Abstract
A detailed understanding of the mechanisms by which tumors acquire resistance to targeted anticancer agents should speed the development of treatment strategies with lasting clinical efficacy. RAF inhibition in BRAF-mutant melanoma exemplifies the promise and challenge of many targeted drugs; although response rates are high, resistance invariably develops. Here, we articulate overarching principles of resistance to kinase inhibitors, as well as a translational approach to characterize resistance in the clinical setting through tumor mutation profiling. As a proof of principle, we performed targeted, massively parallel sequencing of 138 cancer genes in a tumor obtained from a patient with melanoma who developed resistance to PLX4032 after an initial dramatic response. The resulting profile identified an activating mutation at codon 121 in the downstream kinase MEK1 that was absent in the corresponding pretreatment tumor. The MEK1C121S mutation was shown to increase kinase activity and confer robust resistance to both RAF and MEK inhibition in vitro. Thus, MEK1C121S or functionally similar mutations are predicted to confer resistance to combined MEK/RAF inhibition. These results provide an instructive framework for assessing mechanisms of acquired resistance to kinase inhibition and illustrate the use of emerging technologies in a manner that may accelerate personalized cancer medicine.
INTRODUCTION
The marked expansion of tumor genomic characterization and new drugs in clinical development has enabled a steady increase in the use of molecular knowledge to guide oncology treatment decisions. Concomitantly, a shifting conceptual framework has emerged wherein salient genetic features may prove at least as decisive as anatomic origins to specify the optimal use and likelihood of response to targeted anticancer therapeutics. Protein kinase inhibitors have proved exemplary in this regard; deployment of these agents is commonly guided by knowledge of tumor genetic alterations that dysregulate cellular signaling mechanisms. Well-known examples include imatinib to treat tumors that contain activating mutations in ABL1 or KIT oncogenes,1–4 gefitinib or erlotinib in the setting of EGFR mutations,5–9 and trastuzumab in ERBB2-amplified cancers.10,11 Newer kinase inhibitors targeting BRAF in melanoma and ALK in lung cancer have shown similarly promising results in clinical trials.12–14
Although therapeutics directed against oncogenic kinases may yield dramatic responses and improved survival in cancers driven by a dominant oncogene, such tumors invariably become resistant to these agents.15–27 Thus, elucidating the mechanisms of acquired resistance (or its presence de novo) is essential to the development of new treatment strategies that improve the clinical benefit. In parallel, diagnostic advances that exploit increasingly powerful genomic technologies may be needed to profile individual tumors for the acquisition of specific resistance mechanisms. Ultimately, a comprehensive knowledge of drug resistance coupled with the ability to diagnose the relevant mechanisms in situ may enable therapeutic combinations capable of engendering prolonged responses in many oncogene-driven cancers.
Principles of Therapeutic Resistance in Kinase-Driven Cancers
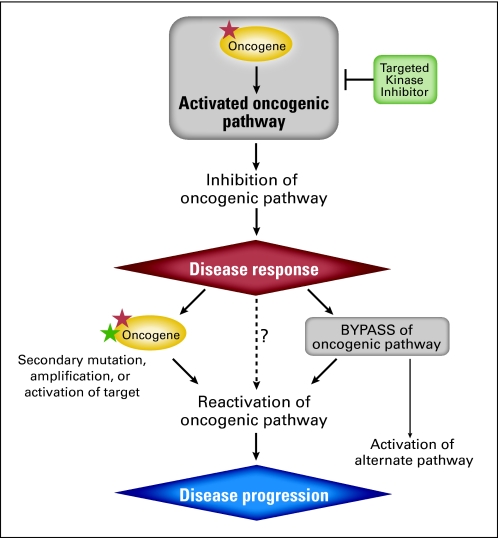
Mechanisms of resistance to anticancer agents may include increased drug efflux, modifications within the target protein(s), activation of downstream or redundant (eg, bypass) pathways, and induction of cell survival pathways.16,17,24,25,27–31 In cancers driven by kinase oncogenes, resistance mechanisms frequently engage the underlying tumor dependency elaborated by these oncogenes. As shown in Figure 1, driver tumor genetic alterations that activate protein kinases may engender an oncogene addiction phenotype, wherein the viability of tumor cells becomes excessively reliant on the cellular signal transduced by the mutated oncogene. This dependency results in a heightened therapeutic index, thereby allowing the relevant oncoprotein or cellular pathway to be intercepted therapeutically with modest (or at least manageable) adverse effects.32–36 As a general rule, oncogene-addicted tumors tend to develop resistance to oncogenic pathway inhibition by reactivating that pathway rather than by engaging completely new oncogenic pathways, although there can be exceptions to this rule (alternative pathway; Fig 1).37 In principle, then, any mechanism that reactivates an oncogenic pathway in the setting of therapeutic inhibition has the potential to engender acquired resistance.
Fig 1.
Kinase oncogene dependence and principles of drug resistance. Tumor genetic alterations (denoted by the red star) may activate protein kinase oncogenes, which in turn dysregulate a cell signaling pathway, resulting in oncogene dependence. Such tumors often respond to treatment using pharmacologic inhibitors of the mutated kinase oncoprotein; however, resistance to such agents is common. Categories of resistance to kinase inhibitors include secondary mutation (denoted by the green star), amplification, or activation of the target kinase, or bypass of the oncogenic pathway, both leading to downstream reactivation and disease progression. Bypass mechanisms activating alternative pathways have also been described (alternate pathway, see Emerging Mechanisms of Resistance to Kinase Oncogene Inhibition). In principle, reactivation of the oncogenic pathway through additional, as yet uncharacterized mechanisms (denoted by question mark) should also confer acquired resistance to targeted therapies.
Accordingly, one common mode of resistance to tyrosine kinase inhibitors involves additional mutations in the target oncogene (Fig 1).18–22,38–47 The oncogene may alternatively undergo gene amplification, which may override the effects of standard therapeutic dosing.18,47,48 A third mechanism involves upstream mutations that activate or upregulate the target oncoprotein.37,49 Thus, characterizing the spectrum of on-target genetic alterations that confer resistance to targeted agents represents an active area of investigation, because this knowledge may facilitate the design of new compounds that show enhanced efficacy against resistant variants.31,50
An alternative resistance mechanism involves elaboration of a bypass signal. In this case, additional genetic or adaptive changes reactivate the downstream pathway in the cancer cell without directly modifying the target oncoprotein (Fig 1). Amplification of the MET oncogene, which has been observed in up to 20% of EGFR-mutant lung cancers treated with erlotinib or gefitinib, provides an instructive example of the bypass phenomenon.51–53 The presence of bypass mechanisms may indicate additional drug targets that might be exploited in future salvage regimens or therapeutic combinations. Theoretically, activating mutations that affect signaling proteins situated downstream of the target oncoprotein might also be predicted to confer acquired resistance (Fig 1); however, such a mechanism has not been described in patients to date.
Melanoma As a Model for Characterizing Resistance to Kinase Inhibition
Recent therapeutic inroads in malignant melanoma highlight both the continuing promise of the driver oncogene-based treatment paradigm and the challenge of resistance to targeted therapeutics. Melanoma is the sixth most common cancer diagnosed in the United States, with 68,130 estimated new cases in 2010.54 Patients with metastatic melanoma exhibit a median survival of only 6 to 15 months. Uncontrolled activity of the MAP kinase (MAPK) pathway engenders an oncogene dependency in the vast majority of melanomas, most commonly through gain-of-function mutations involving codon 600 of the BRAF oncogene (BRAFV600E). More than 50% of metastatic melanomas harbor BRAFV600E mutations,55 which are also recurrent in colon and thyroid cancers.56–58
As noted earlier, clinical trials of drugs targeting mutated BRAF in melanoma have yielded favorable results. In a recent phase I trial of the RAF inhibitor PLX4032, 81% of patients with BRAFV600E melanoma (26 of 32 patients) experienced at least 30% tumor shrinkage by Response Evaluation Criteria in Solid Tumors (RECIST), with a complete response in two patients.14 However, resistance to PLX4032 always emerged, after responses that ranged from 2 to 18 months. Mechanisms of acquired resistance to RAF inhibition in melanoma have recently been described37,59–60a but remain poorly understood. Moreover, the clinical application of genomic approaches to diagnose salient resistance mechanisms in situ remains underdeveloped.
Here, we describe an approach to characterize genetic mechanisms of resistance through systematic mutation profiling. Starting with a patient with metastatic melanoma who developed resistance to PLX4032 after an initial dramatic response, we applied a massively parallel sequencing-based mutation profiling platform followed by experimental validation studies to gain new insights into resistance to RAF inhibition. Our results provide a framework for future targeted clinical trials in BRAF-mutant melanoma, illustrate the use of emerging technologies to characterize clinical resistance mechanisms, and inform an integrated strategy for the implementation of personalized cancer medicine.
METHODS
For complete methodologic details, see the Data Supplement. Sequencing studies were approved by the Broad Institute/Massachusetts Institute of Technology Institutional Review Board, and written informed consent was obtained from the patient.
Exon Capture and Sequencing
We used standard techniques to extract genomic DNA from the tumor and normal skin specimens. Massively parallel sequencing libraries (Illumina, San Diego, CA) were generated according to the manufacturer's directions. Hybrid selection was performed as previously described.61 Briefly, we designed and synthesized approximately 7,000 biotinylated RNA baits (Agilent, Santa Clara, CA) corresponding to the coding sequence of 138 genes known to undergo somatic genomic alterations in cancer (Data Supplement). Genomic DNA libraries were subjected to solution-phase hybrid capture using the RNA baits, followed by massively parallel sequencing. We sequenced 36 bases from both ends of library DNA fragments using an Illumina GAIIx instrument, achieving approximately 60 million purity filtered reads per sample. This yielded target gene haploid coverage of 937-fold and 918-fold from the normal skin and drug-resistant tumor biopsy, respectively. The sequencing data were analyzed for base mutations, insertions, deletions, and copy number alterations (Data Supplement). Mutations detected by massively parallel sequencing were validated using multibase hME extension chemistry by methods described previously.62,63
In Vitro Studies
MEK1 site-directed mutagenesis was performed using standard techniques. Growth inhibition analysis, immunoblot studies, and kinase assays were performed using standard protocols and are described in detail in the Data Supplement. Physical and biologic containment procedures for recombinant DNA followed institutional protocols in accordance with the National Institutes of Health Guidelines for Research Involving Recombinant DNA Molecules.
Random Mutagenesis Screen
Generation of mutagenized libraries was performed using a modification of published methods46,59 and is described in detail in the Data Supplement. MEK1 cDNA was isolated from cells and sequenced using an Illumina GAIIx instrument, as described previously.59
RESULTS
Patient Characteristics
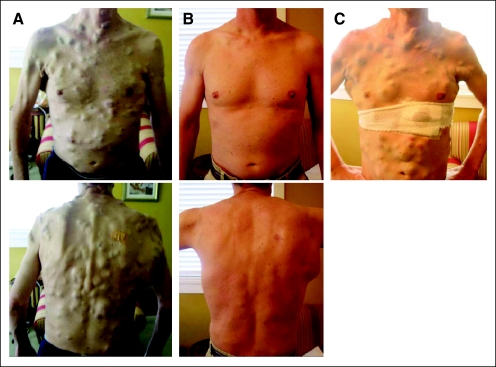
We characterized resistance to RAF inhibition in melanoma tissue obtained from a 38-year-old man who initially presented with a mass in his right axilla. A staging positron emission tomography/computed tomography demonstrated a large lesion in the right latissimus dorsi as well as multiple hypermetabolic foci in the lungs, liver, bones, and several subcutaneous sites. A biopsy of the latissimus dorsi mass revealed malignant melanoma. The tumor was refractory to several therapeutic regimens, including clinical trials of ipilimumab and dacarbazine and a combination of carboplatin, paclitaxel, interferon alfa, and interleukin-2. Disease progression included the interim development of numerous subcutaneous metastatic deposits (Fig 2A). Tumor genotyping studies identified the BRAFV600E mutation, and the patient was enrolled onto a clinical trial of the RAF inhibitor PLX4032. A profound clinical response ensued, including near-complete regression of all subcutaneous tumor nodules at 15 weeks on drug (Fig 2B).
Fig 2.
A 38-year-old man with BRAF-mutant melanoma and miliary, subcutaneous metastatic deposits. Photographs were taken (A) before initiation of PLX4032, (B) after 15 weeks of therapy with PLX4032, and (C) after relapse, after 23 weeks of therapy.
After 16 weeks on PLX4032, the patient experienced widespread disease relapse, which by 23 weeks involved most previous sites of visceral and subcutaneous disease (Fig 2C); PLX4032 was discontinued. A core biopsy of a relapsing subcutaneous thoracic nodule was obtained. The patient continued to have rapid disease progression and died several weeks later.
Mutation Profiling by Targeted, Massively Parallel Sequencing
To determine possible mechanisms of resistance to PLX4032, we isolated genomic DNA from the PLX4032-resistant tumor together with matched normal skin and performed targeted, massively parallel sequencing of all exons corresponding to 138 cancer genes (see Methods and Data Supplement). A total of 14 somatic base substitutions (nine missense and five synonymous mutations) were identified (Table 1). Of these, 79% (11 of 14 substitutions) consisted of CG>TA transitions typical of ultraviolet light exposure, a known environmental risk factor in the development of malignant melanoma. No insertions, deletions, or significant copy number alterations were identified at these loci. As shown in Table 1, missense mutations were seen in the ERBB4, FLT1, MEK1, PTPRD, RET, RUNX1T1, and TERT genes. The BRAFV600E mutation was also detected, as expected, with a mutant allele frequency of 37%. Each of these mutations was confirmed by mass spectrometric genotyping.
Table 1.
Somatic Alterations in the PLX4032-Resistant and PLX4032-Sensitive Tumor Samples in a Patient With Metastatic Melanoma
| Gene | PLX4032-Resistant Tumor |
PLX4032-Sensitive Tumor Protein Change | |||
|---|---|---|---|---|---|
| Genomic Change | Protein Change | Mutation Type | Allele Frequency (%) | ||
| BRAF | g.chr7:140099605A>T | p.V600E | Missense | 37 | p.V600E |
| BRCA1 | g.chr17:38497417C>T | p.E1172E | Synonymous | 75 | p.E1172E |
| BRCA1 | g.chr17:38499682G>A | p.T417T | Synonymous | 77 | p.T417T |
| ERBB4 | g.chr2:211956862C>T | p.G1217E | Missense | 24 | p.G1217E |
| FGFR4 | g.chr5:176454998C>T | p.I527I | Synonymous | 20 | p.I527I |
| FLT1 | g.chr13:27903435C>T | p.A276T | Missense | 66 | p.A276T |
| MEK1 | g.chr15:64516208G>C | p.C121S | Missense | 16 | WT |
| PDGFRB | g.chr5:149477517G>A | p.L998L | Synonymous | 57 | p.L998L |
| PTPRD | g.chr9:8490976C>T | p.E623K | Missense | 55 | p.E623K |
| PTPRD | g.chr9:8497431G>A | p.P503L | Missense | 55 | p.P503L |
| RET | g.chr10:42930184G>C | p.K710N | Missense | 28 | WT |
| RUNX1T1 | g.chr8:93052172C>T | p.D477N | Missense | 76 | p.D477N |
| TERT | g.chr5:1331863C>T | p.E727K | Missense | 58 | p.E727K |
| TERT | g.chr5:1331864C>T | p.T726T | Synonymous | 58 | p.T726T |
NOTE. All the exons from the 138 cancer genes were targeted for sequencing by massively parallel sequencing in the PLX4032-resistant sample. Fourteen somatic base substitutions were found. The original (PLX4032-sensitive) sample was queried for the presence of these mutations using mass spectrometric genotyping, demonstrating WT MEK1 and RET.
Abbreviation: WT, wild type.
MEK1 Mutation Associated With Acquired Resistance to RAF Inhibition
To identify point mutations that may have become enriched during PLX4032 treatment, we used mass spectrometric genotyping to query the original (pretreatment) tumor at these loci. As shown in Table 1, two of the nine missense mutations were undetectable in the original PLX4032-sensitive tumor by this approach. One of these conferred a cysteine-to-serine substitution at codon 121 (C121S) in MEK1, which encodes the kinase immediately downstream from BRAF in the MAPK pathway. The MEK1C121S mutant allele frequency was 16%. The other resistance-associated mutation resulted in a lysine-to-asparagine change at position 710 (K710N) in the RET oncogene, with a mutant allele frequency of 28%. These allele frequencies suggest but do not prove that the mutations were present in a subset of tumor cells in the relapsing specimen. In previous work using small-molecule MEK inhibitors, we discovered specific mutations within or proximal to the N-terminal regulatory domain of MEK1 that conferred modest cross resistance to RAF inhibition, in part through increased MEK kinase activity.59 In contrast, the RETK710N mutation has not previously been observed, and resides in a domain with no known function. Therefore, it seemed most likely that MEK1C121S engendered resistance to PLX4032 in this tumor.
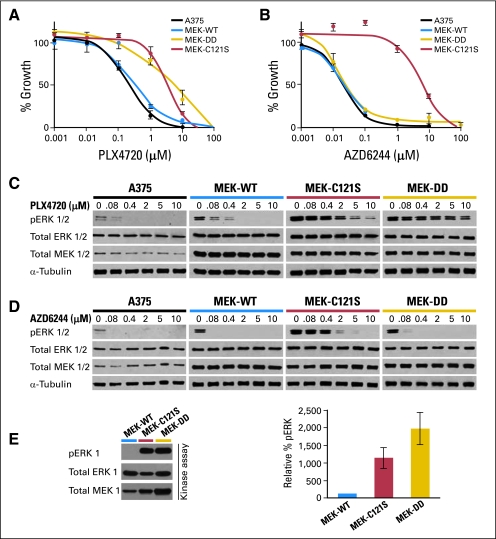
To test this hypothesis, we introduced the C121S mutation into the sequence of wild-type MEK1 (MEK-WT) and expressed the mutant cDNA in the A375 melanoma cell line, which harbors a BRAFV600E mutation and is highly sensitive to RAF and MEK inhibition. As shown in Figure 3A, cells expressing the MEK1C121S mutation were strongly resistant to PLX4720—a compound closely related to PLX4032—with a concentration required to achieve 50% growth inhibition (GI50) 100-fold higher than that observed in wild-type A375 cells or those expressing MEK-WT. This magnitude of resistance was similar to that conferred by a constitutively active variant of MEK1 (MEK-DD) (Fig 3A). In contrast, ectopic expression of the RETK710N allele had no effect on PLX4720 sensitivity in vitro (Data Supplement), further supporting the notion that the MEK1C121S mutation contributed a major genetic mechanism for resistance in this tumor.
Fig 3.
Pharmacologic and biochemical characterization of the MEK1C121S mutation. Growth inhibition curves for (A) the RAF inhibitor PLX4720 and (B) the MEK inhibitor AZD6244 are shown for wild-type A375 (BRAFV600E) melanoma cells (solid black) and A375 cells expressing MEK1C121S (red), wild-type MEK1 (MEK-WT; blue), or a constitutively active MEK1 variant (MEK-DD; gold). Effect of (C) PLX4720 and (D) AZD6244 on ERK1/2 phosphorylation (pERK 1/2) in wild-type A375 cells and those expressing MEK-WT, MEK1C121S, or MEK-DD. The levels of pERK1/2, total ERK1/2, MEK1/2, and α-tubulin are shown for A375 cells expressing MEK1 mutations after a 16-hour incubation at 0, 0.08, 0.4, 2, 5, and 10 μmol/L drug concentrations. (E) In vitro kinase assay measuring pERK in A375 cells expressing MEK1 mutations. Relative levels of pERK compared with total ERK1 and total MEK1 are shown.
Cells expressing MEK1C121S exhibited higher levels of phosphorylated ERK1/2 at baseline and when treated with PLX4720 than wild-type A375 cells or those expressing MEK-WT (Fig 3C), indicative of enhanced MAPK pathway activation. Moreover, MEK1C121S exhibited markedly elevated intrinsic kinase activity compared with MEK-WT (Fig 3E). We also tested the ability of MEK1C121S to confer cross resistance to a class of allosteric MEK inhibitors currently in clinical trials. As shown in Figure 3B, cells expressing MEK1C121S were also strongly resistant to the MEK inhibitor AZD6244, with a nearly 1,000-fold greater GI50 value than that of control A375 cells or cells expressing MEK-DD (which although constitutively active remain sensitive to allosteric inhibition). As with PLX4720, MEK1C121S-expressing cells also showed increased phosphorylated ERK1/2 relative to wild-type A375 cells or MEK-WT cells when treated with AZD6244 (Fig 3D).
MEK1 Mutagenesis Screen Reveals Additional Putative Resistance Mutations
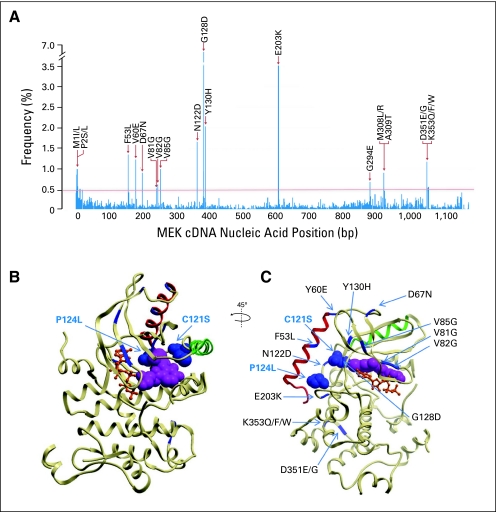
To determine the spectrum of potential mutations in MEK1 associated with resistance to RAF inhibition, we used a random mutagenesis screen together with massively parallel sequencing, as described previously.59 We expressed a saturating cDNA library of MEK1 mutations in A375 cells and cultured them for 4 weeks in the presence of fully inhibitory PLX4720 concentrations (1.5 μmol/L). We recovered and pooled approximately 1,000 resistant clones and characterized these en masse by massively parallel sequencing. A list of the most highly recurrent MEK1 mutations that emerged in the presence of PLX4720 is shown in Figure 4A. (A more complete list is available in the Data Supplement; of note, mutations at C121 were also observed, although they fell below the recurrence threshold used for this experiment).
Fig 4.
MEK1 mutations arising from an in vitro mutagenesis screen for resistance to RAF inhibition. (A) Recurrent mutations across the MEK1 coding sequence from a PLX4720 mutagenesis screen (based on approximately 1,000 sequenced clones) are shown. The corresponding amino acid substitutions from high-scoring mutations (> 0.4%) are indicated. (B and C) Locations of selected resistance alleles are indicated within the crystal structure of MEK1. Adenosine triphosphate (orange) and an allosteric, arylamine MEK inhibitor (PD318088; purple) are shown. Helix C (green) and helix A (red) are indicated. Mutations found to confer clinical resistance to RAF inhibition (C121S) and MEK inhibition (P124L) are indicated (blue spheres). Candidate mutations found in the mutagenesis screen are shown in blue. B and C show alternative views of the same crystal structure, with a 45-degree and slight inferior oblique rotation. bp, base pair.
We mapped these putative resistance mutations within the three-dimensional structure of the full-length MEK1 kinase domain, as shown in Figure 4C. The mutations were largely distinct from those that emerged from a similar screen for resistance to AZD624459; however, the most prevalent mutation (G128D) was observed in the prior screen. Moreover, two additional recurrent MEK1 mutations (N122D and Y130H) were located near the resistance alleles observed clinically (codon 121, seen here, and codon 124, reported previously59). Additional mutations mapped to the N terminus (eg, codons 53, 60, 67, and 81 to 85; Fig 4), some of which were located at or near residues implicated in the cardio-facio-cutaneous syndrome (a disorder characterized by germline mutations in RAS, RAF, and MEK proteins64–66). Other mutations localized to the C terminus of the protein, as noted previously (eg, codons 351 and 353; Fig 4)59; the functional significance of these mutations remains unknown. Taken together, these results suggest additional candidate mutations within MEK1 that may confer resistance to RAF inhibition. Some of these mutations would be predicted to emerge in future sequencing studies of patients with relapsing melanoma.
DISCUSSION
Using targeted, massively parallel sequencing, we identified an activating MEK1 mutation in a patient who developed resistance to a selective RAF inhibitor. Although the tumor was initially highly responsive to PLX4032 treatment, rapid disease progression ensued after 4 months of treatment. The MEK1C121S mutation results in increased kinase activity and confers resistance to both RAF and MEK inhibition in vitro. Taken together, these results indicate one likely mechanism by which this patient's tumor became resistant to RAF inhibition.
The MEK1C121S allele was not detected in the pretreatment biopsy sample by mass spectrometric genotyping; we speculate that it was present at low levels and underwent selection during the course of treatment. Indeed, studies of BCR-ABL, EGFR, and MET suggest that resistance mutations are commonly present at low levels in treatment-naive tumors and undergo clonal selection during treatment.19,51,67–71 Conceivably, this phenomenon might also help explain the development of simultaneous pan-resistance seen in this patient. Metastatic lesions have been shown to cross seed via circulating tumor cells, which may in principle distribute small numbers of resistant cells across many metastatic foci.72 In such a case, treatment-refractory clones could emerge at multiple sites simultaneously, resulting in a widespread resistance phenotype. Alternatively, multiple resistance mechanisms (genetic, epigenetic, and/or others) may have arisen independently in this patient.
Emerging Mechanisms of Resistance to Kinase Oncogene Inhibition
As noted earlier, genetic mechanisms of acquired resistance to targeted kinase inhibitors typically fall within one of the following two categories: mutations affecting the target kinase, or alterations of other genes within the target signaling pathway that may compensate for or bypass target oncoprotein inhibition (Fig 1 and Table 2). Resistance mutations in the target kinase commonly populate the catalytic domain, where they sterically impede drug binding while maintaining (or in some cases increasing) catalytic activity. This mechanism has been well described for several tyrosine kinases, including ABL, KIT, PDGFRA (imatinib), EGFR (erlotinib or gefitinib), FLT3 (PKC412), and ALK (crizotinib).18–22,38,40–47 Of particular importance are the gatekeeper mutations that occur in a conserved threonine residue near the kinase active site.19–21,45 These mutations substitute a larger hydrophobic residue for the conserved threonine, thereby increasing the kinase affinity for adenosine triphosphate (ATP). Because most kinase inhibitors in clinical use are ATP-competitive agents, the increased ATP affinity that results from gatekeeper substitutions provides a kinetic means of drug resistance.
Table 2.
Exemplary Mechanisms of Acquired Resistance to Kinase Inhibitors
| Targeted Agent | Target Gene | Acquired Resistance via Secondary Mutation, Amplification, or Activation of Target | Acquired Resistance via Bypass | Acquired Resistance via Downstream Mutation |
|---|---|---|---|---|
| Imatinib | ||||
| ABL | T315I | IGF1R amplification | ||
| Y253F/H | AXL overexpression*† | |||
| E255K/V | ||||
| ABL amplification | ||||
| T670I | ||||
| V654A | ||||
| D816A/G/H/V | ||||
| D820A/E/G/Y | ||||
| KIT | Y823D | |||
| KIT amplification | ||||
| PDGFRA | T674I | |||
| Gefitinib or erlotinib | EGFR | T790M | MET amplification | |
| D761Y | HGF overexpression*† | |||
| L747S | IGFBP3 loss*† | |||
| T854A | ||||
| EGFR amplification* | ||||
| Trastuzumab | HER2 | |||
| Lapatinib | HER2/EGFR | |||
| PKC412 | FLT3 | N676K | ||
| FGFR | ||||
| AZD6044 | MEK1 | MEK1 P124L | ||
| BRAF amplification* | ||||
| PLX4032 | BRAF | NRAS Q61K | COT overexpression† | MEK1 C121S |
| PDGFRβ overexpression† | ||||
| CRAF overexpression*† | ||||
| AXL overexpression*† | ||||
| HER2 overexpression*† | ||||
| Crizotinib | ALK/MET | L1196M | ||
| C1156Y | ||||
| F1174L |
Abbreviations: IGF1R, insulin-like growth factor 1 receptor; HGF, hepatocyte growth factor; IGFBP3, insulin-like growth factor receptor binding protein-3; PDGFRβ, platelet-derived growth factor β; HER2, human epidermal growth factor receptor 2.
Mechanisms that have been described in vitro.
Nongenetic mechanisms.
In addition to gatekeeper mutations, other types of second mutations in the target oncogene have also been reported, albeit less commonly.38–40,42,46 Some of these mutations destabilize the autoinhibitory (inactive) protein conformation bound by certain targeted drugs. Alternatively, gene amplification of the target kinase oncogene may override the ability of the drug to fully extinguish oncoprotein activity. Amplification as a means for resistance has been described for BCR-ABL and KIT in patients with chronic myeloid leukemia and GI stromal tumor (GIST), respectively, as well as for EGFR in lung cancer cell lines.18,47,48 Genetic alterations upstream of the target oncogene may provide an additional mechanism; these events cause resistance through upregulation or activation of the target oncoproteins. Toward this end, some BRAF-mutant melanoma tumors and cell lines that are resistant to RAF inhibition have been found to harbor NRAS mutations.37 Furthermore, melanoma cell lines cultured in vitro in the presence of MEK inhibitors may exhibit BRAF amplification.49 In the PLX4032-resistant melanoma examined here, we did not observe additional mutations or amplifications involving the BRAF or NRAS loci.
Bypass resistance mechanisms may involve genomic alterations that dysregulate a cellular effector acting in parallel to the drug target. As described in the introduction, the MET oncogene, which is amplified in approximately 20% of EGFR-mutant lung cancers after treatment with erlotinib or gefitinib, can activate PI3K/AKT and ERK signaling even in the presence of EGFR inhibition.51–53,73 Other bypass resistance mechanisms, including activation of IGF-1Rβ/IRS-1 signaling and signaling via the MET ligand HGF, have also been described in cell lines with acquired resistance to inhibition by erlotinib, gefitinib, and/or trastuzumab.51,73–77
In melanoma, several bypass mechanisms resulting in resistance to PLX4032 have been recently described. Elevated expression of the kinase COT (MAP3K8) drives resistance to PLX4032 in melanoma cell lines and, apparently, in some tumor samples.60 CRAF activation also results in resistance to PLX4032 in cell lines.60,78 Both COT and CRAF dysregulation reactivate the MAPK pathway. Another recently described bypass mechanism—upregulation of PDGFRβ—may activate a MAPK-independent pathway.37 Receptor tyrosine kinases such as AXL, ERBB2, and IGF1R may also confer resistance to RAF inhibition in a MAPK-independent manner, at least in vitro.60,60a In the current study, we observed mutations in the receptor tyrosine kinases ERBB4 and RET. Although ERBB4 mutations have been implicated in the pathogenesis of melanoma,79 the presence of the ERBB4 mutation in both pretreatment and postrelapse tumor DNA argues against a specific role in acquired resistance in this patient. Similarly, our in vitro analysis suggests that the RET mutation observed here is unlikely to contribute a major mechanism of resistance, although an additive role in vivo cannot be excluded.
The emergence of a somatic MEK1 mutation in the setting of clinical RAF inhibition represents the first reported example, to our knowledge, of an acquired resistance mechanism in which the tumor develops an activating mutation downstream of the target kinase. We previously described a MEK1 mutation involving codon 124 (MEK1P124L) arising in a patient with metastatic melanoma that developed resistance to the MEK inhibitor AZD6244.59 Like MEK1C121S, this mutation was proximal to the C helix and conferred resistance to AZD6244 as well as a modest cross resistance to PLX4720 (Fig 4B). The key difference here is that the MEK1C121S mutation arose in the setting of RAF inhibition rather than MEK inhibition. Moreover, the pharmacologic and biochemical effect of MEK1C121S was substantially greater than that of MEK1P124L. Although downstream resistance mutations have also been described in clinical and preclinical studies of de novo resistance to trastuzumab and lapatinib through activation of the PI3K pathway80–83 and to anti-EGFR therapy via KRAS activation,84–86 a role for such mechanisms in acquired resistance has not previously been demonstrated.
Therapeutic Implications of Emerging Resistance Mechanisms
Mechanisms of acquired resistance often have important therapeutic implications. Overcoming resistance mutations in kinase oncogenes may sometimes be achieved simply by increasing the dose of the targeted agent. Toward this end, several studies have demonstrated increased efficacy of elevated imatinib doses in patients with chronic myeloid leukemia who experience relapse at standard dose levels.87–92 Similarly, one phase III study of imatinib in GIST showed that 33% of patients who experienced progression on 400 mg of imatinib experienced a second response and/or disease stabilization when the dose was increased to 800 mg.93 Dose escalation might also be an option in the setting of resistance as a result of gene amplifications or the setting of altered pharmacokinetics resulting from certain types of resistance mutations.
However, dose escalation may be limited by adverse drug effects and has proved largely ineffective in the presence of gatekeeper mutations. Thus, ongoing discovery efforts are geared toward the development of new drugs to circumvent on-target kinase resistance mechanisms. One promising avenue involves agents engineered to maintain efficacious kinase inhibition despite the presence of resistance mutations. Dasatinib and nilotinib are examples of newer agents that exhibit clinical activity in the setting of various resistance mutations in ABL1.89,94–106 Similarly, sunitinib, an inhibitor of KIT and PDGF, is active against imatinib-resistant KIT and has been approved for use in imatinib-refractory GIST.26,107 Although the aforementioned agents suppress many resistance mutations, they typically fail to inhibit the gatekeeper mutations. In particular, the T315I gatekeeper mutation in ABL1 is refractory to dasatinib and nilotinib as well as imatinib. The development of agents that effectively inhibit gatekeeper mutations remains an area of intense study.97,102
New, irreversible inhibitors of EGFR undergoing evaluation in preclinical and clinical studies include neratinib (HKI-272), BIBW-2992, and PF-00299804.108–116 These agents covalently bind Cys-797 of EGFR, allowing them to inhibit erlotinib- and gefitinib-resistant mutant EGFR. Results from phase I and phase II trials suggest that these agents are reasonably well tolerated and may be efficacious in patients whose tumors have developed resistance to gefitinib or erlotinib.40,109,111,114–119 As with the ABL1 gatekeeper mutation described earlier, some of these experimental drugs may have limited efficacy against the EGFRT790M resistance mutation. For example, in vitro studies demonstrated that EGFRT790M conferred acquired resistance to PF-0099804.47,120 A recent phase II study of neratinib also found no benefit in patients with T790M mutations, although dose-limiting toxicity (diarrhea) may have precluded sufficient target inhibition in this case.114 However, neratinib did show evidence of benefit in the setting of a rare G719X EGFR mutation, emphasizing the potential importance of detailed characterization of resistance mechanisms in the clinical setting.
Unlike the aforementioned on-target resistance mechanisms where newer inhibitors may be substituted for the index agents, overriding bypass resistance mechanisms may require combinatorial treatment strategies. Here, the goal is to intercept both the primary oncogene dependency and the bypass mechanism simultaneously. The progression of EGFR-mutant lung cancers that develop resistance to gefitinib or erlotinib through MET amplification may be inhibited by dual treatment with EGFR and MET kinase inhibitors.52 This combination is being investigated in clinical trials of patients with acquired resistance to gefitinib and erlotinib. Similarly, the combination of gefitinib and insulin-like growth factor 1 receptor inhibitors has been shown to inhibit the growth of gefitinib-resistant tumor cells73 and is also under clinical investigation. Concomitant inhibition of downstream pathway reactivation is also being examined in clinical trials. For example, PI3K pathway inhibitors are being tested in combination with EGFR inhibitors, HER2 inhibitors, and inhibitors of the MAPK pathway to overcome both primary and acquired resistance.52,73,82,121–123
The discovery of resistance-associated MEK1 mutations in the setting of BRAF-mutant melanoma predicts that salvage therapies intercepting the MAPK pathway at the level of MEK—or even further downstream—might prove beneficial. Accordingly, the addition of a second MAPK pathway inhibitor to a RAF inhibitor may be required to overcome the effect of the MEK1 resistance mutations and perhaps other bypass mechanisms that reactivate MAPK signaling. More generally, such downstream resistance mutations highlight the potential utility of combination therapy directed against multiple effectors within a driver pathway as opposed to across multiple parallel signaling pathways.
However, MEK1C121S adds an additional layer of complexity; this mutation confers resistance to both RAF inhibition and the allosteric MEK inhibitor chemotype currently in clinical development. Thus, MEK1C121S or similar mutations might confer resistance to both drugs, even when given in combination. This prospect is of particular concern given that clinical trials of combination PLX4032 and AZD6244 are currently under way. Overcoming MEK1C121S or related resistance mechanisms may therefore require inhibition downstream of MEK or alternative mechanisms of inhibiting MEK. Toward this end, ATP-competitive MEK inhibitors or ERK inhibitors represent intriguing possibilities for future therapeutic avenues in BRAF- mutant melanoma.
Challenges to Diagnosis and Management of Targeted Therapeutic Resistance
This study illustrates how knowledge of acquired resistance to kinase inhibitors may guide the conception and deployment of new agents and combination therapies. At the same time, several challenges limit the systematic characterization of resistance mechanisms in the clinical and translational settings. Timely acquisition of high-quality tumor material constitutes one such challenge. Although tissue procurement at the time of relapse would greatly aid the systematic understanding of acquired resistance, such material is only occasionally available in current clinical protocols. Circumventing this barrier will require dedicated multidisciplinary teams and patients to implement rigorous scholarly efforts in this area.
In addition, systematic genetic profiling of cancers remains underdeveloped, even in the research setting. Robust approaches applicable to routine clinical material are needed to procure the salient genetic information from each tumor both before treatment and after relapse. The routine implementation of new genomic technologies in the clinical diagnostic arena, such as the targeted, massively parallel sequencing approach used here, may greatly enable the study of acquired resistance to targeted agents.
Another challenge involves discerning which of the multiple somatic genetic changes that may be observed exerts pivotal resistance mechanisms in any given tumor. This challenge is additionally confounded by the possibility that multiple resistance mechanisms may conceivably occur at multiple tumor foci in the same patient. Here, preclinical models remain crucial as companion experimental avenues. Indeed, multiple resistance mechanisms to imatinib, gefitinib, erlotinib, and PCK412 were first predicted from studies in model systems before they were verified in the clinical setting.41,46,53,124 Our work affirms the importance of such systems in melanoma as well.59 In particular, the dual implementation of in vitro and patient-centered characterization (eg, using massively parallel sequencing) can be highly enabling by streamlining the identification and affirmation of clinically relevant resistance mutations. In addition, dedicated studies that query resistance mechanisms in multiple relapsing tumor sites within a given patient (to the extent possible in the clinical arena) may provide critical information pertaining to the clinical complexity of tumor drug resistance.
Finally, the field of oncology still remains limited in its ability to test rational drug combinations in the clinical trial setting once resistance has developed, even when the mechanisms of resistance are known. As we become increasingly able to genetically characterize individual patients' tumors, there will be a pressing need to develop more efficient ways to test combined therapeutic strategies in defined molecular contexts.
Conclusions
Understanding resistance to targeted anticancer agents has gained increasing importance in light of the success of multiple kinase inhibitors. The experience with RAF inhibition in melanoma offers an example of both the challenges and opportunities inherent in translational studies of cancer drug resistance. Our results illuminate a clinical mechanism of acquired resistance to RAF inhibition that may inform rational therapeutic approaches to target this salient tumor dependency. Profiling BRAF-mutant melanomas for genetic alterations affecting MEK and possibly other downstream effectors when resistance emerges may diagnose the mechanism of resistance and specify optimal salvage therapy.
This work also highlights the power of systematic tumor genomic profiling both before treatment and after relapse to stratify patients based on tumor genotype and to identify clinically relevant resistance mechanisms. In turn, characterizing resistance mechanisms should allow the development of novel therapeutic strategies that achieve more durable responses in cancers driven by a dominant oncogene. Together, these approaches may offer an unprecedented ability to identify druggable genetic changes associated with tumor progression and thereby speed the advent of personalized cancer medicine.
Supplementary Material
Acknowledgment
We thank Brendan Blumenstiel, Matthew DeFelice, and Stacey Gabriel for assistance with hybrid selection and sequencing.
Footnotes
Supported by the National Institutes of Health Director's New Innovator Award, Grant No. DP2OD002750 (L.A.G.), Grants No. R33CA126674 and U24CA143867 from the National Cancer Institute (L.A.G., M.M., W.C.H.), the Starr Cancer Consortium (L.A.G., M.M.), the Melanoma Research Alliance (L.A.G.), the Novartis Institutes for BioMedical Research (L.A.G., M.M.), and the Burroughs-Wellcome Fund (L.A.G.).
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: Caroline Emery, Novartis Institutes for BioMedical Research (C) Consultant or Advisory Role: Nikhil Wagle, Foundation Medicine (C); Michael F. Berger, Foundation Medicine (C); Matthew J. Davis, Foundation Medicine (C); William C. Hahn, Novartis (C); Matthew Meyerson, Foundation Medicine (C), Novartis (C); Levi A. Garraway, Foundation Medicine (C), Novartis (C) Stock Ownership: Nikhil Wagle, Foundation Medicine; Matthew Meyerson, Foundation Medicine; Levi A. Garraway, Foundation Medicine Honoraria: None Research Funding: None Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Financial support: William C. Hahn, Matthew Meyerson, Levi A. Garraway
Manuscript writing: Nikhil Wagle, Caroline Emery, Michael F. Berger, Matthew J. Davis, Allison Sawyer, Panisa Pochanard, Sarah M. Kehoe, Cory M. Johannessen, Laura E. MacConaill, William C. Hahn, Matthew Meyerson, Levi A. Garraway
Final approval of manuscript: Nikhil Wagle, Caroline Emery, Michael F. Berger, Matthew J. Davis, Allison Sawyer, Panisa Pochanard, Sarah M. Kehoe, Cory M. Johannessen, Laura E. MacConaill, William C. Hahn, Matthew Meyerson, Levi A. Garraway
REFERENCES
- 1.Druker BJ. Inhibition of the Bcr-Abl tyrosine kinase as a therapeutic strategy for CML. Oncogene. 2002;21:8541–8546. doi: 10.1038/sj.onc.1206081. [DOI] [PubMed] [Google Scholar]
- 2.Demetri GD, von Mehren M, Blanke CD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 3.Druker BJ, Sawyers CL, Kantarjian H, et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001;344:1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- 4.Druker BJ, Talpaz M, Resta DJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 5.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 6.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 7.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haber DA, Bell DW, Sordella R, et al. Molecular targeted therapy of lung cancer: EGFR mutations and response to EGFR inhibitors. Cold Spring Harb Symp Quant Biol. 2005;70:419–426. doi: 10.1101/sqb.2005.70.043. [DOI] [PubMed] [Google Scholar]
- 9.Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 10.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 11.Vogel CL, Cobleigh MA, Tripathy D, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20:719–726. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- 12.Butrynski JE, D'Adamo DR, Hornick JL, et al. Crizotinib in ALK-rearranged inflammatory myofibroblastic tumor. N Engl J Med. 2010;363:1727–1733. doi: 10.1056/NEJMoa1007056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shannon KM. Resistance in the land of molecular cancer therapeutics. Cancer Cell. 2002;2:99–102. doi: 10.1016/s1535-6108(02)00101-0. [DOI] [PubMed] [Google Scholar]
- 16.Engelman JA, Settleman J. Acquired resistance to tyrosine kinase inhibitors during cancer therapy. Curr Opin Genet Dev. 2008;18:73–79. doi: 10.1016/j.gde.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Engelman JA, Jänne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2008;14:2895–2899. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 18.Gorre ME, Mohammed M, Ellwood K, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 19.Shah NP, Nicoll JM, Nagar B, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 21.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antonescu CR, Besmer P, Guo T, et al. Acquired resistance to imatinib in gastrointestinal stromal tumor occurs through secondary gene mutation. Clin Cancer Res. 2005;11:4182–4190. doi: 10.1158/1078-0432.CCR-04-2245. [DOI] [PubMed] [Google Scholar]
- 23.Ellis LM, Hicklin DJ. Resistance to targeted therapies: Refining anticancer therapy in the era of molecular oncology. Clin Cancer Res. 2009;15:7471–7478. doi: 10.1158/1078-0432.CCR-09-1070. [DOI] [PubMed] [Google Scholar]
- 24.Hammerman PS, Jänne PA, Johnson BE. Resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2009;15:7502–7509. doi: 10.1158/1078-0432.CCR-09-0189. [DOI] [PubMed] [Google Scholar]
- 25.Pohlmann PR, Mayer IA, Mernaugh R. Resistance to trastuzumab in breast cancer. Clin Cancer Res. 2009;15:7479–7491. doi: 10.1158/1078-0432.CCR-09-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gramza AW, Corless CL, Heinrich MC. Resistance to tyrosine kinase inhibitors in gastrointestinal stromal tumors. Clin Cancer Res. 2009;15:7510–7518. doi: 10.1158/1078-0432.CCR-09-0190. [DOI] [PubMed] [Google Scholar]
- 27.Milojkovic D, Apperley J. Mechanisms of resistance to imatinib and second-generation tyrosine inhibitors in chronic myeloid leukemia. Clin Cancer Res. 2009;15:7519–7527. doi: 10.1158/1078-0432.CCR-09-1068. [DOI] [PubMed] [Google Scholar]
- 28.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 29.van der Kuip H, Wohlbold L, Oetzel C, et al. Mechanisms of clinical resistance to small molecule tyrosine kinase inhibitors targeting oncogenic tyrosine kinases. Am J Pharmacogenomics. 2005;5:101–112. doi: 10.2165/00129785-200505020-00003. [DOI] [PubMed] [Google Scholar]
- 30.Camp ER, Summy J, Bauer TW, et al. Molecular mechanisms of resistance to therapies targeting the epidermal growth factor receptor. Clin Cancer Res. 2005;11:397–405. [PubMed] [Google Scholar]
- 31.Jänne PA, Gray N, Settleman J. Factors underlying sensitivity of cancers to small-molecule kinase inhibitors. Nat Rev Drug Discov. 2009;8:709–723. doi: 10.1038/nrd2871. [DOI] [PubMed] [Google Scholar]
- 32.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: Oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. [DOI] [PubMed] [Google Scholar]
- 34.Sharma SV, Settleman J. Oncogene addiction: Setting the stage for molecularly targeted cancer therapy. Genes Dev. 2007;21:3214–3231. doi: 10.1101/gad.1609907. [DOI] [PubMed] [Google Scholar]
- 35.Weinstein IB, Joe AK. Mechanisms of disease: Oncogene addiction—A rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol. 2006;3:448–457. doi: 10.1038/ncponc0558. [DOI] [PubMed] [Google Scholar]
- 36.Weinstein IB. Cancer: Addiction to oncogenes—The Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 37.Nazarian R, Shi H, Wang Q, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med. 2010;363:1734–1739. doi: 10.1056/NEJMoa1007478. [DOI] [PubMed] [Google Scholar]
- 39.Sasaki T, Okuda K, Zheng W, et al. The neuroblastoma associated F1174L ALK mutation causes resistance to an ALK kinase inhibitor in ALK translocated cancers. Cancer Res. 2010;70:10038–10043. doi: 10.1158/0008-5472.CAN-10-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bean J, Riely GJ, Balak M, et al. Acquired resistance to epidermal growth factor receptor kinase inhibitors associated with a novel T854A mutation in a patient with EGFR-mutant lung adenocarcinoma. Clin Cancer Res. 2008;14:7519–7525. doi: 10.1158/1078-0432.CCR-08-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogino A, Kitao H, Hirano S, et al. Emergence of epidermal growth factor receptor T790M mutation during chronic exposure to gefitinib in a non small cell lung cancer cell line. Cancer Res. 2007;67:7807–7814. doi: 10.1158/0008-5472.CAN-07-0681. [DOI] [PubMed] [Google Scholar]
- 42.Balak MN, Gong Y, Riely GJ, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12:6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 43.Heidel F, Solem FK, Breitenbuecher F, et al. Clinical resistance to the kinase inhibitor PKC412 in acute myeloid leukemia by mutation of Asn-676 in the FLT3 tyrosine kinase domain. Blood. 2006;107:293–300. doi: 10.1182/blood-2005-06-2469. [DOI] [PubMed] [Google Scholar]
- 44.Branford S, Rudzki Z, Walsh S, et al. High frequency of point mutations clustered within the adenosine triphosphate-binding region of BCR/ABL in patients with chronic myeloid leukemia or Ph-positive acute lymphoblastic leukemia who develop imatinib (STI571) resistance. Blood. 2002;99:3472–3475. doi: 10.1182/blood.v99.9.3472. [DOI] [PubMed] [Google Scholar]
- 45.Yun CH, Mengwasser KE, Toms AV, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Azam M, Latek RR, Daley GQ. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell. 2003;112:831–843. doi: 10.1016/s0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 47.Ercan D, Zejnullahu K, Yonesaka K, et al. Amplification of EGFR T790M causes resistance to an irreversible EGFR inhibitor. Oncogene. 2010;29:2346–2356. doi: 10.1038/onc.2009.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Debiec-Rychter M, Cools J, Dumez H, et al. Mechanisms of resistance to imatinib mesylate in gastrointestinal stromal tumors and activity of the PKC412 inhibitor against imatinib-resistant mutants. Gastroenterology. 2005;128:270–279. doi: 10.1053/j.gastro.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 49.Corcoran RB, Dias-Santagata D, Bergethon K, et al. BRAF gene amplification can promote acquired resistance to MEK inhibitors in cancer cells harboring the BRAF V600E mutation. Sci Signal. 2010;3:ra84. doi: 10.1126/scisignal.2001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McDermott U, Settleman J. Personalized cancer therapy with selective kinase inhibitors: An emerging paradigm in medical oncology. J Clin Oncol. 2009;27:5650–5659. doi: 10.1200/JCO.2009.22.9054. [DOI] [PubMed] [Google Scholar]
- 51.Turke AB, Zejnullahu K, Wu YL, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 53.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.American Cancer Society: Cancer Facts and Figures 2010. Atlanta, GA: American Cancer Society; 2010. [Google Scholar]
- 55.Fecher LA, Cummings SD, Keefe MJ, et al. Toward a molecular classification of melanoma. J Clin Oncol. 2007;25:1606–1620. doi: 10.1200/JCO.2006.06.0442. [DOI] [PubMed] [Google Scholar]
- 56.Kimura ET, Nikiforova MN, Zhu Z, et al. High prevalence of BRAF mutations in thyroid cancer: Genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res. 2003;63:1454–1457. [PubMed] [Google Scholar]
- 57.Cohen Y, Xing M, Mambo E, et al. BRAF mutation in papillary thyroid carcinoma. J Natl Cancer Inst. 2003;95:625–627. doi: 10.1093/jnci/95.8.625. [DOI] [PubMed] [Google Scholar]
- 58.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 59.Emery CM, Vijayendran KG, Zipser MC, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc Natl Acad Sci U S A. 2009;106:20411–20416. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johannessen CM, Boehm JS, Kim SY, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60a.Villanueva J, Vultur A, Lee JT, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gnirke A, Melnikov A, Maguire J, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.MacConaill LE, Campbell CD, Kehoe SM, et al. Profiling critical cancer gene mutations in clinical tumor samples. PLoS One. 2009;4:e7887. doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomas RK, Baker AC, Debiasi RM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 64.Niihori T, Aoki Y, Narumi Y, et al. Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat Genet. 2006;38:294–296. doi: 10.1038/ng1749. [DOI] [PubMed] [Google Scholar]
- 65.Roberts A, Allanson J, Jadico SK, et al. The cardiofaciocutaneous syndrome. J Med Genet. 2006;43:833–842. doi: 10.1136/jmg.2006.042796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rodriguez-Viciana P, Tetsu O, Tidyman WE, et al. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science. 2006;311:1287–1290. doi: 10.1126/science.1124642. [DOI] [PubMed] [Google Scholar]
- 67.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hofmann WK, Komor M, Wassmann B, et al. Presence of the BCR-ABL mutation Glu255Lys prior to STI571 (imatinib) treatment in patients with Ph+ acute lymphoblastic leukemia. Blood. 2003;102:659–661. doi: 10.1182/blood-2002-06-1756. [DOI] [PubMed] [Google Scholar]
- 69.Michor F, Hughes TP, Iwasa Y, et al. Dynamics of chronic myeloid leukaemia. Nature. 2005;435:1267–1270. doi: 10.1038/nature03669. [DOI] [PubMed] [Google Scholar]
- 70.Roche-Lestienne C, Laï JL, Darré S, et al. A mutation conferring resistance to imatinib at the time of diagnosis of chronic myelogenous leukemia. N Engl J Med. 2003;348:2265–2266. doi: 10.1056/NEJMc035089. [DOI] [PubMed] [Google Scholar]
- 71.Roche-Lestienne C, Soenen-Cornu V, Grardel-Duflos N, et al. Several types of mutations of the Abl gene can be found in chronic myeloid leukemia patients resistant to STI571, and they can pre-exist to the onset of treatment. Blood. 2002;100:1014–1018. doi: 10.1182/blood.v100.3.1014. [DOI] [PubMed] [Google Scholar]
- 72.Kim MY, Oskarsson T, Acharyya S, et al. Tumor self-seeding by circulating cancer cells. Cell. 2009;139:1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guix M, Faber AC, Wang SE, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest. 2008;118:2609–2619. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yano S, Wang W, Li Q, et al. Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor-activating mutations. Cancer Res. 2008;68:9479–9487. doi: 10.1158/0008-5472.CAN-08-1643. [DOI] [PubMed] [Google Scholar]
- 75.Nahta R, Yuan LX, Zhang B, et al. Insulin-like growth factor-I receptor/human epidermal growth factor receptor 2 heterodimerization contributes to trastuzumab resistance of breast cancer cells. Cancer Res. 2005;65:11118–11128. doi: 10.1158/0008-5472.CAN-04-3841. [DOI] [PubMed] [Google Scholar]
- 76.Lu Y, Zi X, Pollak M. Molecular mechanisms underlying IGF-I-induced attenuation of the growth-inhibitory activity of trastuzumab (Herceptin) on SKBR3 breast cancer cells. Int J Cancer. 2004;108:334–341. doi: 10.1002/ijc.11445. [DOI] [PubMed] [Google Scholar]
- 77.Lu Y, Zi X, Zhao Y, et al. Insulin-like growth factor-I receptor signaling and resistance to trastuzumab (Herceptin). J Natl Cancer Inst. 2001;93:1852–1857. doi: 10.1093/jnci/93.24.1852. [DOI] [PubMed] [Google Scholar]
- 78.Montagut C, Sharma SV, Shioda T, et al. Elevated CRAF as a potential mechanism of acquired resistance to BRAF inhibition in melanoma. Cancer Res. 2008;68:4853–4861. doi: 10.1158/0008-5472.CAN-07-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Prickett TD, Agrawal NS, Wei X, et al. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet. 2009;41:1127–1132. doi: 10.1038/ng.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nagata Y, Lan KH, Zhou X, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6:117–127. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 81.O'Brien NA, Browne BC, Chow L, et al. Activated phosphoinositide 3-kinase/AKT signaling confers resistance to trastuzumab but not lapatinib. Mol Cancer Ther. 2010;9:1489–1502. doi: 10.1158/1535-7163.MCT-09-1171. [DOI] [PubMed] [Google Scholar]
- 82.Eichhorn PJ, Gili M, Scaltriti M, et al. Phosphatidylinositol 3-kinase hyperactivation results in lapatinib resistance that is reversed by the mTOR/phosphatidylinositol 3-kinase inhibitor NVP-BEZ235. Cancer Res. 2008;68:9221–9230. doi: 10.1158/0008-5472.CAN-08-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pandolfi PP. Breast cancer: Loss of PTEN predicts resistance to treatment. N Engl J Med. 2004;351:2337–2338. doi: 10.1056/NEJMcibr043143. [DOI] [PubMed] [Google Scholar]
- 84.Takeda M, Okamoto I, Fujita Y, et al. De novo resistance to epidermal growth factor receptor-tyrosine kinase inhibitors in EGFR mutation-positive patients with non-small cell lung cancer. J Thorac Oncol. 2010;5:399–400. doi: 10.1097/JTO.0b013e3181cee47e. [DOI] [PubMed] [Google Scholar]
- 85.Mao C, Qiu LX, Liao RY, et al. KRAS mutations and resistance to EGFR-TKIs treatment in patients with non-small cell lung cancer: A meta-analysis of 22 studies. Lung Cancer. 2010;69:272–278. doi: 10.1016/j.lungcan.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 86.Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol. 2010;28:1254–1261. doi: 10.1200/JCO.2009.24.6116. [DOI] [PubMed] [Google Scholar]
- 87.Jabbour E, Cortes JE, Kantarjian HM. Suboptimal response to or failure of imatinib treatment for chronic myeloid leukemia: What is the optimal strategy? Mayo Clin Proc. 2009;84:161–169. doi: 10.4065/84.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kantarjian HM, Larson RA, Guilhot F, et al. Efficacy of imatinib dose escalation in patients with chronic myeloid leukemia in chronic phase. Cancer. 2009;115:551–560. doi: 10.1002/cncr.24066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kantarjian H, Pasquini R, Lévy V, et al. Dasatinib or high-dose imatinib for chronic-phase chronic myeloid leukemia resistant to imatinib at a dose of 400 to 600 milligrams daily: Two-year follow-up of a randomized phase 2 study (START-R) Cancer. 2009;115:4136–4147. doi: 10.1002/cncr.24504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jabbour E, Kantarjian HM, Jones D, et al. Imatinib mesylate dose escalation is associated with durable responses in patients with chronic myeloid leukemia after cytogenetic failure on standard-dose imatinib therapy. Blood. 2009;113:2154–2160. doi: 10.1182/blood-2008-04-154344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kantarjian HM, Talpaz M, O'Brien S, et al. Dose escalation of imatinib mesylate can overcome resistance to standard-dose therapy in patients with chronic myelogenous leukemia. Blood. 2003;101:473–475. doi: 10.1182/blood-2002-05-1451. [DOI] [PubMed] [Google Scholar]
- 92.Zonder JA, Pemberton P, Brandt H, et al. The effect of dose increase of imatinib mesylate in patients with chronic or accelerated phase chronic myelogenous leukemia with inadequate hematologic or cytogenetic response to initial treatment. Clin Cancer Res. 2003;9:2092–2097. [PubMed] [Google Scholar]
- 93.Blanke CD, Rankin C, Demetri GD, et al. Phase III randomized, intergroup trial assessing imatinib mesylate at two dose levels in patients with unresectable or metastatic gastrointestinal stromal tumors expressing the kit receptor tyrosine kinase: S0033. J Clin Oncol. 2008;26:626–632. doi: 10.1200/JCO.2007.13.4452. [DOI] [PubMed] [Google Scholar]
- 94.Kantarjian H, Shah NP, Hochhaus A, et al. Dasatinib versus imatinib in newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2010;362:2260–2270. doi: 10.1056/NEJMoa1002315. [DOI] [PubMed] [Google Scholar]
- 95.Shah NP, Kim DW, Kantarjian H, et al. Potent, transient inhibition of BCR-ABL with dasatinib 100 mg daily achieves rapid and durable cytogenetic responses and high transformation-free survival rates in chronic phase chronic myeloid leukemia patients with resistance, suboptimal response or intolerance to imatinib. Haematologica. 2010;95:232–240. doi: 10.3324/haematol.2009.011452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Saglio G, Kim DW, Issaragrisil S, et al. Nilotinib versus imatinib for newly diagnosed chronic myeloid leukemia. N Engl J Med. 2010;362:2251–2259. doi: 10.1056/NEJMoa0912614. [DOI] [PubMed] [Google Scholar]
- 97.O'Hare T, Shakespeare WC, Zhu X, et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009;16:401–412. doi: 10.1016/j.ccr.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Weisberg E, Manley PW, Cowan-Jacob SW, et al. Second generation inhibitors of BCR-ABL for the treatment of imatinib-resistant chronic myeloid leukaemia. Nat Rev Cancer. 2007;7:345–356. doi: 10.1038/nrc2126. [DOI] [PubMed] [Google Scholar]
- 99.Quintas-Cardama A, Kantarjian H, Jones D, et al. Dasatinib (BMS-354825) is active in Philadelphia chromosome-positive chronic myelogenous leukemia after imatinib and nilotinib (AMN107) therapy failure. Blood. 2007;109:497–499. doi: 10.1182/blood-2006-07-035493. [DOI] [PubMed] [Google Scholar]
- 100.Jabbour E, Cortes J, Giles F, et al. Drug evaluation: Nilotinib—A novel Bcr-Abl tyrosine kinase inhibitor for the treatment of chronic myelocytic leukemia and beyond. IDrugs. 2007;10:468–479. [PubMed] [Google Scholar]
- 101.Jabbour E, Cortes J, Kantarjian H, et al. Novel tyrosine kinase inhibitor therapy before allogeneic stem cell transplantation in patients with chronic myeloid leukemia: No evidence for increased transplant-related toxicity. Cancer. 2007;110:340–344. doi: 10.1002/cncr.22778. [DOI] [PubMed] [Google Scholar]
- 102.Giles FJ, Cortes J, Jones D, et al. MK-0457, a novel kinase inhibitor, is active in patients with chronic myeloid leukemia or acute lymphocytic leukemia with the T315I BCR-ABL mutation. Blood. 2007;109:500–502. doi: 10.1182/blood-2006-05-025049. [DOI] [PubMed] [Google Scholar]
- 103.Kantarjian H, Giles F, Wunderle L, et al. Nilotinib in imatinib-resistant CML and Philadelphia chromosome-positive ALL. N Engl J Med. 2006;354:2542–2551. doi: 10.1056/NEJMoa055104. [DOI] [PubMed] [Google Scholar]
- 104.Borthakur G, Kantarjian H, Daley G, et al. Pilot study of lonafarnib, a farnesyl transferase inhibitor, in patients with chronic myeloid leukemia in the chronic or accelerated phase that is resistant or refractory to imatinib therapy. Cancer. 2006;106:346–352. doi: 10.1002/cncr.21590. [DOI] [PubMed] [Google Scholar]
- 105.Weisberg E, Manley PW, Breitenstein W, et al. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell. 2005;7:129–141. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 106.Shah NP, Tran C, Lee FY, et al. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 107.Demetri GD, van Oosterom AT, Garrett CR, et al. Efficacy and safety of sunitinib in patients with advanced gastrointestinal stromal tumour after failure of imatinib: A randomised controlled trial. Lancet. 2006;368:1329–1338. doi: 10.1016/S0140-6736(06)69446-4. [DOI] [PubMed] [Google Scholar]
- 108.Engelman JA, Zejnullahu K, Gale CM, et al. PF00299804, an irreversible pan-ERBB inhibitor, is effective in lung cancer models with EGFR and ERBB2 mutations that are resistant to gefitinib. Cancer Res. 2007;67:11924–11932. doi: 10.1158/0008-5472.CAN-07-1885. [DOI] [PubMed] [Google Scholar]
- 109.Wong KK, Fracasso PM, Bukowski RM, et al. A phase I study with neratinib (HKI-272), an irreversible pan ErbB receptor tyrosine kinase inhibitor, in patients with solid tumors. Clin Cancer Res. 2009;15:2552–2558. doi: 10.1158/1078-0432.CCR-08-1978. [DOI] [PubMed] [Google Scholar]
- 110.Zhou W, Ercan D, Chen L, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Eskens FA, Mom CH, Planting AS, et al. A phase I dose escalation study of BIBW 2992, an irreversible dual inhibitor of epidermal growth factor receptor 1 (EGFR) and 2 (HER2) tyrosine kinase in a 2-week on, 2-week off schedule in patients with advanced solid tumours. Br J Cancer. 2008;98:80–85. doi: 10.1038/sj.bjc.6604108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kwak EL, Sordella R, Bell DW, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc Natl Acad Sci U S A. 2005;102:7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rabindran SK, Discafani CM, Rosfjord EC, et al. Antitumor activity of HKI-272, an orally active, irreversible inhibitor of the HER-2 tyrosine kinase. Cancer Res. 2004;64:3958–3965. doi: 10.1158/0008-5472.CAN-03-2868. [DOI] [PubMed] [Google Scholar]
- 114.Sequist LV, Besse B, Lynch TJ, et al. Neratinib, an irreversible pan-ErbB receptor tyrosine kinase inhibitor: Results of a phase II trial in patients with advanced non-small-cell lung cancer. J Clin Oncol. 2010;28:3076–3083. doi: 10.1200/JCO.2009.27.9414. [DOI] [PubMed] [Google Scholar]
- 115.Kelly RJ, Carter C, Giaccone G. Personalizing therapy in an epidermal growth factor receptor-tyrosine kinase inhibitor-resistant non-small-cell lung cancer using PF-00299804 and trastuzumab. J Clin Oncol. 2010;28:e507–e510. doi: 10.1200/JCO.2010.29.3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yap TA, Vidal L, Adam J, et al. Phase I trial of the irreversible EGFR and HER2 kinase inhibitor BIBW 2992 in patients with advanced solid tumors. J Clin Oncol. 2010;28:3965–3972. doi: 10.1200/JCO.2009.26.7278. [DOI] [PubMed] [Google Scholar]
- 117.Janne PA, Schellens JH, Engelman JA, et al. Preliminary activity and safety results from a phase I clinical trial of PF-00299804, an irreversible pan-HER inhibitor, in patients (pts) with NSCLC. J Clin Oncol. 2008;26(suppl):430s. abstr 8027. [Google Scholar]
- 118.Miller VA, Wakelee HA, Lara PN, et al. Activity and tolerance of XL647 in NSCLC patients with acquired resistance to EGFR-TKIs: Preliminary results of a phase II trial. J Clin Oncol. 2008;26(suppl):430s. abstr 8028. [Google Scholar]
- 119.Shih J, Yang C, Su W, et al. A phase II study of BIBW 2992, a novel irreversible dual EGFR and HER2 tyrosine kinase inhibitor (TKI), in patients with adenocarcinoma of the lung and activating EGFR mutations after failure of one line of chemotherapy (LUX-Lung 2) J Clin Oncol. 2009;27(suppl):410s. abstr 8013. [Google Scholar]
- 120.Godin-Heymann N, Ulkus L, Brannigan BW, et al. The T790M “gatekeeper” mutation in EGFR mediates resistance to low concentrations of an irreversible EGFR inhibitor. Mol Cancer Ther. 2008;7:874–879. doi: 10.1158/1535-7163.MCT-07-2387. [DOI] [PubMed] [Google Scholar]
- 121.Junttila TT, Akita RW, Parsons K, et al. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell. 2009;15:429–440. doi: 10.1016/j.ccr.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 122.Rexer BN, Engelman JA, Arteaga CL. Overcoming resistance to tyrosine kinase inhibitors: Lessons learned from cancer cells treated with EGFR antagonists. Cell Cycle. 2009;8:18–22. doi: 10.4161/cc.8.1.7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cools J, Mentens N, Furet P, et al. Prediction of resistance to small molecule FLT3 inhibitors: Implications for molecularly targeted therapy of acute leukemia. Cancer Res. 2004;64:6385–6389. doi: 10.1158/0008-5472.CAN-04-2148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.