnovers [5, 7]. Both of these COX “side-products” indicated that COX-2 activity in each cell type was roughly the same at 8 h and that only the absolute amount of protein expressed dictated total metabolite quantities. Although COX is generally thought of as the rate-limiting step in this pathway, the release of AA by PLA2, as well as AA oxidation and membrane reincorporation, have been considered as including additional factors contributing to differential PG production between various types of macrophages, specifically after stimulation with LPS or zymosan [10, 36–38]. Our own transcript analysis of group IVA cytosolic PLA2 and group V-secreted PLA2 indicated to us that there were differences in these enzymes between the cells (data not shown). Still, it is clear that all of these different macrophages express a distinct level of COX-2 that is saturated with free AA through the course of at least 8 h, and eicosanoid production is solely dependent on COX-2 as the rate-limiting step during the delayed phase. Beyond this point, eicosanoid production greatly diminishes, as evidenced by only a small increase in COX metabolites from 8 h to 24 h (Table 1). When comparing total COX metabolites with 11-HETE, 15-HETE, and COX-2 mRNA and protein at 24 h, all of the correlations seen at 8 h are no longer observed (data not shown). The reason for this slowed metabolite production is likely the result of a combination of decreased expression and suicide inactivation of COX-2 and prostanoid synthases, as well as diminished AA release and/or re-esterification. Ultimately, we can confidently expect a given macrophage to produce a level of COX metabolites using a simplified mechanism in contrast to the previous notion that other factors besides COX-2 expression significantly contribute in a more stochastic scenario.
Correlating enzyme expression with metabolite profiles
We observed (in Fig. 2) that RPM, TGEM, BMDM, and RAW macrophages produce their own unique eicosanoid profiles, so we used an extensive qPCR analysis of the principle enzymes implicated in these macrophages to establish the relative quantitative differences between each cell type. We integrated these results with previously reported findings and accepted enzyme mechanisms to explain the eicosanoid production in each macrophage and to estimate the relative activities of the various prostanoid synthases.
RPMs have previously been shown to produce a balance of all the prostanoids (i.e., PGE2, PGD2, PGI2, and TxA2) during COX-1-dependent metabolism, whereas delayed-phase, COX-2-dominated metabolism generated primarily PGE2 and PGI2 [20]. Using KLA stimulation of RPM cells, we observe this latter profile (Fig. 2). Given that PGIS and mPGES-1 are both expressed in the perinuclear membrane along with COX-2, the production of mostly PGI2 and PGE2 is consistent with the idea of compartmentalization. Interestingly, we found that mPGES-1 transcript expression was vastly greater than that of PGIS (Fig. 4). This would imply that PGIS possesses a much higher capacity to metabolize COX-2-derived PGH2. In fact, PGIS, cotransfected into human embryonic kidney 293 cells, was previously found to have more strict preference for COX-2 than TBXS and PGES [17]. This could be the result of a closer proximity of the enzyme to COX-2 to receive substrate. A recent study demonstrates that when PGIS is coexpressed with COX-1 or COX-2 in COS-7 cells, the topographical distance is shortest between PGIS and COX-2 based on Förster resonance energy transfer analysis [39]. Altogether, these findings describe how compartmentalization and coupling can create a positional advantage for receiving substrate despite low levels of enzyme.
Also regarding RPM is the low level of TxB2 observed, despite the fact that TBXS is localized to the perinuclear membrane and ER, much like PGIS and mPGES-1. The production of very low TxA2 to PGE2 in human monocytes has been proposed to be the result of different kinetics and substrate affinities of PGES and TBXS [16]. We observed between 0 h and 8 h of KLA activation, approximately sixfold higher average mPGES-1 versus TBXS transcript expression compared with approximately fourfold higher PGE2 versus (TxB2+HHT), which suggests that the low TxB2 observed is also due to limited enzyme expression in RPM.
TGEMs express all of the terminal prostanoid synthases like RPM but produce much lower levels of COX metabolites along with a more balanced profile of PGE2, PGD2, PGI2, and TxB2 after KLA stimulation (Fig. 2). This is largely explained by a lower expression of COX-2, which greatly limits the amount of PGH2. The more balanced eicosanoid profile of TGEM observed is qualitatively similar to the aforementioned profile that has been observed for RPM cells when COX-1 metabolism is dominant. Additionally, a more-even expression of TBXS, mPGES-1, and H-PGDS through the course of 24 h is observed in TGEM, whereas RPM cells clearly induce mPGES-1 to a much higher level than the other synthases (Fig. 4).
The BMDM COX eicosanoid profile somewhat resembles the even distribution of TGEM, aside from the absence of PGI2 (Fig. 2). Also, there are comparable levels of TBXS, mPGES-1, and H-PGDS (Fig. 4), which is consistent with the TGEM scenario to suggest roughly similar activities among these enzymes. The total amount of COX-derived eicosanoids is very low at nearly the same level as TGEM, as a result of low expression of induced COX-2. The absence of PGIS and PGI2 is a similarity found uniquely between BMDM and RAW. As the RAW is a widely used model cell for macrophage biology, what primary macrophage it most resembles has been discussed frequently. It has been suggested that as the RAW was originally derived from a pristane-derived cell population, it would better serve as a model of elicited macrophages than resident cells [18]. Our comparisons point to a greater similarity between RAW and BMDM based on their limited sets of prostanoid synthases, whereas RPM and TGEM cells express the full set. There might be a similar cue not present during differentiation of BMDM and RAW, which was available to the TGEM and RPM cells in the peritoneal space. Indeed, peritoneal lavage fluid added to BMDM during in vitro proliferation and differentiation has been shown to significantly alter eicosanoid production, where increased PGI2 production was observed [13]. Differentiating and propagating BM stem cells with M-CSF, as we have done, yields a nearly pure population of macrophages [13]. However, culture in other cytokines, such as GM-CSF, leads to a less pure cell population and can increase AA release and prostanoid production; thus, particular cytokines can certainly affect eicosanoid levels.
We find that RAW cells express the most limited set of prostanoid synthases compared with other macrophages. This results in an ability to examine the competition between mPGES-1 and H-PGDS directly, although a study of all of the different enzymes requires a different cell type. H-PGDS is a cytosolic enzyme that can translocate to the ER during short-term Ca2+-mobilizing events, where predominantly COX-1 eicosanoid metabolism occurs, and to the perinuclear membrane during long-term stimulations, where predominantly COX-2 eicosanoid metabolism occurs [17]. Our analysis shows that along with the absence of PGIS and TBXS, the average expression of H-PGDS after 8 h and 24 h of KLA stimulation was roughly tenfold higher than mPGES-1 expression, which closely matches the ratio of PGD2 versus PGE2 (Fig. 5). Thus, RAW cells produce this profile, as H-PGDS is the dominantly expressed enzyme that can access COX-1 and COX-2 pools of substrate.
Primary cells versus cell lines and the advantage of their differences
The high level of COX-2 expression that we observed in RAW compared with primary cells (especially TGEM and BMDM) and the unique PGD2-dominant eicosanoid profile pose major differences that are important to consider. Since the RAW cells have been cultured and propagated now for several decades, one study addressed this by comparing LPS-stimulated PG production in a stock of RAW cells acquired in the early 1990s with a separate stock acquired in 2001 (both from ATCC, Manassas, VA, USA). They found that the earlier 1990s′ stock produced markedly lower levels of PGs (PGD2 to the greatest extent), as well as lower COX-2 expression after 6 h of stimulation with LPS [10]. Earlier stocks of RAW seem to more closely resemble primary cells, having lower COX-2 expression and a lesser dominance of PGD2. More recently, ecotropic and polytropic MuLVs were reported to be produced in RAW cells obtained from ATCC and that cell-free culture supernatants also induced tumors in mice, contrary to the original report [19]. It is not clear what exact roles the initial transformation and its maintenance have on the RAW cells with regard to eicosanoid production. Still, it must be kept in mind that the presence of viral components may affect signaling aspects of importance here, particularly as they can activate NF-κB [40].
Based on our findings, the RAW cell indeed resembles primary macrophages in terms of basic characterizations (COX-2 and mPGES-1 induction and down-regulation of 5-LOX), and although the specific COX metabolite profile that it produces is quite unique, the same is true for RPM, TGEM, and BMDM. Here, we found these differences to be useful for studying various expression scenarios, where multiple enzymes compete for a common substrate. The more exact details about prostanoid synthase expression in different cell types has received limited attention, which is understandable considering the amount of information it requires. Our analysis allowed us to confirm that in cases where a metabolite was not made at a detectable level, the respective enzyme transcript was barely expressed, if at all. It allowed us to see metabolite ratio shifts that coincided with enzyme transcript ratio shifts and a clear correlation between enzyme message and metabolite levels in several opposing scenarios. Further, we observed a substrate competition, where PGIS appears to take substrate away from PGES rather than PGDS, using two cell types that expressed PGIS and two that did not. Ultimately, the “omic” strategy was ideal for elucidating several nuances in the eicosanoid pathway with regard to specific cells. This work at large is a continuation of our comprehensive study of macrophage lipidomics and highlights our ability to use all of the information about the eicosanoid pathway.
ACKNOWLEDGMENTS
This work was supported by the LIPID Metabolites and Pathways Strategy (MAPS) Large-Scale Collaborative grant U54 GM069338 (E.A.D.) and R01 GM64611 (E.A.D.) from NIH. P.C.N. was supported by the University of California, San Diego, Graduate Training Program in Cellular and Molecular Pharmacology grant from the National Institute of General Medical Sciences T32 GM007752. We thank Dr. Yuan-Hao Hsu for assistance in preparing the manuscript. Protectin D1, Δ15t-Protectin D1, Resolvin D1, and Lipoxin B4 were kind gifts from Prof. Charles N. Serhan (Harvard University).
Footnotes
- 15d PGD2
- 15-deoxy-Δ12,14-PGD2
- 15d PGJ2
- 15-deoxy-Δ12,14-PGJ2
- AA
- arachidonic acid
- AdA
- adrenic acid
- ATCC
- American Type Culture Collection
- BMDM
- bone marrow-derived macrophages
- c-PGES
- cytosolic PGE2 synthase
- CT
- comparative threshold
- CYP
- cytochrome P450
- DGLA
- dihomo-γ linolenic acid
- DHA
- docosahexaenoic acid
- EPA
- eicosapentaenoic acid
- HHT
- heptadecatrienoic acid
- H-PGDS
- hematopoietic PGD2 synthase
- k
- keto
- KLA
- 3-deoxy-d-manno-2-octulosonic acid lipid A
- LC
- liquid chromatography
- L-PGDS
- lipocalin PGD2 synthase
- mPGES
- microsomal PGE2 synthase
- MRM
- multiple reaction monitoring
- MS
- mass spectrometry
- MS/MS
- tandem mass spectrometry
- MuLV
- murine leukemia virus
- PGDS
- PGD2 synthase
- PGES
- PGE2 synthase
- PGIS
- PGI2 synthase
- qRT-PCR
- quantitative RT-PCR
- RAW
- RAW264.7
- RPM
- resident peritoneal macrophage
- solvent A
- water-acetonitrile-acetic acid (70:30:0.02, v/v/v)
- SPE
- solid-phase extraction
- TBXS
- thromboxane A2 synthase
- TGEM
- thioglycollate-elicited macrophage
- Tx
- thromboxane
AUTHORSHIP
P.C.N. and E.A.D. designed the study with critical advice from C.K.G. P.C.N. and D.R. performed the experiments. D.S.D. contributed to data analysis and manuscript preparation. P.C.N. analyzed the data and wrote the manuscript. E.A.D. is the principal investigator and contributed to manuscript preparation. C.K.G. is the coprincipal investigator and contributed to manuscript preparation.
REFERENCES
- 1. Six D. A., Dennis E. A. (2000) The expanding superfamily of phospholipase A(2) enzymes: classification and characterization. Biochim. Biophys. Acta 1488, 1–19 [DOI] [PubMed] [Google Scholar]
- 2. Schaloske R. H., Dennis E. A. (2006) The phospholipase A2 superfamily and its group numbering system. Biochim. Biophys. Acta 1761, 1246–1259 [DOI] [PubMed] [Google Scholar]
- 3. Funk C. D. (2001) Prostaglandins and leukotrienes: advances in eicosanoid biology. Science 294, 1871–1875 [DOI] [PubMed] [Google Scholar]
- 4. Simmons D. L., Botting R. M., Hla T. (2004) Cyclooxygenase isozymes: the biology of prostaglandin synthesis and inhibition. Pharmacol. Rev. 56, 387–437 [DOI] [PubMed] [Google Scholar]
- 5. Smith W. L., DeWitt D. L., Garavito R. M. (2000) Cyclooxygenases: structural, cellular, and molecular biology. Annu. Rev. Biochem. 69, 145–182 [DOI] [PubMed] [Google Scholar]
- 6. Buczynski M. W., Dumlao D. S., Dennis E. A. (2009) Thematic Review Series: Proteomics. An integrated omics analysis of eicosanoid biology. J. Lipid Res. 50, 1015–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fitzpatrick F. A., Soberman R. (2001) Regulated formation of eicosanoids. J. Clin. Invest. 107, 1347–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mosser D. M., Edwards J. P. (2008) Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8, 958–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Van Furth R., Cohn Z. A. (1968) The origin and kinetics of mononuclear phagocytes. J. Exp. Med. 128, 415–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rouzer C. A., Jacobs A. T., Nirodi C. S., Kingsley P. J., Morrow J. D., Marnett L. J. (2005) RAW264.7 cells lack prostaglandin-dependent autoregulation of tumor necrosis factor-α secretion. J. Lipid Res. 46, 1027–1037 [DOI] [PubMed] [Google Scholar]
- 11. Rouzer C. A., Scott W. A., Hamill A. L., Cohn Z. A. (1980) Dynamics of leukotriene C production by macrophages. J. Exp. Med. 152, 1236–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tripp C. S., Unanue E. R., Needleman P. (1986) Monocyte migration explains the changes in macrophage arachidonate metabolism during the immune response. Proc. Natl. Acad. Sci. USA 83, 9655–9659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wenzel S. E., Trudeau J. B., Riches D. W., Westcott J. Y., Henson P. M. (1993) Peritoneal lavage fluid alters patterns of eicosanoid production in murine bone marrow-derived and peritoneal macrophages: dependency on inflammatory state of the peritoneum. Inflammation 17, 743–756 [DOI] [PubMed] [Google Scholar]
- 14. Naraba H., Murakami M., Matsumoto H., Shimbara S., Ueno A., Kudo I., Oh-ishi S. (1998) Segregated coupling of phospholipases A2, cyclooxygenases, and terminal prostanoid synthases in different phases of prostanoid biosynthesis in rat peritoneal macrophages. J. Immunol. 160, 2974–2982 [PubMed] [Google Scholar]
- 15. Jakobsson P. J., Thoren S., Morgenstern R., Samuelsson B. (1999) Identification of human prostaglandin E synthase: a microsomal, glutathione-dependent, inducible enzyme, constituting a potential novel drug target. Proc. Natl. Acad. Sci. USA 96, 7220–7225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Penglis P. S., Cleland L. G., Demasi M., Caughey G. E., James M. J. (2000) Differential regulation of prostaglandin E2 and thromboxane A2 production in human monocytes: implications for the use of cyclooxygenase inhibitors. J. Immunol. 165, 1605–1611 [DOI] [PubMed] [Google Scholar]
- 17. Ueno N., Takegoshi Y., Kamei D., Kudo I., Murakami M. (2005) Coupling between cyclooxygenases and terminal prostanoid synthases. Biochem. Biophys. Res. Commun. 338, 70–76 [DOI] [PubMed] [Google Scholar]
- 18. Raschke W. C., Baird S., Ralph P., Nakoinz I. (1978) Functional macrophage cell lines transformed by Abelson leukemia virus. Cell 15, 261–267 [DOI] [PubMed] [Google Scholar]
- 19. Hartley J. W., Evans L. H., Green K. Y., Naghashfar Z., Macias A. R., Zerfas P. M., Ward J. M. (2008) Expression of infectious murine leukemia viruses by RAW264.7 cells, a potential complication for studies with a widely used mouse macrophage cell line. Retrovirology 5, 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brock T. G., McNish R. W., Peters-Golden M. (1999) Arachidonic acid is preferentially metabolized by cyclooxygenase-2 to prostacyclin and prostaglandin E2. J. Biol. Chem. 274, 11660–11666 [DOI] [PubMed] [Google Scholar]
- 21. Buczynski M. W., Stephens D. L., Bowers-Gentry R. C., Grkovich A., Deems R. A., Dennis E. A. (2007) TLR-4 and sustained calcium agonists synergistically produce eicosanoids independent of protein synthesis in RAW264.7 cells. J. Biol. Chem. 282, 22834–22847 [DOI] [PubMed] [Google Scholar]
- 22. Raetz C. R., Garrett T. A., Reynolds C. M., Shaw W. A., Moore J. D., Smith D. C., Jr., Ribeiro A. A., Murphy R. C., Ulevitch R. J., Fearns C., Reichart D., Glass C. K., Benner C., Subramaniam S., Harkewicz R., Bowers-Gentry R. C., Buczynski M. W., Cooper J. A., Deems R. A., Dennis E. A. (2006) Kdo2-lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4. J. Lipid Res. 47, 1097–1111 [DOI] [PubMed] [Google Scholar]
- 23. Gupta S., Maurya M. R., Stephens D. L., Dennis E. A., Subramaniam S. (2009) An integrated model of eicosanoid metabolism and signaling based on lipidomics flux analysis. Biophys. J. 96, 4542–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blaho V. A., Buczynski M. W., Brown C. R., Dennis E. A. (2009) Lipidomic analysis of dynamic eicosanoid responses during the induction and resolution of Lyme arthritis. J. Biol. Chem. 284, 21599–21612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xiao G., Tsai A. L., Palmer G., Boyar W. C., Marshall P. J., Kulmacz R. J. (1997) Analysis of hydroperoxide-induced tyrosyl radicals and lipoxygenase activity in aspirin-treated human prostaglandin H synthase-2. Biochemistry 36, 1836–1845 [DOI] [PubMed] [Google Scholar]
- 26. Thuresson E. D., Lakkides K. M., Smith W. L. (2000) Different catalytically competent arrangements of arachidonic acid within the cyclooxygenase active site of prostaglandin endoperoxide H synthase-1 lead to the formation of different oxygenated products. J. Biol. Chem. 275, 8501–8507 [DOI] [PubMed] [Google Scholar]
- 27. Hecker M., Ullrich V., Fischer C., Meese C. O. (1987) Identification of novel arachidonic acid metabolites formed by prostaglandin H synthase. Eur. J. Biochem. 169, 113–123 [DOI] [PubMed] [Google Scholar]
- 28. Tripp C. S., Leahy K. M., Needleman P. (1985) Thromboxane synthase is preferentially conserved in activated mouse peritoneal macrophages. J. Clin. Invest. 76, 898–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murakami M., Naraba H., Tanioka T., Semmyo N., Nakatani Y., Kojima F., Ikeda T., Fueki M., Ueno A., Oh S., Kudo I. (2000) Regulation of prostaglandin E2 biosynthesis by inducible membrane-associated prostaglandin E2 synthase that acts in concert with cyclooxygenase-2. J. Biol. Chem. 275, 32783–32792 [DOI] [PubMed] [Google Scholar]
- 30. Murakami M., Nakashima K., Kamei D., Masuda S., Ishikawa Y., Ishii T., Ohmiya Y., Watanabe K., Kudo I. (2003) Cellular prostaglandin E2 production by membrane-bound prostaglandin E synthase-2 via both cyclooxygenases-1 and -2. J. Biol. Chem. 278, 37937–37947 [DOI] [PubMed] [Google Scholar]
- 31. Tanioka T., Nakatani Y., Semmyo N., Murakami M., Kudo I. (2000) Molecular identification of cytosolic prostaglandin E2 synthase that is functionally coupled with cyclooxygenase-1 in immediate prostaglandin E2 biosynthesis. J. Biol. Chem. 275, 32775–32782 [DOI] [PubMed] [Google Scholar]
- 32. Kämpfer H., Brautigam L., Geisslinger G., Pfeilschifter J., Frank S. (2003) Cyclooxygenase-1-coupled prostaglandin biosynthesis constitutes an essential prerequisite for skin repair. J. Invest. Dermatol. 120, 880–890 [DOI] [PubMed] [Google Scholar]
- 33. Joo M., Kwon M., Sadikot R. T., Kingsley P. J., Marnett L. J., Blackwell T. S., Peebles R. S., Jr., Urade Y., Christman J. W. (2007) Induction and function of lipocalin prostaglandin D synthase in host immunity. J. Immunol. 179, 2565–2575 [DOI] [PubMed] [Google Scholar]
- 34. Serhan C. N., Chiang N., Van Dyke T. E. (2008) Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat. Rev. Immunol. 8, 349–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harkewicz R., Fahy E., Andreyev A., Dennis E. A. (2007) Arachidonate-derived dihomoprostaglandin production observed in endotoxin-stimulated macrophage-like cells. J. Biol. Chem. 282, 2899–2910 [DOI] [PubMed] [Google Scholar]
- 36. Humes J. L., Burger S., Galavage M., Kuehl F. A., Jr., Wightman P. D., Dahlgren M. E., Davies P., Bonney R. J. (1980) The diminished production of arachidonic acid oxygenation products by elicited mouse peritoneal macrophages: possible mechanisms. J. Immunol. 124, 2110–2116 [PubMed] [Google Scholar]
- 37. Peres C. M., Procopio J., Costa M., Curi R. (1999) Thioglycolate-elicited rat macrophages exhibit alterations in incorporation and oxidation of fatty acids. Lipids 34, 1193–1197 [DOI] [PubMed] [Google Scholar]
- 38. Watanabe S., Kobayashi T., Okuyama H. (1998) Absence of relation between the expression of cyclooxygenase isoforms and the synthesis of prostaglandin E2 in resident and thioglycollate-elicited macrophages in rats. Prostaglandins Other Lipid Mediat. 56, 7–18 [DOI] [PubMed] [Google Scholar]
- 39. Ruan C. H., So S. P., Ruan K. H. (2011) Inducible COX-2 dominates over COX-1 in prostacyclin biosynthesis: mechanisms of COX-2 inhibitor risk to heart disease. Life Sci. 88, 24–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gourzi P., Leonova T., Papavasiliou F. N. (2007) Viral induction of AID is independent of the interferon and the Toll-like receptor signaling pathways but requires NF-κB. J. Exp. Med. 204, 259–265 [DOI] [PMC free article] [PubMed] [Google Scholar]