Abstract
An intracellular plant pathogen ‘Candidatus Liberibacter asiaticus,’ a member of the Rhizobiales, is related to Sinorhizobium meliloti, Bradyrhizobium japonicum, nitrogen fixing endosymbionts, Agrobacterium tumefaciens, a plant pathogen, and Bartonella henselae, an intracellular mammalian pathogen. Whole chromosome comparisons identified at least 50 clusters of conserved orthologous genes found on the chromosomes of all five metabolically diverse species. The intracellular pathogens ‘Ca. Liberibacter asiaticus’ and Bartonella henselae have genomes drastically reduced in gene content and size as well as a relatively low content of guanine and cytosine. Codon and amino acid preferences that emphasize low guanosine and cytosine usage are globally employed in these genomes, including within regions of microsynteny and within signature sequences of orthologous proteins. The length of orthologous proteins is generally conserved, but not their isoelectric points, consistent with extensive amino acid substitutions to accommodate selection for low GC content. The ‘Ca. Liberibacter asiaticus’ genome apparently has all of the genes required for DNA replication present in Sinorhizobium meliloti except it has only two, rather than three RNaseH genes. The gene set required for DNA repair has only one rather than ten DNA ligases found in Sinorhizobium meliloti, and the DNA PolI of ‘Ca. Liberibacter asiaticus’ lacks domains needed for excision repair. Thus the ability of ‘Ca. Liberibacter asiaticus’ to repair mutations in its genome may be impaired. Both ‘Ca. Liberibacter asiaticus and Bartonella henselae lack enzymes needed for the metabolism of purines and pyrimidines, which must therefore be obtained from the host. The ‘Ca. Liberibacter asiaticus’ genome also has a greatly reduced set of sigma factors used to control transcription, and lacks sigma factors 24, 28 and 38. The ‘Ca. Liberibacter asiaticus’ genome has all of the hallmarks of a reduced genome of a pathogen adapted to an intracellular lifestyle.
Introduction
Huanglongbing (HLB), arguably the most serious disease of citrus worldwide [1], [2], originated in South Asia [3], [4] and recently became epidemic in Brazil [5] and in Florida [6] homes of the largest orange juice industries in the world. ‘Candidatus Liberibacter asiaticus’ is consistently associated with HLB disease. Using electron microscopy, researchers observed the pathogen within sieve cells of phloem vessels of infected plant hosts [7], [8] or in the salivary glands of citrus psyllids, natural vectors of the pathogen [9]. The pathogen can also be readily detected by PCR-based tests, most based on the 16S rRNA gene [10], [11]. Placement of ‘Ca. Liberibacter asiaticus’ within the α-2 subdivision of the Proteobacteria class [12] was confirmed by sequence analysis of the 16S–23S RNA gene region [13]–[15]. In spite of many efforts [16]–[19], ‘Ca. Liberibacter asiaticus’ has not been artificially cultured. Thus the bacterium seems to have an obligate intracellular lifestyle in either plant or insect hosts.
In their landmark paper, Duan et al. provided a complete genomic sequence of ‘Ca. Liberibacter asiaticus’ strain Psy62 [20]. The sequence was obtained by deep sequencing of DNA obtained from an individual psyllid containing at least 8×108 copies of the ‘Ca. Liberibacter asiaticus’ genome, based on q-PCR of its 16S rRNA gene. This genomic sequence data was confirmed by an independent group who also employed deep sequencing of the contents of phloem cells from infected trees obtained by laser micro dissection [21]. The genome is composed of a circular chromosome of 1.23 MB with no extra chromosomal elements. Analysis of the full genomic sequence enabled Duan et al. (2009) to place ‘Ca. Liberibacter asiaticus,’ within the Rhizobiaceae family of the order Rhizobiales [22]–[23] of the class α-Proteobacteria.
The intracellular bacterium ‘Ca. Liberibacter asiaticus’ and other intracellular members of the α-Proteobacteria, such as members of the genera Rickettsia and Bartonella, have reduced genome size when compared to free-living related bacteria [20], [24]. Intracellular bacteria also typically have genomes with remarkably low mol% G+C content [25], but there are also examples of free-living bacteria with larger genomes but equally low mol% G+C in their genomic sequence [26]. Forces that drive the reduction in genome size and the decrease in mol% G+C content remain obscure but are likely to result ultimately from intense selection for managing scarce resources and the lower energy costs associated with AT vs GC base pairs [27]. Intracellular organisms may have an enhanced rate of genomic evolution [28], and indeed ‘Ca. Liberibacter asiaticus’ occupies by far the longest branch in a phylogram of members of the Rhizobiales [20]. Other generalizations made about characteristics of reduced genomes include the reduction of ribosomal gene operons, the loss of some genes encoding enzymes needed for DNA repair, the loss of sigma factors needed to regulate transcription, and the loss or retention of different sets of genes that tend to be either dispensable or indispensable [29], [30].
When chromosomes are compared, phylogenetically-related bacteria typically share clusters of orthologous genes (COGs). We propose the term MOG (Microsyntenous Orthologous Genes) to avoid confusion with proteins described in the COG database meaning clusters of orthologous groups of proteins. We compared MOGs in ‘Ca. Liberibacter asiaticus’ with those in Sinorhizobium meliloti, Bradyrhizobium japonicum and Agrobacterium tumefaciens. These bacteria, capable of free-living, symbiotic nitrogen fixing, and pathogenic lifestyles are highly adaptable and thus have large genomes particularly when compared to the reduced genome of ‘Ca. Liberibacter asiaticus’. We therefore also compared the genome of ‘Ca. Liberibacter asiaticus’ to that of Bartonella henselae, a flea-vectored pathogen of cats and, opportunistically, of humans. Although B. henselae can be artificially cultured, it has complex nutritional requirements and thus it can be considered a semi-obligate intracellular pathogen of cats, reproducing in erythrocytes [31]. It possesses a severely reduced genome, not much larger than that of ‘Ca. Liberibacter asiaticus’ [31], [32].
We have studied these genomes in detail, within regions of conserved microsynteny between reduced and large genomes, and within signature regions of orthologous proteins. We have also performed global comparisons of gene sets whose members are often reduced in the genomes of intracellular pathogens. This analysis provides insight into the remarkable evolutionary pathway leading to a reduced genome in ‘Ca. Liberibacter asiaticus’ as compared to the larger genomes of other members of the Rhizobiales with free-living and plant-associated lifestyles.
Results and Discussion
Length and isoelectric point of orthologous proteins
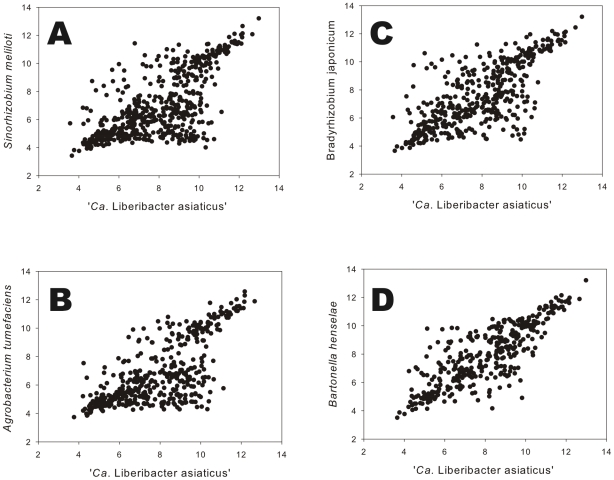
We identified orthologous protein pairs, and hence, conserved orthologous genes, based on BlastP e values of less than 10−15, a relatively conservative criterion. Manual curation of the resulting datasets increased the size of conserved clusters of genes by 10–15%. We compared orthologous protein pairs encoded by corresponding genes from microsyntenous orthologous groups. Isoelectric points (pIs) and the number of amino acids predicted for orthologous protein pairs were plotted for ‘Ca. Liberibacter asiaticus’ vs S. meliloti, B. japonicum, A. tumefaciens, and B. henselae. Although there seems to be a general tendency for the pIs of orthologous protein pairs to correlate, the correlation was weak and substantial variation was observed (Fig. 1). Nonetheless, we observed generally higher pI's for proteins of ‘Ca. Liberibacter asiaticus’ as compared to their orthologs in the free living Rhizobiales. For example, the mean pI for 552 orthologous proteins for ‘Ca. Liberibacter asiaticus vs S. meliloti was 7.90 vs 6.91. Similar results were found for 426 orthologous proteins from ‘Ca. Liberibacter asiaticus’ vs A. tumefaciens. However, when 362 orthologous proteins from the intracellular pathogens were compared, the pIs were nearly the same, 8.02 for ‘Ca. Liberibacter asiaticus’ and 7.90 for B. henselae.
Figure 1. Isoelectric points of orthologous protein pairs.
Orthologous proteins from ‘Ca. Liberibacter asiaticus’ and Sinorhizobium meliloti (A), Agrobacterium tumefaciens (B), Bradyrhizobium japonicum (C) and Bartonella henselae (D).
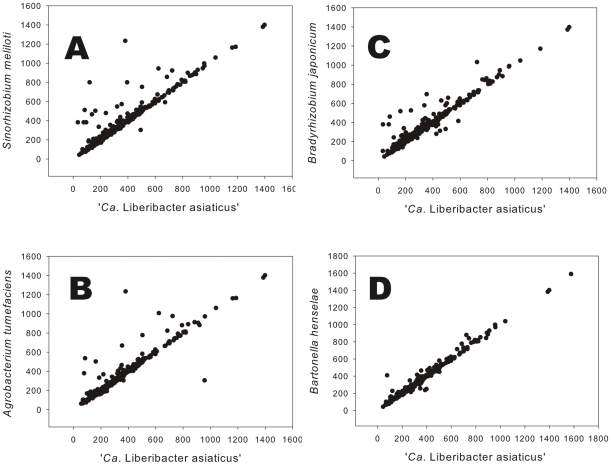
We also plotted the number of amino acids in orthologous protein pairs in microsyntenous orthologous groups and found that the correlation in length of orthologous protein pairs was much stronger (Fig. 2). Amino acid plots of orthologous protein pairs shared between S. meliloti and ‘Ca. Liberibacter asiaticus’ and orthologous protein pairs shared between B. japonicum and ‘Ca. Liberibacter asiaticus’ revealed several proteins pairs well separated from the diagonal. For the most part, these fell above the diagonal, indicating that the orthologous protein in ‘Ca. Liberibacter asiaticus’ was shorter in length than the corresponding protein in S. meliloti or B. japonicum. In contrast, the correlation between the lengths of each member of orthologous protein pairs was very high for ‘Ca. Liberibacter asiaticus’ and B. henselae, the two bacteria with similar intracellular parasitic lifestyles and reduced genomes (Fig. 2).
Figure 2. Number of amino acids in orthologous protein pairs.
Orthologous proteins from ‘Ca. Liberibacter asiaticus’ and Sinorhizobium meliloti (A), Agrobacterium tumefaciens (B), Bradyrhizobium japonicum (C) and Bartonella henselae (D).
The relatively high mean isoelectric points of proteins from ‘Ca. Liberibacter asiaticus’ as compared to their orthologs from free living bacteria, is similar to other intracellular pathogens [33]. The mean of the isoelectric points of proteins from ‘Ca. Liberibacter asiaticus’ were not different from that of orthologs in B. henselae, consistent with both bacteria being intracellular pathogens with reduced genomes. The isoelectric points (pIs) of orthologous protein pairs are not under stringent selection since conservative substitutions of amino acids can retain function while changing the pI [34], [35].
When the pIs of orthologous protein pairs from the closely related S. meliloti and A. tumefaciens were individually compared graphically [34], a great deal of dispersion away from the diagonal was observed and subsets of protein pairs were considered to be differentially grouped away from the diagonal [34]. Such grouping of protein pairs away from the diagonal was not observed in any of the graphical comparisons made here between ‘Ca. Liberibacter asiaticus’ and the other members of the Rhizobiales; instead the dispersion from the diagonal was continuous but was noticeably less when B. henselae and ‘Ca. Liberibacter asiaticus’ were compared. The lack of correlation of pIs between orthologous proteins of ‘Ca. Liberibacter asiaticus’ when compared to those of other members of the Rhizobiales is consistent with phylogenetic distance [20] and the extreme shift in codon usage between ‘Ca. Liberibacter asiaticus’ and free-living members of the Rhizobiales. However, the relative lengths of orthologous proteins was well conserved despite a consistent trend toward overall reduction in the obligate intracellular microorganism, extending data from other systems [34], [35] to intracellular pathogens with extremely reduced genomes.
GC content in microsyntenous orthologous groups of paired genomes
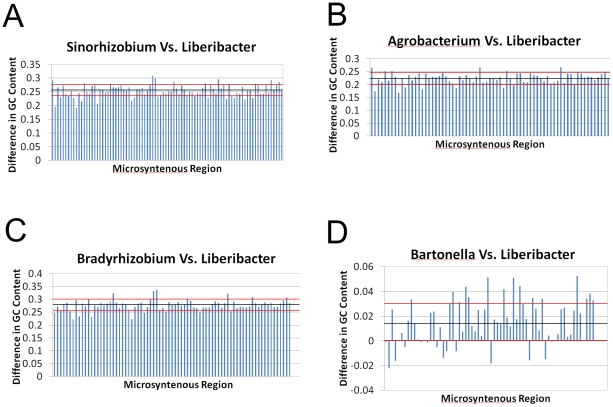
Ninety-one microsyntenous orthologous groups, (MOGs), were identified when the genomes of ‘Ca. Liberibacter asiaticus’ and S. meliloti were compared. The mol% GC content of the microsyntenous MOG regions in S. meliloti was 25.5±2.1 percent greater than the corresponding regions of ‘Ca. Liberibacter asiaticus (Fig. 3). The MOGs of B. japonicum had 27.8±2.1 mol% higher GC content than their orthologous MOGs in ‘Ca. Liberibacter asiaticus’ (Fig. 3). In contrast, when the GC content of 66 MOGs conserved between ‘Ca. Liberibacter asiaticus and B. henselae were compared, the differences in mol% GC content were small, only 1.5±0.2 percentage points. These data are consistent with the overall GC content of the respective genomes, and the fact the B. henselae, like ‘Ca. Liberibacter asiaticus’ has a highly reduced genome.
Figure 3. Differences in GC content in microsyntenous orthologous groups.
The GC content of microsyntenous orthologous groups of ‘Ca. Liberibacter asiaticus’ compared with other members of the Rhizobiales. The dark bar and red lines denote the mean and one standard deviation of the mean.
To determine if the preference for AT-rich codons held in the functionally and structurally conserved motifs of the encoded proteins, five arbitrarily selected sets of MOGs from ‘Ca. Liberibacter asiaticus’ and S. meliloti were chosen for comparative study. The percentage of GC base pairs in the genomic sequences encoding the functional domains of the proteins was consistent with the overall GC usage for each genome, with ‘Ca. Liberibacter asiaticus’ and S. meliloti having GC content of 35–40% and 60–65% respectively in the functional domains of these orthologous proteins.
Orthologous proteins have been retained in ‘Ca. Liberibacter asiaticus’ without conservation of the coding sequences, but with a low GC bias for the genome as a whole. This low G+C codon bias is often observed in intracellular bacteria, and mathematical analysis has shown that such synonymous codon usage bias is dominated by selection at the third codon position [36] . The evolutionary driver of the low GC bias may very likely be related to the higher energy cost to synthesize a GC base pair relative to an AT base pair in the genome [27], as a part of a larger competition for limited metabolic resources from the host. In addition to the energy saved in the synthesis of AT vs GC base pairs, the thermodynamic stability of AT rich sequences is lower than that of GC rich sequences. DNA helicase consumes ATP in order to separate DNA strands [37]. Therefore we hypothesize that replication and transcription of an AT rich genome will be less energetically expensive than that of a GC rich genome, based on a lower ATP cost for strand separation. This contributes to the evolution of AT rich genomes in intracellular parasites where competition for resources with the host is intense.
Codon preference and amino acid usage in proteins of ‘Ca. Liberibacter asiaticus’ vs other members of the Rhizobiales
Amino acid codon usage by ‘Ca. Liberibacter asiaticus’ was distinctly different from that of A. tumefaciens, B. japonicum and S. meliloti, consistent with an overall drastic reduction in GC content of the genome (Table S1). For example, across the entire ORFeome, ‘Ca. Liberibacter asiaticus’ the codon ‘CAA’ for glutamine was used 73% of the time while A. tumefaciens, B. japonicum and S. meliloti used the codon ‘CAG’ for glutamine more than 80% of the time. ‘Ca. Liberibacter asiaticus’ ‘preferred’ the codon ‘TGT’ for cysteine (75%) while A. tumefaciens, B. japonicum and S. meliloti used the codon ‘TGC’ for cysteine more than 80% of the time. Similarly, codons ‘TTA’, ‘CTA’, or ‘CTT’ were used by ‘Ca. Liberibacter asiaticus’ to encode leucine 65% of the time but were used only about 15% of the time by A. tumefaciens, B. japonicum and S. meliloti. These bacteria instead used codons ‘CTG,’ ‘TTG’ or ‘CTC’ about 88% of the time. Reduced usage of G and C nucleotides in triplet codons determining a given amino acid incorporated into protein is consistent throughout the ‘Ca. Liberibacter asiaticus’ genome. B. henselae shared the strong preference for AT-rich amino acid codons in the ORFeome with ‘Ca. Liberibacter asiaticus’ (Table S1). The preference for A or T codons at the third position of amino acid codons in intracellular bacteria has been observed previously [36]. This is consistent with rapid evolution of genes in bacterial endosymbionts relative to those of free living bacteria, a phenomenon reflected in amino acid sequences of proteins [28].
Genome-wide comparisons also showed that usage of the amino acids proline, arginine, valine, glycine and alanine was lower in B. henselae and ‘Ca. Liberibacter asiaticus’ than in the free-living Rhizobiales. Usage of the amino acids phenylalanine, serine, cysteine, glutamine, isoleucine, asparagine, lysine was higher in B. henselae and ‘Ca. Liberibacter asiaticus’ as compared with that of the free-living Rhizobiales (Table S2). The tRNA synthetases corresponding to these amino acids are present in the proteome. Thus, the differences in usage are not likely to be due to an inability of ‘Ca. Liberibacter asiaticus’ to synthesize amino acids de novo due to loss of metabolic capability with the reduced genome. Instead, we observe that there is a strong correlation between the AT content of the codons for amino acids and the frequency of occurrence of the amino acids in the ‘Ca. Liberibacter asiaticus’ and B. henselae genomes (Table S2).
Purine and pyrimidine metabolism
‘Ca. Liberibacter asiaticus’ is markedly deficient in its ability to carry out purine and pyrimidine metabolism as compared to S. meliloti (Figures S1 and S2). ‘Ca. Liberibacter asiaticus’ retains only 47 genes annotated for purine metabolism compared with the 95 genes annotated for purine metabolism in the genome of S. meliloti. Although it can produce the nucleotides GTP and ATP from their respective nucleoside diphosphates, ‘Ca. Liberibacter asiaticus’ does not appear able to synthesize guanine, xanthine, hypoxanthine or adenine. Likewise, ‘Ca. Liberibacter asiaticus’ retains only 38 genes for pyrimidine metabolism compared with the 60 genes for pyrimidine metabolism in the genome of S. meliloti. Although ‘Ca. Liberibacter asiaticus’ can produce UTP and CTP from their respective nucleoside diphosphates it does not appear to be able to produce uracil, cytosine or thymine.
‘Ca. Liberibacter asiaticus,’ likely has lost the ability to synthesize and metabolize both purines and pyrimidines, and these may have to be obtained from the host as di- or tri-nucleosides. The loss of these pathways would be consistent with a general bias towards deletion of genes in the process leading to reduced genomes if the gene products become redundant in the intracellular environment [38]. The incorporation of uracil into DNA by either replication error or cytosine to uracil deamination has been proposed as a driver towards the low GC/high AT genome, particularly if DNA repair if ineffective [33], as is likely to be the case here. In this context it is notable that E.C. 3.5.4.1, used to interconvert cytosine and uracil, is missing in ‘Ca. Liberibacter asiaticus’. This defect has been noted previously in Ureaplasma urealyticum, another intracellular bacterium [25]. A caveat is necessary here. The lack of annotated orthologous proteins to perform a particular task in ‘Ca. Liberibacter asiaticus’ does not mean that the organism cannot perform that task. Several examples have been described in Mycoplasma spp. where relaxed substrate specificity for enzymes compensated for loss of function [39]. Examples of enzymes with relaxed specificities included kinases [40] and malate/lactate dehydrogenase [41]. Still, the apparent loss of metabolic capability is striking.
DNA replication and repair
Atypical and reduced DNA replication complements are typically observed in reduced genomes [30]. Two critical components of the DNA polymerase III complex, the alpha and gamma/tau subunits, were annotated as pseudogenes in the original description of the ‘Ca. Liberibacter asiaticus’ genome [20]. We have conducted a complete bioinformatic analysis of the DNA replication complex in ‘Ca. Liberibacter asiaticus’ (Table 1). The DNA polymerase III alpha subunit (COG0587) is located between genes CLIBASIA_03630 and CLIBASIA_03645 in the Liberibacter genome and thus occurs between positions (797277–801416). The protein length is 1000+ amino acids long and is characteristic of other DNA polymerase III alpha subunits. SMART analysis reveals a pFAM DNA_pol3_alpha domain. There is an open reading frame using a triplet codon encoding leucine as the starting amino acid (797696–800797) with a strong blast hit to Rhizobium etli DNA polymerase III alpha subunit (YP_001977851.1, E-value 0.0). This protein is thus presumed to be functional. DNA polymerase III subunit gamma/tau (COG2812) is located between genes CLIBASIA_03275 and CLIBASIA_03260 in the Liberibacter genome. The protein is 370 amino acids long, shorter than other DNA Pol III gamma/tau subunits. However Smart analysis shows an ATPase domain and there is an open reading frame from nucleotide position 499568 to 500677. This protein has an e-value of 9.0E-151 when compared to its ortholog in S. meliloti, and the domains of the ‘Ca. Liberibacter asiaticus’ and S. meliloti proteins show similar e-values when compared to the pFAM models (Table 1). The rest of the DNA replication complex is also present with low e-values values to both orthologs in S. meliloti and functional domains in pFAM. The major difference between S. meliloti and ‘Ca. Liberibacter asiaticus’ in the DNA replication complex is that S. meliloti encodes three RNase H orthologs but ‘Ca. Liberibacter asiaticus’ encodes only two orthologs of RNase H (Table 1).
Table 1. Proteins involved in the DNA Polymerase III replication complex in ‘Ca. Liberibacter asiaticus’ and Sinorhizobium meliloti.
| ANNOTATION | Protein in | Protein in | E value to | L. asiaticus | L. asiaticus | S. meliloti | S. meliloti |
| L. asiaticus | S.meliloti | L. asiaticus to | pFAM | E-value to | pFAM | E-value to | |
| S. meliloti | Domains | pFAM model | Domains | pFAM model | |||
| Epsilon | ACT57291 | CAC41392 | 6.00E-67 | EXO III | 5.91E-39 | EXO III | 9.44E-47 |
| Alpha | N/Aa | CAC45868 | 0 | DNA Pol 3 α | 3.40E-267 | DNA Pol 3 α | 8.20E-302 |
| Pol IIIAc | 1.15E-18 | Pol IIIAc | 4.60E-18 | ||||
| Gamma/Tau | N/Ab | CAC41667 | 9.00E-151 | AAA | 1.54E-08 | AAA | 3.52E-12 |
| Delta' | ACT56690 | CAC46182 | 2.00E-78 | Nonec | None | None | None |
| Beta | ACT56927 | CAC41772 | 2.00E-99 | Pol 3Bc | 8.67E-87 | Pol3Bc | 1.76E-119 |
| Chi | ACT56864 | CAC45735 | 5.00E-21 | DNA Pol3 chi | 4.60E-08 | DNA Pol3 chi | 2.50E-43 |
| Delta | ACT57277 | CAC47912 | 4.00E-45 | DNA Pol3 δ | 3.60E-04 | DNA Pol3 δ | 3.30E-05 |
| DNAB | ACT56805 | CAC45710 | 0 | DNAB | 9.30E-35 | DNAB | 1.40E-32 |
| DNAG | ACT57307 | CAC46897 | 3.00E-164 | ZnF CHCC | 3.34E-23 | ZnF CHCC | 2.76E-24 |
| Toprim N | 1.90E-57 | Toprim N | 1.20E-61 | ||||
| SSB | ACT56656 | CAC46137 | 3.00E-46 | SSB | 3.60E-32 | SSB | 1.90E-31 |
| RNaseH | ACT57269 | CAC45486 | 5.00E-49 | RNase H | 3.00E-47 | RNase H | 8.40E-58 |
| ACT56750 | CAC45411 | 2.00E-53 | RNase HII | 4.20E-63 | RNase HII | 2.60E-62 | |
| ------------ | CAC45163 | ----------- | ---------- | ---------- | RNase H | 5.50E-40 | |
| DNA Pol I | CAC41562 | 0 | 53EXOc | 7.69E-111 | 53EXOc | 2.40E-121 | |
| HhH2 | 4.15E-11 | HhH2 | 1.10E-15 | ||||
| POLAc | 2.19E-106 | POLAc | 2.91E-120 | ||||
| Ligase | ACT57645 | CAC46743 | 0 | LIGANc | 9.68E-172 | LIGANc | 1.30E-227 |
Protein list compiles from the Kyoto Encyclopedia of Genomes and Genes.
These proteins were originally annotated as pseudogenes and so a protein designation is not available.
This protein does not match any pFAM domain.
The process leading to genome reduction such as has occurred in ‘Ca. Liberibacter asiaticus’ is thought to be facilitated by deficient DNA repair capability [29], [33]. We therefore searched the ‘Ca. Liberibacter asiaticus’ genome for the suite of enzymes active in DNA repair by comparison with the suite of DNA repair enzymes present in S. meliloti. Most such enzymes present in S. meliloti were also present in ‘Ca. Liberibacter asiaticus’, and are likely to be in functional condition based on the e-values between orthologs in S. meliloti and ‘Ca. Liberibacter asiaticus. We also found similar matches of functional domains of the proteins from these two bacteria to the pFAM database (Table S3). The noteworthy exceptions were for DNA PolI and DNA ligase. In the case of DNA PolI, the ortholog in ‘Ca. Liberibacter asiaticus’ was lacking domains 53EXOc and 35EXOc, which are expected to be particularly important in DNA excision repair. Both bacteria had three DNA ligase enzymes annotated on their chromosome, but two of these proteins were apparently not functional in ‘Ca. Liberibacter asiaticus’, as the e-values to S. meliloti orthologs were poor and there was no match at all to the model proteins in pFAM. It is important to note that we found an additional eight ligases encoded on pSymA and pSymB of S. meliloti, and 2 ligases encoded on the A. tumefaciens chromosome as well as five more ligases encoded on the linear chromosome and the At and Ti plasmids. B. japonicum has 10 ligases encoded on its chromosome (Table S4).
The accelerated rate of evolution observed within obligate intracellular bacteria [20], [28] may be facilitated by inefficient repair of mutations caused by loss of genes encoding DNA repair enzymes. ‘Ca. Liberibacter asiaticus’ is likely to be deficient in DNA repair as compared to S. meliloti. DNA ligase participates in all three DNA repair pathways, and so having only one ligase rather than eleven ligases as found in S. meliloti or 10 ligases as found in B. japonicum very likely limits the efficiency of DNA repair processes. Likewise, the lack of the EXO53 and EXO35 domains in the DNA PolI enzyme of ‘Ca. Liberibacter asiaticus’ is very likely to impede the excision repair process, and is also an example of an enzyme evolving relaxed substrate specificity concurrent with genome reduction [39], [40].
Sigma factors
The genome of ‘Ca. Liberibacter asiaticus’ encodes only three sigma subunit factors, which enable differential transcription of genes by RNA polymerase (Table 2). These include one sigma 70 for genes expressed during exponential growth, one sigma 54 for genes involved in nitrogen metabolism and one sigma 32, involved in the heat shock or other stress responses. In contrast, the S. meliloti encodes 11 sigma factors, including three sigma 54 subunits and four sigma 24 subunits which enable expression of factors needed for the heat shock response as well as for factors exported from the cell. S. meliloti also has a special sigma factor, FecI for the expression of genes needed for the uptake of iron.
Table 2. Sigma factors present in the genomes of Sinorhizobium meliloti and ‘Ca. Liberibacter asiaticus.’.
| Sinorhizobium | Liberibacter | Identification | E. coli | Liberibacter vs | Function |
| Protein | Protein | gene | Sinorhizobium | ||
| E value | |||||
| CAC46893 | ACT56762 | sigma 70 | rpoD | 0 | Exponential growth |
| CAC47374 | ACT57389 | sigma 54 | rpoN | 9.00 E-80 | Nitrogen metabolism |
| CAC41817 | N/A | sigma 54 | rpoN | ___________ | Nitrogen metabolism |
| CAC41818 | N/A | sigma 54 | rpoN | ___________ | Nitrogen metabolism |
| N/A | N/A | sigma 38 | rpoS | ___________ | stationary phase |
| CAC47305 | ACT57084 | sigma 32 | rpoH | 6.00 E-124 | heat shock |
| CAC47835 | N/A | sigma 32 | rpoH | ___________ | heat shock |
| N/A | N/A | sigma 28 | rpoF | ___________ | flagella |
| CAC46681 | N/A | sigma 24 | rpoE | ___________ | heat shock and export |
| CAC47413 | N/A | sigma 24 | rpoE | ___________ | heat shock and export |
| CAC46954 | N/A | sigma 24 | rpoE | ___________ | heat shock and export |
| CAC47014 | N/A | sigma 24 | rpoE | ___________ | heat shock and export |
| CAC46610 | N/A | FecI | FecI | ___________ | Iron uptake |
It is notable that ‘Ca. Liberibacter asiaticus’ has no recognizable sigma factors 24 or 38, which in E. coli are used for the expression proteins intended for export or for expression in the stationary phase of growth, respectively [42]. Neither S. meliloti nor ‘Ca. Liberibacter asiaticus’ encodes a sigma factor 28 which is required for expression of flagellar genes in E. coli [42]. S. meliloti produces flagellae so one of the other sigma subunits is apparently responsible for this activity in S. meliloti. Flagellae have not been observed in ‘Ca. Liberibacter asiaticus’, though an extensive set of flagellar genes is present [20].
The early workers who created the proposed genus ‘Candidatus Liberibacter’ placed it within the α-subdivision of the Proteobacteria based on sequence analysis of the 16S rRNA gene [12], [14]. Duan et al. [20] created a phylogenetic tree of the α-Proteobacteria based on concatenated sequence alignments of 94 proteins. Their overall alignment of the α-Proteobacteria was as previously demonstrated [43], with the addition of ‘Ca. Liberibacter asiaticus’ firmly within the Rhizobiales as an “early branching member” of the Rhizobiaceae. We took advantage of the relationship of ‘Ca. Liberibacter asiaticus’ with other members of the Rhizobiales to design the experiments described herein. Although the large regions of synteny previously observed in other similar comparisons [34] has been largely obscured by the process of genomic reduction, we have identified numerous microsyntenous orthologous groups of proteins that are conserved across all five species studied (Kuykendall et al., in preparation). The existence of so many MOGs in these inter specific genomic comparisons reflects the underlying evolutionary relationships among these species.
The ‘Ca. Liberibacter asiaticus’ genome is typical of reduced genomes adapted for the niche of intracellular parasitism [24], [29]–[30], [39] and in this regard is very similar to the genome of Bartonella henselae [32]. Orthologous protein pairs were identified and plotted against each other based on size and isoelectric points. The lengths of proteins in orthologous pairs, as well as the isoelectric points of the orthologous proteins, were very well correlated between the two intracellular pathogens. This is consistent with the greatly reduced genome sizes of ‘Ca. Liberibacter asiaticus’ and Bartonella henselae as compared with the other species. Both bacteria use a specialized genetic code and the amino acid composition in proteins is altered to minimize the occurrence of guanine and cytosine in their genomes, presumably because guanine and cytosine are more energetically expensive to make than are adenine and thymidine [27]. We further suggest that because the unwinding of the double helix to allow replication or transcription is done at the expense of ATP [37] , a genome rich in AT base pairs should be relatively less costly to replicate because of the lower thermodynamic stability of AT as compared with GC base pairs.
The GC content of the chromosomal DNA of ‘Ca. Liberibacter asiaticus’ and B. henselae is 26–27 mol% less than that of the other members of the Rhizobiales, consistent with other obligate intracellular pathogens with reduced genomes [24]. We show that the low GC content is achieved by a global and systematic reordering of the transcription process to favor AT vs GC rich codons for amino acids, such as phenylalanine and proline, when such a choice is available in the universal code. This AT preference extends to the preferred usage of TAA as the ‘stop’ codon over TGA in the other Rhizobiales. In any case GC content ought not to be a taxonomic criterion, as members of the well accepted order Rhizobiales within the α-Proteobacteria are shown here to have widely divergent GC content.
Although the DNA replication machinery appears to be intact in ‘Ca. Liberibacter asiaticus’, the relative efficiencies of orthologs from ‘Ca. Liberibacter asiaticus’ and S. meliloti have not been compared, so the relative efficiency of DNA replication between the two bacteria can not be accurately estimated.
The complete and uniform revision of the genome to favor AT rich codons over GC rich codons requires a massive accumulation of mutations. The capacity of ‘Ca. Liberibacter asiaticus’ to repair DNA appears to be limited, which would likely contribute to the rapid evolution of the genomic sequence observed [20], [28]. DNA PolI of ‘Ca. Liberibacter asiaticus’ lacks both of the exonuclease domains used for excision repair, and ‘Ca. Liberibacter asiaticus’ has only one ligase enzyme used more generally in DNA repair. The lack of capacity for DNA repair may represent a potential vulnerability for ‘Ca. Liberibacter asiaticus.
Likewise the lack of endogenous metabolic capability of ‘Ca. Liberibacter asiaticus’, as shown by its apparent inability to synthesize purines or pyrimidines may also represent vulnerabilities to ‘Ca. Liberibacter asiaticus’. The repertoire of sigma subunit factors is also greatly reduced in ‘Ca. Liberibacter asiaticus’ as compared to S. meliloti. Loss of sigma subunits is common in reduced genomes of bacteria adapted to an intracellular lifestyle [30]. Because of the relative richness of sigma factor subunits encoded by the S. meliloti genome, gene expression in S. meliloti is apparently under much more nuanced control than is the case with ‘Ca. Liberibacter asiaticus’, which is adapted to a stable host cell environment requiring few sigma factors. The apparent lack of nuanced control of genetic expression may present another weakness for ‘Ca. Liberibacter asiaticus’. We have noted a number of reductions in the ‘Ca. Liberibacter asiaticus’ genome that lead to a markedly reduced metabolic capability as well as reduced capabilities for DNA repair and fine control of genetic expression. Nonetheless, this reduced genome provides ‘Ca. Liberibacter asiaticus’ with the means to successfully infect and multiply to high levels in several plant species [7], [44] as well as within the insect vector Diaphorina citri [20] with whom it is also highly adapted. Thus the reduced genome is very subtly adapted to the intracellular environment, which is perhaps surprisingly similar between plant and insect hosts.
Materials and Methods
Orthologous protein pairs were identified by P-BLAST alignments from genomic sequence data using a cut off e-value of less than 10−15. Microsyntenous regions that are conserved between the ‘Ca. Liberibacter asiaticus’ chromosome and those of four other members of the Rhizobiales were identified by using criteria that included (1) at least three orthologous genes in succession, (2) in the same order and (3) with predicted protein products that shared Blast alignment e values of less than 10−15 (Kuykendall et al., in preparation).
Identification of orthologous proteins in microsyntenous orthologous groups
To identify orthologous genes and characterize them in some detail, amino acid sequences (‘Ca. Liberibacter asiaticus’ strain psy62, CP001677; Bartonella henselae strain Houston-1, BX897699; Sinorhizobium meliloti strain 1021, AL591688; Bradyrhizobium japonicum strain USDA 110, BA000040 and Agrobacterium tumefaciens strain C58, AE007869) were downloaded from NCBI and used with the author's original annotation. ‘Ca. Liberibacter asiaticus’ strain psy62 does not have plasmid DNA, and extra chromosomal DNA sequences were not included in this study except as noted. Using default BLAST parameters, each amino acid sequence from a chromosome of this group of strains was blasted against the amino acid sequences of the other chromosomes of this group of phylogenetically related strains. Perl scripts and Excel spreadsheets were created to easily identify hits between genomes with low, negative e-values.
Calculation of GC content and amino acid and codon usage
A perl script was written to calculate the GC content of each gene in each syntenic block. The GC composition of syntenic blocks were compared between the genome of ‘Ca. Liberibacter asiaticus’ and the other members of the Rhizobiales and summarized graphically. Another perl script was written calculating codon usage for each gene in all 5 genomes. The percentage of occurrence of each amino acid and each amino acid codon were also calculated for each genome. The amino acid sequences of signature domains of proteins in selected syntenic blocks were determined using the SMART program (http://smart.embl-heidelberg.de/). The nucleotide sequence encoding these domains was extracted and the codon usage and GC content was calculated.
We also calculated the isoelectric points and the number of amino acids of orthologous proteins encoded in microsyntenous blocks of each genome. Isoelectric points of each protein in each syntenic block were calculated with the mw iep program (http://gchelpdesk.ualberta.ca/). The isoelectric points and number of amino acids in orthologous protein pairs in microsyntenous regions identified in the ‘Ca. Liberibacter asiaticus’ proteome as compared to the other members of the Rhizobiales were plotted against each other (Sigma Plot 11, Systat, San Jose, CA).
The functional capabilities of proteins involved in DNA repair and DNA replication pathways were estimated by comparing the proteins from ‘Ca. Liberibacter asiaticus to orthologous proteins from S. meliloti. Comparisons included e-values of the proteins to each other as well as e values of each protein to the pFAM models for each protein. Sigma factors were identified by reciprocal searches of the genomes of S. meliloti and ‘Ca. Liberibacter asiaticus’. The purine and pyrimidine metabolic networks were taken from the Kyoto Encyclopedia of Genomes and Genes. Orthologous genes in S. meliloti and ‘Ca. Liberibacter asiaticus’ were mapped on to these networks.
Supporting Information
Purine metabolism in Sinorhizobium meliloti and ‘ Ca . Liberibacter asiaticus’. Enzymes annotated as present in both ‘Ca. Liberibacter asiaticus’ and S. meliloti are colored green. Enzymes annotated as present in S. meliloti but not in ‘Ca. Liberibacter asiaticus are colored orange. The metabolic pathway is from the Kyoto Encyclopedia of Genomes and Genes.
(TIF)
Pyrimidine metabolism in Sinorhizobium meliloti and ‘ Ca . Liberibacter asiaticus’. Enzymes annotated as present in both ‘Ca. Liberibacter asiaticus’ and S. meliloti are colored green. Enzymes annotated as present in S. meliloti but not in ‘Ca. Liberibacter asiaticus are colored orange. The metabolic pathway is from the Kyoto Encyclopedia of Genomes and Genes.
(TIF)
Frequency (%) of usage of amino acid codons in genomes of intracellular and free living members of the Rhizobiales.
(RTF)
Amino acids over- and under-represented in the ‘ Ca . Liberibacter asiaticus’ and Bartonella henselae proteomes as compared to the proteomes of free-living members of the Rhizobiales .
(RTF)
Proteins involved in DNA repair pathways in ‘ Ca . Liberibacter asiaticus and Sinorhizobium meliloti .
(RTF)
Proteins that contain Ligase domains annotated on chromosomal and extra chromosomal elements of ‘ Ca . Liberibacter asiaticus’ and other members of the Rhizobiales.
(RTF)
Acknowledgments
We thank Larissa Higginbotham and Felicia Davenport for outstanding technical support.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was funded by the United States Department of Agriculture, Agricultural Research Service. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.da Graça J, Korsten L. Citrus Huanglongbing: Review, present status and future strategies. In: Naqvi SAMH, editor. Diseases of Fruits and Vegetables Vol. I. Kluwer, The Netherlands: 2004. pp. 229–245. [Google Scholar]
- 2.Bové JM. Huanglongbing: A destructive, newly-emerging, century old disease of citrus. J Plant Pathol. 2006;88:7–37. [Google Scholar]
- 3.Nath D, Husain MA. The citrus psylla (Diaphorina citri, Kuw.) [Psyllidae:Homoptera]. Memoirs of the Department of Agriculture in India. 1927;10:1–27. [Google Scholar]
- 4.Lin KH. Observations on yellow shoot disease. Acta Phytopathologica Sinica. 1956;2:1–42. [Google Scholar]
- 5.Teixeira DC, Danet JL, Eveillard S, Martins EC, Jesus de WC., Jr Citrus huanglongbing in Sao Paulo state, Brazil: PCR detection of the ‘Candidatus’ Liberibacter species associated with the disease. Molecular and Cellular Probes. 2005;19:173–179. doi: 10.1016/j.mcp.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Florida Department of Agriculture and Consumer Services. US Department of Agriculture and Florida Department of Agriculture confirm detection of citrus greening. Department Press Release; 2005. 30 [Google Scholar]
- 7.Hartung JS, Paul C, Achor D, Brlansky RH. Colonization of dodder, Cuscuta indecora, by ‘Candidatus Liberibacter asiaticus’ and ‘Ca. L. americanus’. Phytopathology. 2010;100:756–762. doi: 10.1094/PHYTO-100-8-0756. [DOI] [PubMed] [Google Scholar]
- 8.Kim JS, Sagaram US, Burns JK, Li JL, Wang N. Response of sweet orange (Citrus sinensis) to ‘Candidatus Liberibacter asiaticus’ infection: Microscopy and microarray analyses. Phytopathology. 2009;99:50–57. doi: 10.1094/PHYTO-99-1-0050. [DOI] [PubMed] [Google Scholar]
- 9.Moll JN, Martin MM. Electron microscope evidence that citrus psylla (Trioza erytreae) is a vector of greening disease in South Africa. Phytophylactica. 1973;5:41–44. [Google Scholar]
- 10.Jagoueix S, Bové JM, Garnier M. PCR detection of the two ‘Candidatus’ liberobacter species associated with greening disease of citrus. Molecular and Cellular Probes. 1996;10:43–50. doi: 10.1006/mcpr.1996.0006. [DOI] [PubMed] [Google Scholar]
- 11.Li W, Hartung JS, Levy LE. Quantitative real time PCR for detection and identification of Candidatus Liberibacter species associated with citrus huanglongbing. J Microbiological Methods. 2006;66:104–115. doi: 10.1016/j.mimet.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 12.Jagoueix S, Bové J-M, Garnier M. The Phloem-Limited Bacterium of Greening Disease of Citrus is a Member of the alpha Subdivision of the Proteobacteria. Int J System Bacteriol. 1994;44:379–386. doi: 10.1099/00207713-44-3-379. [DOI] [PubMed] [Google Scholar]
- 13.Planet P, Jagoueix S, Bové JM, Garnier M. Detection and characterization of the African citrus greening Liberobacter by amplification, cloning, and sequencing of the rplKAJL-rpoBC operon. Current Microbiology. 1995;30:137–141. doi: 10.1007/BF00296198. [DOI] [PubMed] [Google Scholar]
- 14.Jagoueix S, Bové JM, Garnier M. Comparison of the 16S/23S Ribosomal Intergenic Regions of “Candidatus Liberobacter asiaticum” and “Candidatus Liberobacter africanum”, the Two Species Associated with Citrus Huanglongbing (Greening) Disease. Int J Syst Bacteriol. 1997;47:224–227. doi: 10.1099/00207713-47-1-224. [DOI] [PubMed] [Google Scholar]
- 15.Subandiyah S, Iwanami T, Kondo Y, Kobayashi M, Tsuyumu S, et al. Comparison of 16S rDNA and 16S/23S Intergenic Region Sequences Among Citrus Greening Organisms in Asia: Plant Disease. 2000;84:15–18. doi: 10.1094/PDIS.2000.84.1.15. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh SK, Raychaudhuri SP, Varma A, Nariani TK. Isolation and culture of mycoplasma associated with citrus greening disease. Current Science. 1971;40:299–300. [Google Scholar]
- 17.Garnett HM. Isolation of the greening organism. Symposium of citrus greening. Nelspruit, South Africa: 1984. pp. 4–6. [Google Scholar]
- 18.Davis MJ, Mondal SN, Chen H, Rogers ME, Brlansky RH. Co-cultivation of ‘Candidatus Liberibacter asiaticus’ with actinobacteria from citrus with huanglongbing. Plant Disease. 2008;92:1547–1550. doi: 10.1094/PDIS-92-11-1547. [DOI] [PubMed] [Google Scholar]
- 19.Sechler A, Schuenzel EL, Cooke P, Donnua S, Thaveechai N, et al. Cultivation of ‘Candidatus Liberibacter asiaticus’, ‘Ca. L. africanus’, and ‘Ca. L. americanus’ associated with Huanglongbing. Phytopathology. 2009;99:480–486. doi: 10.1094/PHYTO-99-5-0480. [DOI] [PubMed] [Google Scholar]
- 20.Duan Y, Zhou L, Hall DG, Li W, Doddapaneni H, et al. Complete genome sequence of citrus huanglongbing bacterium, ‘Candidatus Liberibacter asiaticus’ obtained through metagenomics. Molecular Plant-Microbe Interactions. 2009;22:1011–1020. doi: 10.1094/MPMI-22-8-1011. [DOI] [PubMed] [Google Scholar]
- 21.Tyler HL, Roesch LFW, Gowda S, Dawson WO, Triplett EW. Confirmation of the sequence of ‘Candidatus Liberibacter asiaticus’ and assessment of microbial diversity in Huanglongbing-infected citrus phloem using a metagenomic approach. Molecular Plant-Microbe Interactions. 2009;22:1624–1634. doi: 10.1094/MPMI-22-12-1624. [DOI] [PubMed] [Google Scholar]
- 22.Kuykendall LD. Order Rhizobiales, Type Family Rhizobiaceae Genus Rhizobium. In: Brenner DJ, Kreig NR, Staley JT, Garrity GM, editors. Bergey's Manual of Systematic Bacteriology, 2nd. Edition, Vol. II. The Proteobacteria. New York: Springer; 2005. 324 [Google Scholar]
- 23.Kuykendall LD. List of new names and new combinations previously effectively, but not validly, published. Validation List no. 107. Int J of Syst and Evol Microbiol. 2006;56:1–6. doi: 10.1099/ijs.0.64188-0. [DOI] [PubMed] [Google Scholar]
- 24.Sallstrom B, Andersson SGE. Genome reductionin the alpha-proteobacteria. Curr Opinion in Microbiol. 2005;8:579–585. doi: 10.1016/j.mib.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Glass JL, Lefkowitz EJ, Glass JS, Hlener CR, Chen EY, et al. The complete sequence of the mucosal pathogen Ureaplasma urealyticum. Nature. 2000;407:757–762. doi: 10.1038/35037619. [DOI] [PubMed] [Google Scholar]
- 26.Miller WG, Parker CT, Rubenfield M, Mendz GL, Wosten MM. The complete genome sequence and analysis of the epsilon proteobacterium Arcobacter butzleri: PLoS One. 2007;2:e1358. doi: 10.1371/journal.pone.0001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rocha EPC, Danchin A. Base composition bias might result from competition for metabolic resources: TRENDS in Genetics. 2002;18:291–294. doi: 10.1016/S0168-9525(02)02690-2. [DOI] [PubMed] [Google Scholar]
- 28.Itoh T, Martin W, Nei M. Acceleration of genomic evolution caused by enhanced mutation rate in endocellular symbionts. PNAS USA. 2002;99:12944–12948. doi: 10.1073/pnas.192449699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merhej V, Royer-Carenzi M, Pontarotti P, Raoult D. Massive comparative genomic analysis reveals convergent evoluiton of specialized bacteria: Biology Direct. 2009;4:13. doi: 10.1186/1745-6150-4-13. doi: 10.1186/1745-6150-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moran NA. Microbial minimalism: Genome reduction in bacterial pathogens. Cell. 2002;108:583–586. doi: 10.1016/s0092-8674(02)00665-7. [DOI] [PubMed] [Google Scholar]
- 31.Alsmark CM, Frank AC, Karlberg EO, Legault B-A, Ardell DH, et al. The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae. PNAS USA. 2004;101:9716–9721. doi: 10.1073/pnas.0305659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engel P, Dehio C. de Reuse H, Bereswill S, editors. Genomicsof host-restricted pathogens of the genus Bartonella. 2009. pp. 158–169. Microbial Pathogenomics: Genome Dynamics: Karger, Basel. [DOI] [PubMed]
- 33.Carbajal D, Morano KA, Morano LD. Indirect immunofluorescence microscopy for direct detection of Xylella fastidiosa in xylem sap. Current Microbiology. 2004;49:372–375. doi: 10.1007/s00284-004-4369-5. [DOI] [PubMed] [Google Scholar]
- 34.Guerrero G, Peralta H, Aguilar A, Diaz R, Villalobos MA, et al. Evolutionary, structural and functional relationships revealed by comparative analysis of syntenic genes in Rhizobiales. BMC Evolutionary Biology. 2005;5:55. doi: 10.1186/1471-2148-5-55. doi: 10.1186/1471-2148-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiraga J, Mackiewicz P, Mackiewicz D, Kowalczuk M, Biecek P, et al. The relationships between the isoelectric point and: length of proteins, taxonomy and ecology of organisms: BMC Genomics. 2007;8:163. doi: 10.1186/1471-2164-8-163. doi: 10.1186/1471-2164-8-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wan X-F, Xu D, Kleinhofs A, Zhou J. Quantitative relationship between synonymous codon usage bias and GC composition across unicellular genomes. BMC Evolutionary Biology. 2004;4:19. doi: 10.1186/1471-2148-4-19. doi: 10.1186/1471-2148/4/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dillingham MS, Soultanos P, Wigley DB. Site-directed mutagenesis of motif III in PcrA helicase reveals a role in coupling ATP hydrolysis to strand separation. Nucleic Acids Research. 1999;27:3310–3317. doi: 10.1093/nar/27.16.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andersson JOA, Andersson SGE. Molecular Biology and Evolution. 2001;18:829–839. doi: 10.1093/oxfordjournals.molbev.a003864. [DOI] [PubMed] [Google Scholar]
- 39.Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, et al. Essential genes of a minimal bacterium, PNASUSA. 2006;103:425–430. doi: 10.1073/pnas.0510013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pollack JD, Myers MA, Dandekar T, Herrmann R. Suspected Utility of Enzymes with Multiple Activities in the Small Genome Mycoplasma Species: The Replacement of the Missing “Household” Nucleoside Diphosphate Kinase Gene and Activity by Glycolytic Kinases. OMICS. 2002;6:247–258. doi: 10.1089/15362310260256909. [DOI] [PubMed] [Google Scholar]
- 41.Cordwell SJ, Basseal DJ, Pollack JD, Humphery-Smith I. Malate/lactate dehydrogenase in mollicutes: evidence for a multienzyme protein. Gene. 1997;197:113–120. doi: 10.1016/s0378-1119(97)00063-2. [DOI] [PubMed] [Google Scholar]
- 42.Jishage M, Iwata A, Ueda S, Ishihama A. Regulation of RNA Polymerase sigma subunit synthesis in Escherichia coli: Intracellular levels of four species of sigma subunit under various growth conditions. J Bacteriol. 1996;178:5447–5451. doi: 10.1128/jb.178.18.5447-5451.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams KP, Sobral BW, Dickerman AW. A robust species tree for the Alpha proteobacteria: J Bacteriol. 2007;189:4578–4586. doi: 10.1128/JB.00269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li W, Levy L, Hartung JS. Quantitative distribution of ‘Candidatus Liberibacter asiaticus’ in citrus plants with citrus huanglongbing. Phytopathology. 2009;99:139–144. doi: 10.1094/PHYTO-99-2-0139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Purine metabolism in Sinorhizobium meliloti and ‘ Ca . Liberibacter asiaticus’. Enzymes annotated as present in both ‘Ca. Liberibacter asiaticus’ and S. meliloti are colored green. Enzymes annotated as present in S. meliloti but not in ‘Ca. Liberibacter asiaticus are colored orange. The metabolic pathway is from the Kyoto Encyclopedia of Genomes and Genes.
(TIF)
Pyrimidine metabolism in Sinorhizobium meliloti and ‘ Ca . Liberibacter asiaticus’. Enzymes annotated as present in both ‘Ca. Liberibacter asiaticus’ and S. meliloti are colored green. Enzymes annotated as present in S. meliloti but not in ‘Ca. Liberibacter asiaticus are colored orange. The metabolic pathway is from the Kyoto Encyclopedia of Genomes and Genes.
(TIF)
Frequency (%) of usage of amino acid codons in genomes of intracellular and free living members of the Rhizobiales.
(RTF)
Amino acids over- and under-represented in the ‘ Ca . Liberibacter asiaticus’ and Bartonella henselae proteomes as compared to the proteomes of free-living members of the Rhizobiales .
(RTF)
Proteins involved in DNA repair pathways in ‘ Ca . Liberibacter asiaticus and Sinorhizobium meliloti .
(RTF)
Proteins that contain Ligase domains annotated on chromosomal and extra chromosomal elements of ‘ Ca . Liberibacter asiaticus’ and other members of the Rhizobiales.
(RTF)