Abstract
The humoral immune response is a highly specific and adaptive sensor for changes in the body's protein milieu, which responds to novel structures of both foreign and self antigens. Although immunoglobulins represent a major component of human serum and are vital to survival, little is known about the response specificity and determinants that govern the human immunome. Historically, antigen-specific humoral immunity has been investigated using individually-produced and purified target proteins, a labor-intensive process that has limited the number of antigens that have been studied. Here, we present the development of methods for applying self-assembling protein microarrays and a related method for producing 96-well formatted macroarrays for monitoring the humoral response at the proteome scale. Using plasmids encoding full-length cDNAs for over 850 human proteins and 1700 pathogen proteins, we demonstrate that these microarrays are highly sensitive, specific, reproducible, and can simultaneously measure immunity to thousands of proteins without a priori protein purification. Using this approach, we demonstrate the detection of humoral immunity to known and novel self-antigens, cancer antigens, autoimmune antigens, as well as pathogen-derived antigens. This represents a powerful and versatile tool for monitoring the immunome in health and disease.
Introduction
The humoral immune system, which is one branch of adaptive immunity, plays a potent role in a vast array of human diseases. Antibodies are produced by B cells, and have highly mutagenic variable regions that result in reversible noncovalent interactions with target antigens with high affinities. The antigen binding regions of antibodies have aromatic amino acids to enhance van der Waals and hydrophobic interactions [1]. Due to genetic recombination and somatic hypermutation, it is estimated that the immunoglobulin repertoire may contain at least 1011 unique antibodies [1].
The induction of antibodies is a primary mechanism of protection against pathogens, by triggering a cascade of immune activation of complement and opsonization. In contrast, antibodies to self antigens, called autoantibodies, can lead to devastating autoimmune diseases in some cases, but may offer protection against diseases like cancer in others. Whereas the specificity of antibody responses to pathogens with relatively few proteins, such as HIV, are well-defined, the breadth of antibody specificities to most pathogens, especially those with large protein complements, is not well known [2, 3]. Even less is known about the autoimmune repertoire in health and disease [4]. An integrated “systems immunology” approach is needed for the global analysis of immune responses [5].
Modern vaccine development relies on defining the humoral immune response against microbial pathogens. Only a small fraction of a pathogen's proteome triggers humoral immunity and only a fraction of these induces protective immunity [6]. Identification of these immunoprotective antigens enables the design of targeted vaccines and limits toxicities due to non-protective antigens. For example, the initial vaccine developed against the causative agent for whooping cough, Bordetella pertussis, contained inactivated bacteria. This vaccine was protective but concerns about its safety limited its use. Of the 3,816 genes in the Bordetella proteome, only four proteins were necessary to induce effective immunity, which are now the components of the recently developed acellular pertussis vaccine [7, 8]. Another example is vaccinia, used worldwide for smallpox vaccination. Vaccinia contains approximately 200 proteins, but little is known about the key targets of humoral immunity [9]. The use of vaccinia is associated with clinical complications, including severe generalized vaccinia in immunocompromised hosts, so that identification of the targets of effective antibody immunity to tailor vaccine design would have profound clinical consequences.
In contrast to pathogen-specific antibodies, which can be advantageous, autoimmune disease represents the inappropriate development of both humoral and cellular immunity against self-derived antigens, leading to tissue damage. The incidence of autoimmunity is about 90 per 100,000 persons per year, with a prevalence of approximately 3% of the population [10]. In diabetes, as well as other autoimmune diseases such as rheumatoid arthritis and systemic lupus, there exists a direct relationship between autoantibody titer to specific antigens and disease severity [11]. For example, type 1 diabetes mellitus (IDDM) is an autoimmune disease targeting pancreatic insulin-secreting beta cells, leading to inadequate insulin secretion, hyperglycemia and ketoacidosis [12, 13]. IDDM is associated with the autoantibodies to islet cell autoantigens (ICAs) including GAD65, IA2 and insulin. Antibodies to GAD65 and IA2 can be detected in as many as 70% of newly diagnosed IDDM patients. The appearance of these autoantibodies can be detected years prior to the clinical presentation [14, 15] and may predict disease progression. Multiplexed testing for all three autoantibodies increases the sensitivity of disease detection to >85% and the specificity to 98% at the onset of the disease [16, 17]. These findings strongly suggest that the identification of new autoantigens would improve the early detection of at-risk individuals and predict therapeutic outcomes.
Cancer also induces autoantibodies directed to self antigens, in this case referred to as tumor antigens [18]. These autoantibodies can be detected at early cancer stages and prior to cancer diagnosis revealing their potential as biomarkers [18–20]. Autoantibodies against individual antigens may be prevalent in only a fraction of a specific disease cohort, limiting the sensitivity of single autoantibodies as cancer biomarkers. This can be improved by simultaneously testing for antibodies to multiple tumor antigens [18–20], and several recent studies using proteomic technologies point to the utility of these antibodies as biomarkers of disease [21–23].
There are multiple methods for detecting antibodies in patient sera. In the enzyme-linked immunosorbent assay (ELISA), the gold standard used in clinical testing laboratories, purified recombinant proteins are adhered onto 96-well plastic plates and probed with patient or control sera added in dilutions and then detected with standard detection reagents. Significant effort is required to produce the protein for this method, limiting its application to proteins that are either known antigens or strong candidates. The development of phage expression systems (SEREX and phage display) has led to the identification of multiple autoantigens [24]. In these methods, cDNA libraries are randomly expressed and probed with serum to find antigenic proteins. However, cDNA libraries are limited in their proteome coverage, skewed by gene expression abundance, and produce non-natural peptides from aberrant reading frames.
A more recent approach entails probing microscopic arrays of full-length proteins. This has the advantage that the protein identity at each feature is known, that proteins are represented with the same frequency and that protein products are in the cognate reading frame. These arrays lend themselves well to probing with serum to detect antigens [25, 26]. However, historically spotted protein arrays are produced by expressing, purifying and printing proteins, which become a costly and laborious process when done at scale. Moreover, the manipulations and storage of the arrays may affect protein stability, which may then impact the detection of proteins by the serum antibodies.
We have exploited an alternative method for generating protein microarrays in which printed and anchored full-length expression cDNAs encoding target proteins fused to a common epitope are expressed and translated locally to produce proteins in situ on the array surface [27]. The fusion proteins are captured using anti-tag antibodies that are co-printed on the arrays, enabling the presentation of over 3,000 individual antigens per slide. This results in a highly reproducible, stable, and scientifically flexible protein microarray. Here, we demonstrate that these protein microarrays can be adapted for the highly sensitive, reproducible, and specific detection of antibodies to established and novel antigens, including pathogen antigens, autoimmune antigens, and tumor antigens. To enable high-throughput serologic screening of select target antigens, we have adapted this approach for a 96-well macroarray format that is as sensitive and specific as a standard recombinant protein ELISA.
Material and methods
Patient Sera
Sera used in these analyses were obtained from multiple sources: Breast cancer: the Lurie Breast Cancer Tissue and Blood Repository and the Specialized Program of Research Excellence (SPORE) in Breast Cancer at the Dana-Farber Cancer Institute, and Fox Chase Cancer Center. Diabetes sera from patients and controls from the University of Florida. Prostate cancer serum samples were collected at the Harvard University and University of Michigan. Serum from patients infected with P.aeruginosa was obtained from the Stephen Lory (Harvard Medical School). Serum from F.tularensis infected mice was obtained from Gerald Beltz and Dennis Kasper, Harvard Medical School. Written consent was obtained from all human subjects under institutional review board approval.
RAPID ELISA
GST 96-well detection plates coated with anti-GST antibody (GE Healthcare, Piscataway, NJ) were blocked overnight at 4°C with blocking buffer (5% milk/0.2% Tween20 in phosphate buffered saline (PBS-T). On the next day, the desired genes were expressed using the TNT T7 Coupled Reticulocyte Lysate System (Promega, Madison, WI) per manufacturer's recommendations. To each tube of lysate was added: 16 μl reaction buffer, 8 μl polymerase, 4 μl minus-methionine, 4 μl minus-leucine, 8 μl RNaseOUT (Invitrogen, Carlsbad, CA), 500 ng DNA, to a final volume of 400 μl. The DNA-lysate mixture was incubated at 30°C for 90 minutes. After incubation, PBS was added to the mixture and 50 μl transferred to each well. The plate was incubated for 2 hours at room temperature (rt.) while shaking at 800 rpm, and then washed 5 times with blocking buffer. Human serum samples were diluted 1:300 in blocking buffer and 100 μl added to each well. To test gene expression, mouse anti-GST monoclonal antibody (Cell Signaling Technology, Danvers, MA) was diluted 1:1000 in SuperBlock blocking buffer (Pierce, Rockford, IL) and 100 μl added to each well. Secondary antibodies were diluted at 1:1000 and were HRP-conjugated goat anti-human IgG antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) or anti-mouse IgG secondary antibody (GE Healthcare, Piscataway, NJ). For detection, equal volumes of solutions 1 and 2 from the SuperSignal ELISA Femto Max Sensitivity (Pierce, Rockford, IL) were mixed and 100 μl added to each well. The solutions were mixed in the plate for 1 minute by shaking at 800 rpm and the luminescence read using a Wallac plate reader.
P53 antibody ELISA
The p53 protein ELISA assay was performed per manufacturer's recommendations (p53 ELISAPLUS Autoantibody Kit, Calbiochem, Gibbstown, NJ). 100 μl each of calibrators, controls, and serum samples were added in duplicate. The calibrators were used neat, 1:1.5, 1:2, 1:3, 1:4, and 1:6 to obtain a calibration curve. Serum samples were diluted 1:100 and bound for 1 hour rt. The samples were removed and the wells were washed, then incubated with 100 μl of detector antibody conjugate for 1 hour at rt then washed. To develop the wells, 100 μl of substrate solution was added and incubated in the dark at rt. After 30 minutes, 50 μl of stop solution was added. The absorbance was determined at 450 nm using a Wallac plate reader.
Generation of NAPPA protein microarrays
The expression plasmid DNA (pANT7_cGST) encoding for the gene of interest was grown in 1.5 mL culture of DH5alpha cells for 24 hrs and purified using an anion exchange resin[27]. The plasmid DNA (1.5 μg/μL) was supplemented with capture antibody (50 μg/mL, anti-GST antibody, General Electric), protein crosslinker (2 mM, BS3, Pierce) and BSA (3 mg/mL, Sigma) to the DNA prior to printing onto the array surface (manuscript submitted). All samples were printed using a Genetix QArrayer2 with 300 μm solid tungsten pins. Amine treated glass slides were used in all arrays except for the F.tularensis proteome array which was printed on hydrogel slides (Nexterion).
Detecting serum antibodies on NAPPA arrays
The printed DNA was transcribed and translated in situ using previously published protocols [27]. For detecting serum antibodies, the arrays were incubated with serum diluted 1:600 in 5% PBS milk with 0.2% Tween 20. All incubations were carried out at 4°C overnight with mixing (Corning hybridization chambers) unless indicated otherwise. Detection was carried out using either an anti-human IgG or anti-mouse IgG conjugated with HRP. The slides were developed for fluorescent detection by treating the slides with Tyramide Signal Amplification reagent (PerkinElmer) as per manufacturer's instructions. Slides were scanned with a Perkin Elmer ProScanArray HT and the images were quantitated using MicroVigene software (Vigene Tech version 2.9.9.2).
Statistical Analysis
For the commercial standard ELISA, internal positive and negative controls were included with each assay and titrated per manufacturer's recommendations. For Rapid ELISA, interassay variability was determined using luminescent measurements of p53 autoantibodies of the same sera repeated on 4 different days. These signals were averaged, and the coefficient of variation [(SD/mean)×100] was calculated.
Success of protein signal from the expression, capture, and detection of the target proteins on the array was calculated as the percent of printed genes that had signal (average of replicate spots) above a background level, which was defined as 10% above the average signal for pairs of spots where no antigen was present. To compare serum signals from array to array, the signals were normalized to the 50% of the trimmed mean (25–75%) of all protein spots on the slide.
Results
Development of RAPID ELISA
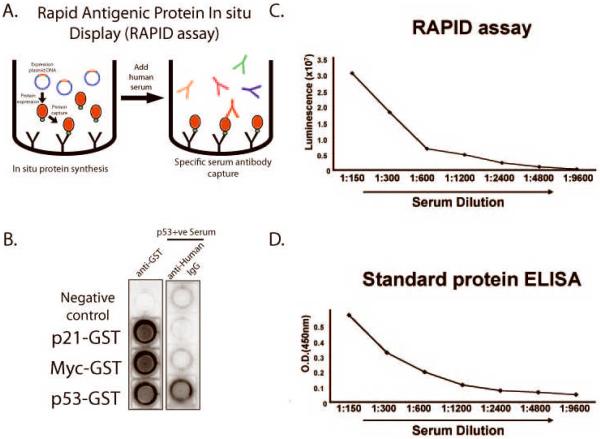
We had developed a system of in situ antigen expression/capture for protein microarray generation that exploits the ready availability of large collections of ORF clones without a priori protein purification. This approach, termed NAPPA (Nucleic Acid Programmable Protein Microarray), uses reticulocyte lysate for in vitro transcription/translation (IVTT) of recombinant proteins that can be captured in situ using anti-tag antibodies [27]. However, to process a larger number of clinical samples against a limited number of proteins, we adapted this technology to a 96-well format ELISA, termed RAPID ELISA (Rapid Antigenic Protein In Situ Display) that is reminiscent of the PISA method [28, 29]. In RAPID ELISA, antigens are expressed from full-length C-terminal tagged fusion cDNA constructs (i.e. GST) with reticulocyte lysate, and specifically captured onto 96-well plates using anti-tag antibodies. This approach replaces amplifying the coding sequences by PCR with preparing quantified and supercoiled plasmid DNA. In addition, the optimized vector provides promoter, termination, IRES and poly adenine tail sequences to optimize protein expression. Finally, this approach uses a GST capture tag at the carboxyl terminus that we have found is more compatible with reticulocyte translation and allows confirmation of full length protein. Serum is added and detected with anti-human IgG secondary antibodies and a highly sensitive horseradish peroxidase (HRP)-based luminescence detection system (Fig. 1A).
Figure 1.
Development of RAPID 96-well format ELISA assay for detection of autoantibodies in serum. A. Schematic of RAPID ELISA. cDNA encoding full-length antigens are expressed as tagged fusion proteins using rabbit reticulocyte lysate. The proteins are captured by anti-tag antibodies in individual wells. Human serum containing antigen-specific antibodies is added and detected with HRP-conjugated secondary anti-IgG antibodies. B. Specific detection of anti-p53 autoantibodies by RAPID-NAPPA. Three C-terminal GST-tagged antigens, p21, myc, and p53, were expressed and captured in wells with anti-GST antibodies. The GST fusion protein was detected with an anti-GST detection antibody (left), demonstrating protein expression. Human sera positive for anti-p53 antibodies were added (right) and human IgG was detected. No added DNA was the negative control. C and D. RAPID ELISA shows comparable sensitivity to standard protein ELISA for the detection of anti-p53 antibodies. P53 antibody-positive breast cancer patient sera were titrated, and antibodies were detected by RAPID-NAPPA (C) and a standard p53 recombinant protein ELISA (D). Internal positive and negative control sera were titrated in duplicate on every standard ELISA assay, with the negative control O.D.(450nm)<0.3.
To validate RAPID ELISA, we tested three full-length proteins (p21, myc, and p53) fused to c-terminal GST protein (Fig. 1B). All three antigens were strongly expressed, captured and displayed, as detected with an anti-GST antibody. Using a commercially-available ELISA displaying recombinant p53 protein, we identified human breast cancer patient serum that contained autoantibodies to the p53 protein (data not shown). Application of this serum to the GST- fusion proteins shows specific detection of p53 protein but not the control proteins. Testing over multiple days we found the interassay coefficient of variation for detection of protein expression by RAPID NAPPA was 3.9%. We compared detection by the RAPID ELISA assay to standard ELISA employing recombinant purified p53 protein after titrating this serum on each. The RAPID ELISA showed comparable sensitivities over a wide serum dilution range (Figures 1C and 1D).
Evaluating NAPPA protein microarray and RAPID ELISA
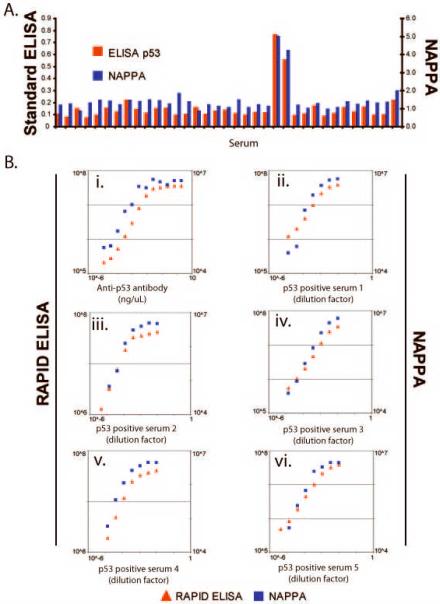
To expand the detection of autoantibodies to a wide array of protein antigens, we have adapted the NAPPA protein microarray system for the detection of antibodies within human serum. First, we chose to determine the concordance of signal obtained by a standard ELISA well assay to the NAPPA microarrays. To evaluate concordance of signal between NAPPA and ELISA, we tested 35 early-stage breast cancer patient sera and found 2 of the 35 to be positive for p53 by both methods. P53 positive sera were defined on NAPPA and on standard ELISA as those that have fold change of 3 or more when compared to the P53 negative sera. We assumed that majority of sera was P53 negative, and computed fold changes as ratio of the serum signal to the median signal of all sera assayed. We found a significant concordance between NAPPA and standard ELISA (Kappa=1.0, p-value=0.0017).
Next, to determine the detection limits and the linearity of the serum response as detected by our RAPID ELISA and NAPPA microarrays, we titrated known amounts of a well-characterized monoclonal antibody to p53 (Fig. 2B). Using a range of anti-p53 antibody concentrations (10 fg/μL to 2 ng/μL), we found that both methods detected p53 antibodies at concentrations as low as 10 fg/μL, which is comparable to existing methods [30–33]. The linear range of detection covered 3 orders of magnitude (30 fg/μL and 25 pg/μL) for RAPID ELISA and NAPPA microarrays. We also tested the sensitivity of detection of p53 autoantibodies in several breast cancer patient sera by NAPPA and by RAPID ELISA. In Figure 2B ii-vi, patient sera were titrated over a broad range (1:100 to >1:70, 000) and the p53 autoantibodies were detected by RAPID ELISA or NAPPA ELISA. For all five sera, both methods showed a broad linear range of detection of autoantibodies in breast cancer sera.
Figure 2.
Autoantibodies to p53 are detected by protein microarray (NAPPA) and RAPID (96-well format) ELISA. A. Detection of p53-positive antibodies by p53 standard ELISA and by NAPPA in cancer patient sera. 35 breast cancer patient sera were screened for anti-p53 autoantibodies by p53 standard ELISA using recombinant p53 protein, and by NAPPA. The signals on the NAPPA arrays were normalized to the 50% trimmed mean (mean of the middle 50% of protein spots). Antibodies to p53 antigen were detected in two patient sera by both assays. B. i. Quantification of p53 antigen detection by RAPID ELISA and NAPPA microarrays. P53-GST fusion protein was expressed by RAPID ELISA and NAPPA, and detected with titrations of an anti-p53 MAb (D01, Sigma). Both methods detected 10 fg/μL of antibody, with a linear range of 20 pg/μL-30 fg/μL. Serum signals greater than 110% of a feature with no antigen were considered positive on the NAPPA arrays. B. ii–vi. Detection of anti-p53 antibodies by RAPID ELISA and NAPPA microarrays from 5 human cancer patient sera containing p53 autoantibodies.
Optimization of conditions for detecting autoantibodies
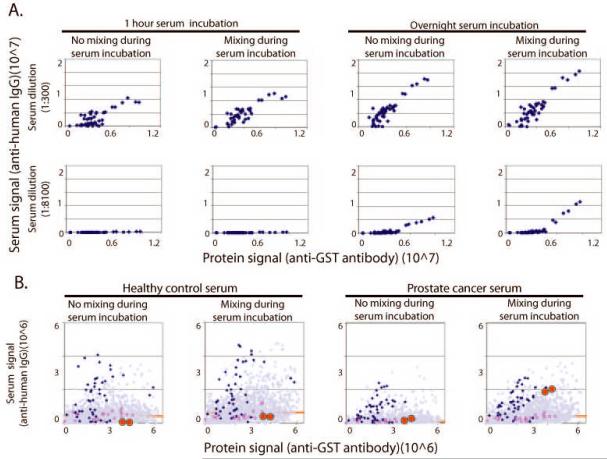
We predicted that certain autoantibodies, in particular anti-tumor antibodies, may occur at low abundance or with low affinity. The detection of these antibodies would rely on optimized binding conditions. Altering both incubation time and the mixing of serum sample during incubation were critical factors to detect select autoantibodies (Fig. 3). To determine antibody detection as a function of antigen concentration, different concentrations of DNA template (0.1 μg/μL to 3 μg/μL) encoding a fragment of the Epstein-Barr virus antigen EBNA-1 (positive control antigen) were printed on the arrays. In Figure 3A, the serum response (Y-axis) is shown as a function of the amount of each antigen displayed (X-axis) on the array, as measured by anti-GST signal.
Figure 3.
Optimizing the detection of bona fide but rare antibody signals on NAPPA. Because many autoantibodies can be of low concentration and low affinity in patient sera, we tested the effect of serum mixing, incubation time, and incubation temperature on the detection of the known viral antigen EBNA-1 and to an antigen, NAP1L3, in prostate cancer sera. A. At both 1:300 (top) and 1:8100 (bottom) serum dilutions, bound IgG antibodies are more readily detected with an overnight incubation on NAPPA (right) than with a one hour incubation (left). Antibody detection correlates with protein expression, but only during extended incubations and with mixing does the reaction achieve equilibrium. B. Mixing of the sera increases detection of tumor antigen-specific antibodies. Healthy control sera (left two panels) and prostate cancer serum (right two panels) were diluted at 1:600 and incubated on the protein microarrays overnight at 4°C, with or without mixing as shown. EBNA-specific responses are shown in blue, p53 specific response are shown in red squares, and the prostate tumor antigen NAP1L3 is shown in red circles. Response to other genes on the array is shown as purple triangles and the trimmed mean (25–75%) signal is shown as a red line. By mixing the sera, detection of NAP1L3 is specifically enhanced over background.
As expected, there was as direct relationship between the amount of EBNA1 DNA printed and the amount of EBNA1 protein displayed. Under short, static incubations (1 hour at room temperature without mixing), the system clearly had not reached equilibrium as indicated by the plateau at the higher antigen concentrations. Increasing the incubation time and mixing the sample revealed a linear response to the amount of antigen present on the array implying that equilibrium was achieved. The anti-EBNA response in this serum was both abundant and high affinity. To simulate weaker responses, we diluted the EBNA positive serum 1:8100. Under dilute antibody conditions, the effect of longer incubation times and mixing were even more pronounced and were required for antibody detection.
To determine if these conditions enabled detection of a tumor antigen-specific autoantibody, we tested sera from a prostate cancer patient and a healthy control. Both sera had detectable antibodies to EBNA and no response to tumor antigen p53. We designed an array that included EBNA, p53, and 648 unique genes printed in duplicate. Included in this gene set was the antigen, NAP1L3 (Fig 3B, red circles) [34]. NAP1L3 is highly homologous to cell division autoantigen-1 (CDA-1), which has been detected by autoimmune sera in SEREX. Responses to EBNA antigen and p53 on the array agreed with results from standard ELISA. NAP1L3 was not detected in the normal sera (Fig. 3B, left panels) under any condition. However, in the cancer patient sera, when the sample was mixed (right panel), this specific response was selectively detected compared to the majority of the other proteins on the array.
Reproducibility of detecting autoantibodies
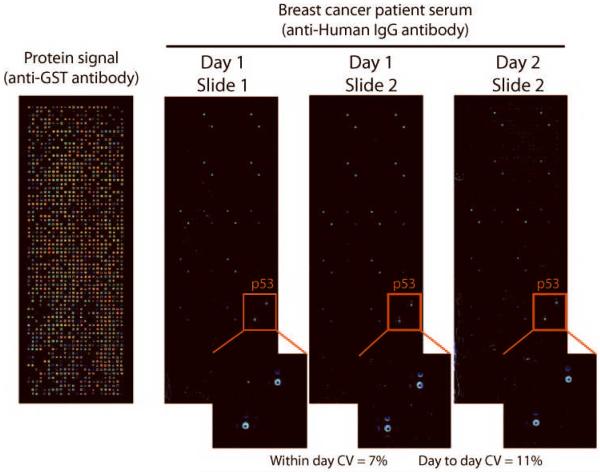
As all clinical assays require a high degree of reliability and reproducibility, coefficients of variation (CVs) of NAPPA antibody detection were determined. Replicate arrays with 624 genes (Supplemental table 1) were printed and expressed in duplicate pairs with an overall success of 94% for protein display. These were probed with serum positive for p53 antibodies (Fig. 4). The normalized serum signals for p53 (normalized to 50% trimmed mean (25–75%) signal) among the arrays processed on the same day showed excellent reproducibility with a CV of 7%. Serum signals for arrays processed on different days had a comparable CV of 11%.
Figure 4.
Detection of autoantibodies by NAPPA ELISA is highly reproducible. Over 600 individual c-terminal GST tagged proteins were expressed in microarray format, Antigen expression across the slide was confirmed by anti-GST antibody (left). Slides containing anchored cDNAs were stored at room temperature, and proteins were expressed on consecutive days. Human IgG registration spots are visible as diagonal duplicates across the slide. P53 antigen was specifically detected in duplicate using human sera containing anti-p53 antibodies. The coefficients of variation for antibody detection were 7% (within day) and 11% (between days). The CV's were calculated based on the means of the replicate spots on each slide.
Multiplexed detection of humoral responses
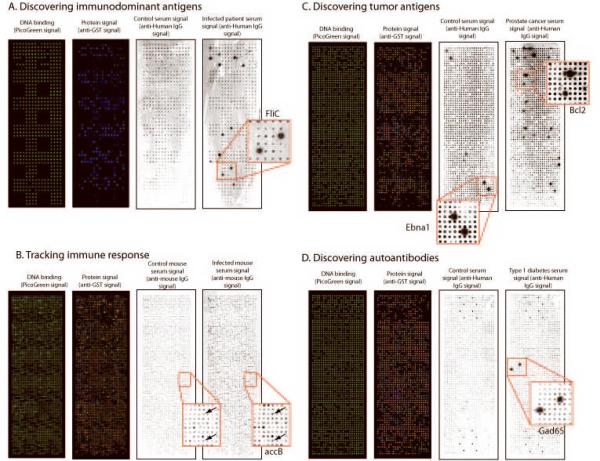
With the growing number of humoral immune responses that have been identified against antigens from infectious agents and self antigens shown to have clinical utility, there is a need for platforms to test multiple antigens simultaneously. We considered whether the versatility of programming the NAPPA arrays to readily produce proteins of varying size and type from different organisms might lend itself as a tool for multiple antigen testing. In the proof of concept study summarized in Fig. 5, the NAPPA approach specifically detected IgG antibodies from sera to a wide variety of known target antigens.
Figure 5.
Proof of concept detection of antibodies in several disease states. A. Detection of immunodominant pathogen-derived antigens. A selected set of 264 genes predicted to be outer membrane proteins from the pseudomonas genome was expressed by NAPPA and probed with control serum or serum derived from an infected patient. A well documented response to the pseudomonal antigen, fliC, was specifically detected in the patient sera. B. The Francisella tularensis proteome was expressed on a NAPPA array. The array was probed with sera from a mouse infected with a live attenuated strain and from a control mouse. The immune responses were detected with an anti-mouse antibody. C. An array expressing 655 human genes in duplicate was probed with sera from a healthy control and from a patient with prostate cancer. Antibodies to the common viral antigen EBNA-1 are readily detected in both sera, but antibodies to Bcl2 are only detected in the prostate cancer patient serum. D. An array expressing 659 human genes was probed with sera from a diabetic patient and a nondiabetic control. Serum response to a well known antigen related to IDDM, GAD65, was observed only in the patient sera.
Infection or co-infection caused by an opportunistic pathogen such as Pseudomonas aeruginosa is well documented, especially for chronically ill or immunocompromised patients [35]. To test patients' immune responses against this pathogen, we printed a subset of genes from P.aeruginosa (Supplemental table 2) and probed them with sera from a healthy control and an infected patient (Fig. 5A). We detected specific serum response from the patient serum to a handful of antigens including the well known immunodominant antigen, fliC [36]. Identifying immune responses to pathogens like P. aeruginosa will be important for developing specific vaccines.
Vaccination using attenuated or killed organisms can lead to humoral immune responses to various pathogen antigens, the identities of which are often unknown. The ability to employ protein microarrays to vaccinated animal models would enable the rapid identification and tracking of the immune response. To demonstrate this, we chose to identify immunogenic antigens within the Francisella tularensis proteome that elicit an immune response in infected mice. A NAPPA array containing 94% (1511/1603 genes, Supplemental table 3) of the F.tularensis genome was expressed yielding 100% success for protein display as detected with anti-GST antibodies (Fig. 5B). The arrays were probed with serum from a healthy mouse and a mouse infected with a live attenuated F. tularensis strain, and anti-mouse IgG antibodies were detected with secondary antibodies. Immune responses to several antigens were identified with the infected serum including known responses to antigens accB and pcp [37]. We found a broad humoral immune response to F. tularensis antigens in sera from the infected mouse, as measured by signals greater than 3 fold over control. 5% of antigens showed over 5-fold changes in serum antibody responses.
To determine if NAPPA could also be used to detect autoantibodies in cancer patient sera, a protein array containing genes for 655 potential tumor antigens (Supplemental table 4) was expressed with 91% success and probed with serum from a healthy control and a prostate cancer patient (Fig. 5C,). Although both individuals responded to the viral EBNA1 antigen (positive control), only the cancer patient responded to the tumor antigen Bcl2.
Lastly, this approach can be used to detect self antigens associated with autoimmunity. In IDDM, the autoimmune response to the insulin producing beta cells of the pancreas adversely affects insulin production. To detect immune response in diabetic serum samples, we printed an array expressing 659 unique human genes (Supplemental table 5) and detected protein signal for 94%. The arrays were probed with serum from a healthy control and a diabetic serum sample (Fig. 5D). We found a specific response to the known diabetic autoantigen GAD65 from the diabetic serum and none from the control serum.
Discussion
The proteomic detection of antibodies in patient sera requires highly sensitive, reproducible, and flexible assays that can accommodate variations in proteome content, sample collection, and serum components. Here, we demonstrate the use of two approaches, NAPPA microarray and RAPID ELISA, for the sensitive detection of serum antibodies. One key advantage of these approaches is the flexibility of using cDNAs as source antigens. Selecting and re-arraying cDNAs for printing on the arrays is relatively straightforward and allows researchers to assemble arrays as needed for specific experiments.
The NAPPA microarray approach is designed for testing proteome scale collections of antigens in an unbiased manner. There are large collections of human ORF libraries that provide a rich source of content for NAPPA microarrays (e.g., http://plasmid.hms.harvard.edu). In this and other reports, we have demonstrated that thousands of human proteins can be produced on NAPPA microarrays including transmembrane proteins. In addition to human proteins, we demonstrated that over 1,700 pathogen proteins can be produced on a NAPPA microarray with success rates >90%. The ability to screen larger numbers of candidate antigens positions this as a useful discovery tool for finding novel immune responses.
Once identified, the disease specificity of novel immune responses needs confirmation by testing on many clinical samples. The relatively high cost of producing high density protein microarrays of any type makes using high density arrays impractical for such validation studies. Historically, this testing was done by standard ELISA, where a small number of selected antigens are tested against a large number of sera. As commercial ELISA kits are rarely available for most proteins, researchers must develop their own, which is a tedious process of cloning protein expression constructs, optimizing protein expression and purification, and coating the wells of microtiter dishes or for western blotting. The development of RAPID ELISA offers a quick alternative, where cDNAs for the gene of interest can be readily added to the IVTT and the proteins can be synthesized just in time for testing. The sensitivity of detection for RAPID ELISA is comparable to standard ELISAs and it offers the flexibility to produce many genes of interest without having to separately express and purify them.
NAPPA microarrays display sufficient protein (~9 fmoles) to produce a strong signal when probed with serum that contained an abundant anti-EBNA1 response [27]. Signal intensity typically depends on the abundance of antibody, its affinity for the antigen and ensuring that the binding reaction achieves equilibrium. Most immune responses, both antimicrobial and autoantibodies, are likely to be less abundant than the EBNA1 response here. Sensitive detection of antibody-antigen interactions on microarrays have been limited by mass transport due to inadequate processing conditions [31, 38]. We improved the physiochemical properties of the interaction between low concentrations of antibodies and expressed antigens by increasing the time of incubation, and mixing during the assay. Using these enhanced conditions, we observed comparable dynamic range and limit of detection (10 fg/μL) to standard ELISAs and other microarray approaches [30–33].
By doing select testing with known positive controls, this study illustrates proof of concept for the use of high density NAPPA microarrays to detect immune responses. The method is both sensitive and reproducible. What remains is to employ this tool in large controlled studies that compare the responses in patients to controls. The goal here will be to identify new markers that correlate with disease. A major challenge facing such studies will be the development of bioinformatics and biostatistical tools that can handle such large datasets to find significant markers. As biomarkers of infectious, autoimmune, and malignant diseases, identifying novel informative antigens has enormous clinical potential for the early detection of disease and development of targeted immunotherapy.
Supplementary Material
Acknowledgements
We thank Janice Williamson and Mauricio Fernandez from the Harvard Institute of Proteomics, Harvard Medical School for their support. We thank David Lane and Alison Sparks (University of Dundee) for the p53 antibodies. We thank Dr. Andrew Godwin at Fox Chase Cancer Center for providing us with serum from Breast cancer patients. We thank Drs. Stephen Lory, Dennis Kasper, and Gerald Beltz from the Harvard Medical School for providing serum from patients with infected with P.aeruginosa, and mice infected with F.tularensis. This study was supported by a research grants from the Early Detection Research Network (EDRN, NCI, Grant 5U01CA117374-02), the National Institute of Allergy and Infectious Diseases (NIAID, Contract HHSN266200400053C) and the Juvenile Diabetes Research Foundation (Grant 17-2007-1045).
References
- [1].Janeway CA. Immunobiology: the immune system in health and disease. 6th edition 2005. [Google Scholar]
- [2].Dorner T, Radbruch A. Antibodies and B cell memory in viral immunity. Immunity. 2007;27:384–392. doi: 10.1016/j.immuni.2007.09.002. [DOI] [PubMed] [Google Scholar]
- [3].Huber M, Trkola A. Humoral immunity to HIV-1: neutralization and beyond. Journal of internal medicine. 2007;262:5–25. doi: 10.1111/j.1365-2796.2007.01819.x. [DOI] [PubMed] [Google Scholar]
- [4].Notkins AL. New predictors of disease. Molecules called predictive autoantibodies appear in the blood years before people show symptoms of various disorders. Tests that detected these molecules could warn of the need to take preventive action. Scientific American. 2007;296:72–79. [PubMed] [Google Scholar]
- [5].Benoist C, Germain RN, Mathis D. A plaidoyer for `systems immunology'. Immunological reviews. 2006;210:229–234. doi: 10.1111/j.0105-2896.2006.00374.x. [DOI] [PubMed] [Google Scholar]
- [6].Amanna IJ, Slifka MK, Crotty S. Immunity and immunological memory following smallpox vaccination. Immunological reviews. 2006;211:320–337. doi: 10.1111/j.0105-2896.2006.00392.x. [DOI] [PubMed] [Google Scholar]
- [7].Pertussis Vaccination Use of Acellular Pertussis Vaccines Among Infants and Young Children Recommendations of the Advisory Committee on Immunization Practices (ACIP) CDC Recommendations and Reports. 1997 March 28;46:1–25. [PubMed] [Google Scholar]
- [8].Poland GA. Acellular pertussis vaccines: new vaccines for an old disease. Lancet. 1996;347:209–210. doi: 10.1016/s0140-6736(96)90398-0. [DOI] [PubMed] [Google Scholar]
- [9].Harrop R, Ryan MG, Golding H, Redchenko I, Carroll MW. Monitoring of human immunological responses to vaccinia virus. Methods in molecular biology (Clifton, NJ. 2004;269:243–266. doi: 10.1385/1-59259-789-0:243. [DOI] [PubMed] [Google Scholar]
- [10].Cooper GS, Stroehla BC. The epidemiology of autoimmune diseases. Autoimmunity reviews. 2003;2:119–125. doi: 10.1016/s1568-9972(03)00006-5. [DOI] [PubMed] [Google Scholar]
- [11].Betterle C, Volpato M, Rees Smith B, Furmaniak J, Chen S, Zanchetta R, Greggio NA, Pedini B, Boscaro M, Presotto F. II. Adrenal cortex and steroid 21-hydroxylase autoantibodies in children with organ-specific autoimmune diseases: markers of high progression to clinical Addison's disease. J Clin Endocrinol Metab. 1997;82:939–942. doi: 10.1210/jcem.82.3.3849. [DOI] [PubMed] [Google Scholar]
- [12].Atkinson MA, Eisenbarth GS. Type 1 diabetes: new perspectives on disease pathogenesis and treatment. Lancet. 2001;358:221–229. doi: 10.1016/S0140-6736(01)05415-0. [DOI] [PubMed] [Google Scholar]
- [13].Mathis D, Benoist C. Back to central tolerance. Immunity. 2004;20:509–516. doi: 10.1016/s1074-7613(04)00111-6. [DOI] [PubMed] [Google Scholar]
- [14].Achenbach P, Warncke K, Reiter J, Naserke HE, Williams AJ, Bingley PJ, Bonifacio E, Ziegler AG. Stratification of type 1 diabetes risk on the basis of islet autoantibody characteristics. Diabetes. 2004;53:384–392. doi: 10.2337/diabetes.53.2.384. [DOI] [PubMed] [Google Scholar]
- [15].Bingley PJ, Bonifacio E, Williams AJ, Genovese S, Bottazzo GF, Gale EA. Prediction of IDDM in the general population: strategies based on combinations of autoantibody markers. Diabetes. 1997;46:1701–1710. doi: 10.2337/diab.46.11.1701. [DOI] [PubMed] [Google Scholar]
- [16].Bingley PJ, Bonifacio E, Ziegler AG, Schatz DA, Atkinson MA, Eisenbarth GS. Proposed guidelines on screening for risk of type 1 diabetes. Diabetes Care. 2001;24:398. doi: 10.2337/diacare.24.2.398. [DOI] [PubMed] [Google Scholar]
- [17].Gale EA, Bingley PJ, Emmett CL, Collier T. European Nicotinamide Diabetes Intervention Trial (ENDIT): a randomised controlled trial of intervention before the onset of type 1 diabetes. Lancet. 2004;363:925–931. doi: 10.1016/S0140-6736(04)15786-3. [DOI] [PubMed] [Google Scholar]
- [18].Anderson KS, LaBaer J. The sentinel within: exploiting the immune system for cancer biomarkers. J Proteome Res. 2005;4:1123–1133. doi: 10.1021/pr0500814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Koziol JA, Zhang JY, Casiano CA, Peng XX, Shi FD, Feng AC, Chan EK, Tan EM. Recursive partitioning as an approach to selection of immune markers for tumor diagnosis. Clin Cancer Res. 2003;9:5120–5126. [PubMed] [Google Scholar]
- [20].Qiu J, Madoz-Gurpide J, Misek DE, Kuick R, Brenner DE, Michailidis G, Haab BB, Omenn GS, Hanash S. Development of natural protein microarrays for diagnosing cancer based on an antibody response to tumor antigens. J Proteome Res. 2004;3:261–267. doi: 10.1021/pr049971u. [DOI] [PubMed] [Google Scholar]
- [21].Chatterjee M, Mohapatra S, Ionan A, Bawa G, Ali-Fehmi R, Wang X, Nowak J, Ye B, Nahhas FA, Lu K, et al. Diagnostic markers of ovarian cancer by high-throughput antigen cloning and detection on arrays. Cancer research. 2006;66:1181–1190. doi: 10.1158/0008-5472.CAN-04-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hudson ME, Pozdnyakova I, Haines K, Mor G, Snyder M. Identification of differentially expressed proteins in ovarian cancer using high-density protein microarrays. Proc Natl Acad Sci U S A. 2007;104:17494–17499. doi: 10.1073/pnas.0708572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang X, Yu J, Sreekumar A, Varambally S, Shen R, Giacherio D, Mehra R, Montie JE, Pienta KJ, Sanda MG, et al. Autoantibody signatures in prostate cancer. N Engl J Med. 2005;353:1224–1235. doi: 10.1056/NEJMoa051931. [DOI] [PubMed] [Google Scholar]
- [24].Chen YT, Scanlan MJ, Sahin U, Tureci O, Gure AO, Tsang S, Williamson B, Stockert E, Pfreundschuh M, Old LJ. A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc Natl Acad Sci U S A. 1997;94:1914–1918. doi: 10.1073/pnas.94.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Joos TO, Schrenk M, Hopfl P, Kroger K, Chowdhury U, Stoll D, Schorner D, Durr M, Herick K, Rupp S, et al. A microarray enzyme-linked immunosorbent assay for autoimmune diagnostics. Electrophoresis. 2000;21:2641–2650. doi: 10.1002/1522-2683(20000701)21:13<2641::AID-ELPS2641>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- [26].Robinson WH, DiGennaro C, Hueber W, Haab BB, Kamachi M, Dean EJ, Fournel S, Fong D, Genovese MC, de Vegvar HE, et al. Autoantigen microarrays for multiplex characterization of autoantibody responses. Nature medicine. 2002;8:295–301. doi: 10.1038/nm0302-295. [DOI] [PubMed] [Google Scholar]
- [27].Ramachandran N, Hainsworth E, Bhullar B, Eisenstein S, Rosen B, Lau AY, Walter JC, LaBaer J. Self-assembling protein microarrays. Science. 2004;305:86–90. doi: 10.1126/science.1097639. [DOI] [PubMed] [Google Scholar]
- [28].He M, Taussig M. Single step generation of protein arrays from DNA by cell-free expression and in situ immobilisation (PISA method) Nucleic Acids Res. 2001;29:E73–73. doi: 10.1093/nar/29.15.e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].He M, Taussig MJ. DiscernArray technology: a cell-free method for the generation of protein arrays from PCR DNA. J Immunol Methods. 2003;274:265–270. doi: 10.1016/s0022-1759(02)00521-5. [DOI] [PubMed] [Google Scholar]
- [30].Haab B, Dunham M, Brown P. Protein microarrays for highly parallel detection and quantitation of specific proteins and antibodies in complex solutions. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-2-research0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kusnezow W, Syagailo YV, Ruffer S, Baudenstiel N, Gauer C, Hoheisel JD, Wild D, Goychuk I. Optimal design of microarray immunoassays to compensate for kinetic limitations: theory and experiment. Mol Cell Proteomics. 2006;5:1681–1696. doi: 10.1074/mcp.T500035-MCP200. [DOI] [PubMed] [Google Scholar]
- [32].Pawlak M, Grell E, Schick E, Anselmetti D, Ehrat M. Functional immobilization of biomembrane fragments on planar waveguides for the investigation of side-directed ligand binding by surface-confined fluorescence. Faraday Discuss. 1998;111:273–288. doi: 10.1039/a806704j. [DOI] [PubMed] [Google Scholar]
- [33].Pawlak M, Schick E, Bopp MA, Schneider MJ, Oroszlan P, Ehrat M. Zeptosens' protein microarrays: a novel high performance microarray platform for low abundance protein analysis. Proteomics. 2002;2:383–393. doi: 10.1002/1615-9861(200204)2:4<383::AID-PROT383>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- [34].Chai Z, Sarcevic B, Mawson A, Toh BH. SET-related cell division autoantigen-1 (CDA1) arrests cell growth. The Journal of biological chemistry. 2001;276:33665–33674. doi: 10.1074/jbc.M007681200. [DOI] [PubMed] [Google Scholar]
- [35].Saiman L. Microbiology of early CF lung disease. Paediatric respiratory reviews. 2004;5(Suppl A):S367–369. doi: 10.1016/s1526-0542(04)90065-6. [DOI] [PubMed] [Google Scholar]
- [36].Doring G, Meisner C, Stern M. A double-blind randomized placebo-controlled phase III study of a Pseudomonas aeruginosa flagella vaccine in cystic fibrosis patients. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11020–11025. doi: 10.1073/pnas.0702403104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Eyles JE, Unal B, Hartley MG, Newstead SL, Flick-Smith H, Prior JL, Oyston PC, Randall A, Mu Y, Hirst S, et al. Immunodominant Francisella tularensis antigens identified using proteome microarray. Crown Copyright 2007 Dstl. Proteomics. 2007;7:2172–2183. doi: 10.1002/pmic.200600985. [DOI] [PubMed] [Google Scholar]
- [38].Kusnezow W, Syagailo YV, Goychuk I, Hoheisel JD, Wild DG. Antibody microarrays: the crucial impact of mass transport on assay kinetics and sensitivity. Expert Rev Mol Diagn. 2006;6:111–124. doi: 10.1586/14737159.6.1.111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.