Abstract
The shape of a plant is largely determined by regulation of lateral branching. Branching architecture can vary widely in response to both genotype and environment, suggesting regulation by a complex interaction of autonomous genetic factors and external signals. Tillers, branches initiated at the base of grass plants, are suppressed in response to shade conditions. This suppression of tiller and lateral branch growth is an important trait selected by early agriculturalists during maize domestication and crop improvement. To understand how plants integrate external environmental cues with endogenous signals to control their architecture, we have begun a functional characterization of the maize mutant grassy tillers1 (gt1). We isolated the gt1 gene using positional cloning and found that it encodes a class I homeodomain leucine zipper gene that promotes lateral bud dormancy and suppresses elongation of lateral ear branches. The gt1 expression is induced by shading and is dependent on the activity of teosinte branched1 (tb1), a major domestication locus controlling tillering and lateral branching. Interestingly, like tb1, gt1 maps to a quantitative trait locus that regulates tillering and lateral branching in maize and shows evidence of selection during maize domestication. Branching and shade avoidance are both of critical agronomic importance, but little is known about how these processes are integrated. Our results indicate that gt1 mediates the reduced branching associated with the shade avoidance response in the grasses. Furthermore, selection at the gt1 locus suggests that it was involved in improving plant architecture during the domestication of maize.
Plants have evolved complex mechanisms to sense environmental changes and respond by modulating developmental programs to maximize their productivity. Plants develop from meristems, groups of stem cells that continually produce new organs throughout their life cycle, a major contrast to animal development. This prolonged developmental program facilitates enormous plasticity in plant architecture in response to environmental stimuli. Key to this plasticity is the control of meristem activity. Shoot growth is initiated by the shoot apical meristem, which produces leaf primordia on its flanks, and the tissues of the stem beneath. Axillary meristems initiate near the position where the leaf attaches to the stem. In species with strong apical dominance, these axillary meristems arrest after producing a few protective leaves, forming a lateral bud. Therefore, branching architecture is determined by regulating the switch between lateral bud dormancy and outgrowth.
The control of lateral bud dormancy or outgrowth is complex, involving both intrinsic genetic and hormonal cues as well as extrinsic signals, such as shading and nutrient availability (1). The antagonistic actions of the hormones auxin and cytokinin are major regulators of this switch (2, 3); basipetal transport of auxin from the apex promotes dormancy (3), whereas acropetal movement of cytokinin from the roots promotes bud outgrowth (4, 5). Recently, it has been shown that strigolactones, a new class of hormones, move from the roots to promote bud dormancy (6, 7). Several mutants in auxin (8–10), cytokinin (11, 12), and strigolactone (13–16) biosynthesis or signaling affect lateral bud dormancy, and the interaction of these signals is thought to regulate axillary bud outgrowth (17). In addition to hormonal regulation, at least one transcription factor, teosinte branched1 (tb1), plays a key role in lateral bud dormancy (18) and might inhibit bud growth directly by controlling cell cycle regulators (19–23). Environmental signals also strongly affect lateral bud fate; for example, plants grown at high density develop fewer branches (24–26). This response appears to result from competition for limiting nutrient resources (27) as well as from specific signals induced by shading (28–30). Plants perceive shade as a decrease in the red/far red (R/FR) light ratio, because photosynthetic pigments preferentially absorb light in the red and blue regions of the spectrum. This perception occurs via the phytochrome photoreceptor to initiate multiple developmental changes known as the shade avoidance response. In many plants, including the grasses, suppression of lateral bud outgrowth is an important part of the shade avoidance response (24–26).
It is not yet clear how the diverse intrinsic pathways and external signals controlling bud growth are integrated, and this provides an interesting system to study developmental plasticity in response to a changing environment. Furthermore, as a key determinant of plant architecture, lateral branch growth has important implications for productivity and yield in grain and bioenergy crops. In particular, the domestication of maize from its wild ancestor, teosinte (Zea mays ssp. parviglumis) (31, 32), involved a strong selection for suppression of branches (tillers) at the base of the plant and of lateral inflorescence branch (ear) elongation further up the plant. A similar reduction of tillering accompanied domestication in foxtail millet (Setaria italica) (33). Branching is more prolific in Pooideae and Erhartoideae grain crops, such as rice (Oryza sativa), wheat (Triticum aestivum), and barley (Hordeum vulgare), although increased yield in rice also results from a reduction in tiller number (34, 35). Thus, branching architecture has been a target of domestication and crop improvement in diverse cereal crops.
Quantitative trait loci (QTLs) controlling the reduction in tillering during maize domestication have also been mapped, and one major locus, tb1, has been cloned (36). tb1 is the best characterized regulator of branch architecture in maize, and homologs play similar functions in other grasses (18) and in Arabidopsis (37, 38). Selection for increased expression of tb1 during maize domestication is associated with decreased branching, and, correspondingly, maize tb1 loss-of-function mutants are highly branched (36). Interestingly, the Sorghum (Sorghum bicolor) tb1 ortholog, SbTb1, accumulates to higher levels in shade conditions, suggesting that tb1 could mediate the branching component of the shade avoidance response (39). However, additional genetic factors that regulate branching in response to shading have not been identified.
Here, we describe the isolation of grassy tillers1 (gt1), a class I homeodomain leucine zipper (HD-Zip) gene that controls lateral branching in maize. The expression of gt1 is dependent on tb1, indicating that they act in a common pathway. Furthermore, gt1 transcripts accumulate in response to shade, suggesting that this gene negatively regulates axillary bud outgrowth in response to shade signals. gt1 also maps within the interval of a major domestication QTL and shows reduced sequence diversity in maize compared with teosinte, suggesting that it was a target of selection during crop domestication.
Results
Isolation of gt1.
In a screen for mutants affecting floral development in maize, we identified a mutant in which carpel growth in the male inflorescence (tassel) was derepressed (Fig. 1 A and B). WT tassel florets abort carpels early in development (40), whereas tassel florets of the mutant frequently contained carpel-like organs and, occasionally, silks would protrude from tassel spikelets. However, carpels of the mutant were never fertile and were usually misshapen, suggesting either partial abortion or a partial lack of carpel identity.
Fig. 1.
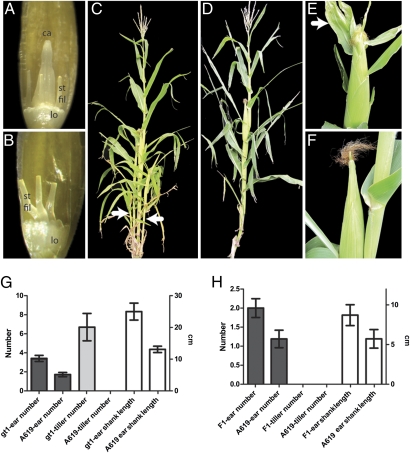
Phenotypic characterization of gt1. (A) gt1-1 tassel floret, with anthers removed to reveal the growth of a deformed carpel-like organ surrounded by three stamen filaments and two lodicules. ca, carpel-like organ; lo, lodicule; st fil, stamen filament. (B) WT tassel floret with anthers removed shows no carpel growth; only stamens filaments and lodicules are present. (C) gt1-1 mutant in B73 background with tillers (arrows). (D) WT B73 with no tillers. (E) Ear from a gt1-1 mutant, with the arrow indicating a prominent blade on a husk leaf. (F) Ear from WT B73 has no blades on husk leaves. (G) Graph showing a comparison of ear number, tiller number, and ear shank length between gt1-1 and the isogenic WT A619. The gt1-1 mutants have significantly more tillers and ears, with longer ear branches. Error bars indicate the 95% confidence interval. The scale to the left (number) is for ear and tiller number, whereas the scale to the right (cm) is for ear shank length. (H) Graph as in G showing a comparison of an F1 between A619 and gt1-1 (gt1-1/+) with WT A619 (+/+). Although the heterozygote has no tillers, it does have a significant increase in ear number and ear shoot length, indicating that gt1-1 is not fully recessive.
In addition to the floral phenotype, mutants showed increased tiller growth during vegetative development (Fig. 1 C and D), increased ear number, and elongation of ear branches (Fig. 1G). All these phenotypes indicate a failure to initiate or maintain axillary bud dormancy. Under our growing conditions, the WT A619 inbred produced no tillers, whereas the mutants (in the same genetic background) produced, on average, six to seven tillers. In addition, the mutants produced approximately twice the number of ear branches, which were significantly longer (P < 0.001) because of elongation of ear shank internodes beneath the ear proper. Husk leaves covering the ear were also abnormal, because gt1 mutants in both the A619 and B73 backgrounds frequently had blade tissue extending from the sheath region of the leaf, whereas A619 and B73 husks are primarily composed of sheath (Fig. 1 E and F). Some of these phenotypes were not completely recessive, because heterozygotes had a slight but significant increase in ear number (P < 0.001) and length of ear shanks (P = 0.002), although they had no tillers or floral phenotype (Fig. 1H). The mutant phenotypes and semidominance are similar to those reported for the classic maize mutant gt1 (41, 42). All the F1 (n = 18) and F2 progeny (n = 25) of a cross between the newly isolated mutant and gt1-ref (obtained from the Maize Genetics Coop Stock Center) were strongly tillered, indicating that they are allelic; thus, we designated our mutant gt1-1.
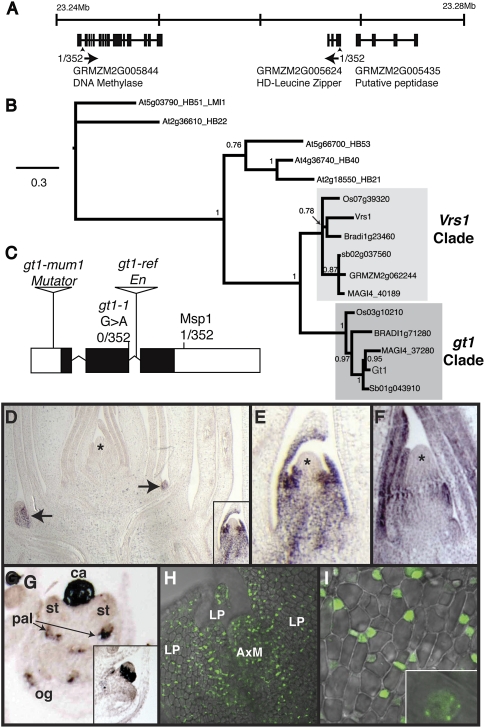
We mapped gt1-1 by bulked segregant analysis (43) to the short arm of chromosome 1, and fine mapping localized the mutation to a region containing two predicted genes, a putative HD-Zip gene and a putative DNA methylase (Fig. 2A). Sequencing of the HD-Zip gene revealed a G > A transition mutation at a putative splice donor site relative to the A619 progenitor allele (Fig. 2C). RT-PCR of the HD-Zip transcript showed that an alternative splice donor site was used in the gt1-1 transcript, causing a frameshift within the conserved homeodomain. Furthermore, we were unable to amplify a transcript from gt1-ref tissues by RT-PCR, and sequencing of the locus from this allele revealed the presence of an Enhancer/Supressor-mutator (En/Spm) (44, 45) insertion in the second intron. A third allele (gt1-mum1) carrying a Mutator (46) insertion in the 5′-untranslated region was identified by reverse genetic screening (47). Together, these data indicate that gt1 encodes an HD-Zip putative transcriptional regulatory gene.
Fig. 2.
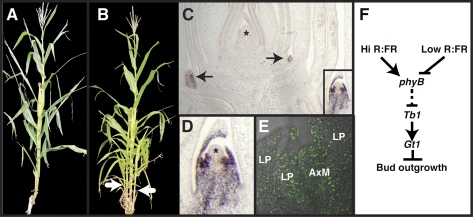
Cloning and expression of gt1. (A) Genomic region on maize chromosome 1S containing gt1-1 as determined by positional cloning. Most closely linked markers are indicated by arrowheads, with the recombination frequency and direction of recombinants underneath. (B) Phylogenetic analysis of gt1 and closely related genes from grasses (sorghum, Brachypodium, rice, and maize) and Arabidopsis. The paralogous gt1 and Vrs1 grass clades are indicated. Nodal support is indicated as Bayesian posterior probability. (C) Gene model for gt1, with position of lesions in mutant alleles gt1-1, gt1-mum1, and gt1-ref indicated. A CAPS marker for the G > A splice site mutation in gt1-1 was completely linked in our mapping population of 352 chromosomes, whereas a nearby intragenic MspI polymorphism showed recombination, indicating that the gt1 locus is in a region of high recombination. (D) In situ RNA hybridization of gt1 on a maize shoot apex. Short exposure revealed strong expression in leaves and provasculature of lateral buds (arrows and Inset) but no expression in the shoot apical meristem (*) or surrounding leaves. (E) Higher magnification of boxed lateral bud in D showing gt1 expression in the adaxial domain of surrounding leaf primordia but absent from the meristem (*). (F) Longer exposure revealed gt1 expression in the leaf primordia surrounding the shoot apex, apparently attributable to a lower level of expression than that present in the lateral buds (* indicates apical meristem). (G) gt1 is strongly expressed in degenerating carpel primordium of young tassel florets and weakly expressed in the palea and outer glume but is absent from stamen primordia. ca, carpel primordium; pal, palea; og, outer glume; st, stamen primordia. (Inset) Transverse section of a tassel floret with gt1 expression in a ring at the base of the carpel. (H) GT1-YFP expression in both the leaf primordia and the axillary meristem of a lateral tiller bud. AxM, axillary meristem; LP, leaf primordia. (I) GT1-YFP nuclear localization in the leaf primordium of lateral bud. (Inset) Magnified view.
BLAST searches and a phylogenetic analysis (Fig. 2B) indicate that gt1 is a class I HD-Zip, belonging to the ∂-subfamily (48). Although no Arabidopsis member of this subfamily has been functionally characterized, some of them are regulated by the hormones abscisic acid and auxin (48, 49). Within the grasses, there are two paralogous clades of class I HD-Zips in the ∂-subfamily, one containing gt1 and the other containing the barley (H. vulgare) six-rowed spike 1 (Vrs1) gene that controls spikelet row number. Vrs1 loss-of-function alleles were selected during barley domestication to generate six-rowed barley varieties from the ancestral two-rowed state (50).
gt1 Is Expressed in Developing Buds and Flowers.
To determine the pattern of gt1 expression, we performed in situ RNA hybridizations. The gt1 transcripts were detected in shoot axillary buds (Fig. 2 D and F) and were limited to leaf primordia and provascular tissue subtending the meristem but were absent from the meristem itself (Fig. 2E). With longer exposures, weaker expression was also detected in the leaf primordia surrounding the shoot apical meristem (Fig. 2F). In tassel primordia, gt1 transcripts were strongly expressed in the gynoecial ridge of young carpel primordia (Fig. 2G). Weaker expression was also detected in the palea and outer glume (Fig. 2G) but was apparently absent from the lemma and inner glume. The expression of gt1 in lateral buds and carpels is consistent with its function inferred from the mutant phenotype, namely, the suppression of lateral bud growth and carpel development in male florets.
To examine GT1 protein localization, we transformed maize plants with a construct expressing a C-terminal YFP fusion (GT1-YFP) under the control of the native gt1 promoter. GT1-YFP was localized to the nucleus and expressed in the leaves of axillary buds (Fig. 2 H and I), confirming the in situ pattern. Some nuclei showed subnuclear foci of GT1-YFP fluorescence, typical of transcriptional regulators. GT1-YFP fluorescence was also detected in the nuclei of cells in the meristem, suggesting that gt1 mRNA levels in the meristem might be under the level of detection by in situ hybridization or that the GT1 protein might traffic cell to cell, as described for other transcription factors in plants (51), from the leaf primordia into the meristem.
gt1 Expression Is Regulated by Light Signals in Sorghum and Teosinte.
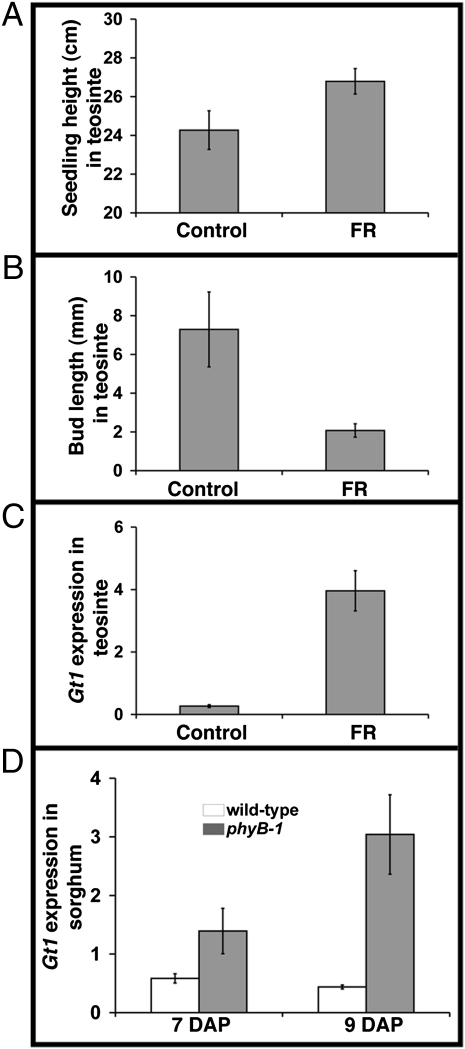
Maize gt1 is expressed, and presumably acts, within the axillary buds to repress bud outgrowth, a typical shade avoidance trait. To ask if gt1 might act in a shade avoidance pathway, we analyzed its expression in axillary buds from grasses grown with supplemental FR light to simulate shade conditions. We used teosinte and S. bicolor (sorghum) for these studies, because axillary bud growth in domesticated maize varieties, including the B73 WT strain, is constitutively repressed, whereas teosinte and sorghum display a robust shade avoidance response (39, 52, 53). Teosinte seedlings grown with supplemental FR light displayed normal shade avoidance responses, including increased plant height and inhibition of bud outgrowth (Fig. 3 A and B). We then measured teosinte gt1 transcript levels, and consistent with the idea that they act in a shade avoidance pathway, gt1 transcripts accumulated to higher levels in axillary buds following FR light treatment (Fig. 3 C). To confirm that this regulation acts through phytochrome, we examined the branching response and gt1 expression in a closely related grass, sorghum, where the appropriate mutants are available. A loss-of-function allele of the primary red light photoreceptor, phyB-1, in sorghum results in plants with few or no axillary branches, whereas the WT plants branch prolifically (39, 53). As observed in teosinte plants treated with FR light, the inhibition of bud outgrowth in sorghum phyB-1 mutants was correlated with an increased accumulation of S. bicolor Gt1 (SbGt1) transcripts in axillary buds (Fig. 3D). Together, these results suggest that the gt1 ortholog in both sorghum and teosinte regulates apical dominance of axillary buds in a shade avoidance pathway and that this response is under the control of phytochrome signal transduction.
Fig. 3.
Regulation of gt1 by light in teosinte and sorghum. Plant height (A) and length (B) of buds in the first leaf axil of teosinte seedlings at 11 d after planting grown without supplemental FR light (Control) and with supplemental FR light for 2 d starting at 9 days after planting. Error bars represent SE of seedling height and bud length of 9 or 10 seedlings. (C) Relative expression level of gt1 in axillary buds of FR-treated or control teosinte seedlings determined by quantitative real-time PCR. Error bars represent SE of three biological replicates, each from at least 3 axillary buds. (D) Relative expression level of the sorghum Gt1 (SbGt1) in WT and phyB-1 mutant axillary buds in the first leaf axil. DAP, days after planting. The expression level of SbGt1 was measured using quantitative RT-PCR. Error bars represent SE of two biological replicates, each from at least 10 axillary buds.
gt1 Expression Is Regulated by tb1.
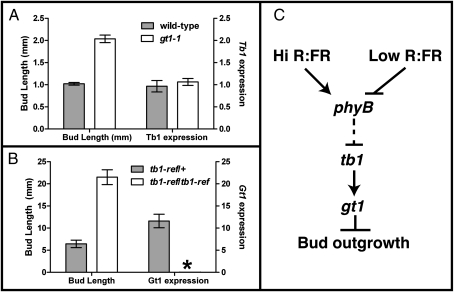
Because both tb1 and gt1 encode transcription factors that suppress bud outgrowth, we asked if they might act in a common pathway. We first examined the expression of tb1 in gt1-1 mutants. As expected, bud outgrowth in the WT (A619) is repressed, whereas buds grow out in gt1-1 mutants (Fig. 4A). However, expression of tb1 in the growing axillary buds of gt1-1 mutants was similar to that in the repressed buds of WT, suggesting that normal levels of tb1 transcripts are insufficient to mediate bud repression in the absence of gt1 (Fig. 4A). Buds also grew out in tb1-ref mutant seedlings (Fig. 4B), as expected. However, the expression of gt1 in axillary buds of homozygous tb1-ref mutants was extremely low compared with that in the WT (Fig. 4B). Together, these results suggest that gt1and tb1 act in a common pathway to control bud outgrowth and that gt1 expression is under the control of tb1 (Fig. 4C).
Fig. 4.
Interactions between gt1 and tb1. (A) Bud length and relative tb1 expression level were measured in WT (A619) and homozygous gt1-1 mutant seedlings. (B) Bud length and relative gt1 expression level were measured in heterozygous tb1-ref/+ and homozygous tb1-ref mutants. Error bars represent SE of 30 axillary buds for WT and gt1-1 mutant seedlings and three biological replicates for the expression of tb1, each from at least four axillary buds. For the tb1 mutants, error bars represent SE of the length and expression of gt1 in eight and seven axillary buds of heterozygous tb1-ref/+ and homozygous tb1-ref mutants, respectively. No detectable gt1 expression above background is indicated by an asterisk for tb1-ref/tb1-ref. (C) Model for light regulation of axillary bud growth in grasses. Perception of shading (low R/FR) via the phytochromeB photoreceptor (phyB) initiates a signaling cascade in the leaves, which ultimately transports a signal to the axillary bud that promotes tb1 transcription. tb1 expression promotes gt1 expression, leading to suppression of lateral bud outgrowth in the shade.
gt1 Shows Evidence of Selection During Maize Domestication.
A major domestication QTL for reduced tillering in maize maps to the short arm of chromosome 1 in an interval that includes gt1 (54), raising the possibility that this trait resulted from selection at the gt1 locus during domestication. To detect molecular signature(s) that should accompany selection at gt1, we sequenced both regulatory and coding sequences from diverse maize and teosinte lines. Neither the 5′-untranscribed region nor the coding sequence showed any evidence of selection; however, a region in the 3′-untranscribed region, ∼1.2–1.9 kb downstream of the stop codon, showed significant evidence of selection using both a Hudson–Kreitman–Augadé test of neutrality and a coalescent simulation (P << 0.001 for both) (SI Materials and Methods). Despite the significant evidence of selection, there was no fixed polymorphism unique to the maize haplotypes. This might mean that the causative difference lies outside the region that we sequenced, is caused by an epigenetic imprint, or occurs in several teosinte lines and is not unique to maize. Although the apparent selection and strong tillering phenotype of gt1 mutants are consistent with gt1 as the domestication QTL, further confirmation awaits the positional cloning of the locus.
In summary, we have identified gt1 as a putative HD-Zip transcription factor that regulates axillary bud dormancy and integrates external (light) signals with intrinsic (developmental) signals acting downstream of tb1 to control the shade avoidance response, a critical selected trait in crop domestication.
Discussion
Here, we show that gt1 encodes a protein with homology to class I HD-Zip transcription factors (55). None of the Arabidopsis homologs to gt1 have been characterized genetically; however, other class I genes have been associated with functions in development (56, 57) or are regulated by light (48, 58). The most closely related gene that has been functionally characterized is Vrs1, which regulates the growth of lateral spikelets in barley (50). Wild relatives of barley produce an inflorescence that is composed of a two-rowed spike, and each row produces a central fertile spikelet with two sterile lateral spikelet buds that fail to grow. During domestication, multiple loss-of-function alleles for Vrs1 were selected that allowed the growth of these lateral spikelets, producing a higher yielding six-rowed spike. It is interesting to note that gt1 and Vrs1 appear to play similar developmental roles in the suppression of lateral meristems, although they use this function in distinct developmental contexts, with Vrs1 functioning in inflorescence development and gt1 functioning during vegetative growth. This conserved function suggests that the ancestral function of the Gt1/Vrs1 clade is to suppress growth of lateral buds. We present evidence that like Vrs1, gt1 was selected during domestication to improve crop plant architecture. However, it is interesting that the effect of selection on these related genes appears to be opposite: Selection on Vrs1 was for loss of function, to promote axillary meristem growth, and selection on gt1 appears to be for increased function, to repress growth.
Another contrasting finding relates to expression. Vrs1 transcripts are detected throughout lateral spikelet meristems, whereas gt1 transcripts are found only in leaf primordia and not in developing lateral meristems. However, GT1 protein, detected as a native-expressed YFP fusion, was observed in the meristem itself. This potential non–cell-autonomous activity of gt1 could result from movement of GT1 protein from young leaf primordia into the meristem, where it presumably functions to inhibit growth. This growth inhibition is reversible, because the dormant buds can be reactivated under appropriate conditions. Gt1 expression in teosinte and sorghum axillary buds is regulated by shade. However, these buds are unlikely to be directly exposed to light because they are tightly enclosed in the axil of a large vegetative leaf. Gt1 response to the shade signal is thus likely to be uncoupled from the perception of the signal. It is possible that shading (low R/FR light ratio) induces a signal in the mature leaves and that this signal moves to the axillary bud, where it induces Gt1 expression. A similar signal has been described in Arabidopsis, where FR light perceived by the cotyledons induces the expression of a reporter gene in hypocotyls (59); however, the identity of the mobile signal is unknown.
The growth of carpels in gt1 mutants represents a similar phenomenon of derepressed growth. The unisexual male tassel florets of maize are produced by the specific abortion of carpel primordia early in the development of male floret primordia (40). This abortion is not simply an arrest of carpel growth but involves programmed cell death of the carpel primordia (60). How then does gt1 contribute to carpel abortion? One possibility is that gt1 inhibits growth during the early stages of carpel development and that this is required for complete abortion by the programmed cell death pathway. Thus, gt1 mutants produce deformed and partially aborted carpel-like organs. Another possibility is that gt1 has a distinct role in the carpel abortion pathway that is distinct from its bud growth inhibition activity. Further work will be necessary to understand the role of gt1 in floral development.
gt1 as a Mediator of the Shade Avoidance Response.
Low R/FR light ratio, or shade, is perceived by phytochromes and induces a set of responses, including increased plant height, enhanced apical dominance, and early flowering, known collectively as the shade avoidance syndrome (61). The enhanced apical dominance response to shade has been investigated for the past several decades (25, 26), although the molecular mechanisms have remained elusive. Using a phyB-1 mutant of sorghum, Kebrom et al. (39) showed that the inhibition of axillary bud outgrowth by FR light, perceived by phyB, was associated with increased expression of tb1, suggesting a molecular link between the shade avoidance pathway and a known regulator of bud dormancy. Here, we show that the inhibition of bud outgrowth by FR light is also associated with enhanced expression of gt1. Thus, both tb1 and gt1 appear to inhibit bud outgrowth in response to shade signals perceived by phyB. Our expression analyses suggest that tb1 acts upstream of gt1, because gt1 expression is dependent on tb1 activity, although it remains to be determined whether this regulation is direct.
gt1 Regulates Agronomically Important Traits.
gt1 regulates tiller growth and the shade avoidance response, making it a promising candidate for the modification of agronomically important traits. For example, production of increased biomass is particularly important for improvement of proposed bioenergy crops, such as switchgrass (Panicum virgatum) (62). Maintaining branching at high planting densities, by decreasing Gt1 activity, could therefore provide a mechanism to improve yield in bioenergy crops. In contrast, reduced branching has been associated with increased yields in rice (34, 35), suggesting that the fine-tuning of axillary bud development is central to regulating grain yield as well.
In addition to its possible utility for crop improvement, gt1 activity appears to have been a target of selection during domestication and is a candidate for a QTL for reduced lateral branch growth in maize. This result suggests that maize domestication involved modification of a developmental pathway that integrates environmental cues and shows how modification of such responses is critical for growth of plants under changing environmental conditions.
Materials and Methods
Genetic Materials and Sources.
gt1-1 was isolated from a screen of M2 families generated by ethyl methanesulfonate (EMS) mutagenesis of the A619 maize inbred (63). gt1-ref was reportedly isolated from seed mutagenized during nuclear bomb tests on the Bikini Atoll, and stock was obtained from the Maize Genetics Coop Stock Center. gt1-mum1 was isolated by reverse genetic screening for Mutator transposon insertions in the gt1 locus. Newly developed materials described in this article may be available for noncommercial research purposes on acceptance and signing of a material transfer agreement. Obtaining any permissions will be the sole responsibility of the requestor.
Phenotypic Characterization.
Measurement of tiller number, ear number, and shank length was performed on field-grown plants in San Diego, CA. The gt1-1 and A619 plants were planted in separate rows, with individuals spaced ∼8–12 inches apart, and measured at maturity. All branches originating from nodes at or below ground level were considered tillers, whereas those originating from nodes further up were considered ear branches. Ear shank length was measured as the distance from the base of the ear (last kernel) to the point where the branch originated on the main stem. Similar measurements were taken for the F1 plants of a cross between A619 and gt1-1 (gt1-1/+), but these were grown in Molokai, HI, at a higher density (∼3–5 inches between individuals).
Cloning of gt1.
Bulked-segregant analysis (43) was performed on a pool of 10 homozygous mutants from an F2 population derived from a cross of gt1-1 with the B73 inbred to localize gt1 to the long arm of chromosome 1. Simple sequence repeat markers for bulked-segregant analysis were selected as previously described (64). Markers flanking gt1 (bnlg1614 and PCO139549) were used to screen 176 homozygous gt1-1 mutants for recombinants from the F2 mapping population. Cleaved amplified polymorphic sequence (CAPS) (65) markers were designed for maize genes in this interval by sequencing and identifying polymorphisms between A619 and B73. A CAPS marker to a polymorphic MspI site in the 3′ UTR of gt1 showed a single recombinant with the mutation, whereas a CAPS marker to the G > A splice site mutation of gt1-1 showed complete linkage. The CAPS marker that identifies the gt1-1 mutation uses the primers gt1-CAPs-For (5′-AGGTGGCCGTCTGGTTCCAGAA-3′) and gt1-CAPS-Rev (5′TGGTGCGTCACCGTCGAGAAC-3′) to amplify the sequence by PCR, followed by BsaJI digestion and separation on a 3.5% (wt/vol) MetaPhor agarose (Cambrex Research Products) gel, resulting in fragments of 201, 121, and 39 bp in WT vs. 201 and 160 bp in gt1-1 mutants.
Phylogenetic Analysis.
cDNA sequences for gt1 and its grass and Arabidopsis homologs were aligned (Dataset S1) using ClustalX (66). Phylogeny was inferred using MrBayes (67) with the GTR + G model of nucleotide substitution, 2 million generations, a sample frequency of 100, and a burn-in value of 5,000.
In Situ Hybridization.
The gt1 cDNA was amplified using primers HDLZ-For (5′-CCTAGTCCTAGTACAGGCTACAG-3′) and HDLZ-Rev (5′-CGGTCCATCCATCCATTAACACG-3′) and was cloned into pCRII-TOPO vector (Invitrogen). Antisense digoxygenin-labeled RNA probe was synthesized using T7 RNA polymerase. B73 shoot apices and tassel primordia were prepared, sectioned, and hybridized according to a published protocol (68). Strong signal from the gt1 probe in lateral buds and carpel primordial required a short (3 h) incubation during the detection step, whereas a longer (12–24 h) incubation was required to detect expression in leaves surrounding the apical meristem.
GT1-YFP and Microscopy.
A C-terminal fusion, 10 aa before the stop codon, of YFP to the genomic sequence of gt1, including introns, 3.3 kb 5′ of the start codon, and 1 kb 3′ of the stop codon was created using the Multisite Gateway Three Fragment System (Invitrogen) as described previously (64, 69). The 5′ promoter and coding sequence were amplified using Gt1-attB4 (5′-GGGGACAACTTTGTATAGAAAAGTTGAGTTGATGGCGGTTGAACTC-3′) and Gt1-attB1 (5′-GGGGACTGCTTTTTTGTACAAACTTGCTCCACCGAAGTAGGCGGGCG-3′), whereas the 3′ sequence was amplified using Gt1-attB2 (5′-GGGGACAGCTTTCTTGTACAAAGTGGGAGTCGTCTACGACTACGACC-3′) and Gt1-attB3 (5′-GGGGACAACTTTGTATAATAAAGTTGAGAGAAAAGGCGTGGAGTGA-3′). The underlined sequences in primers denote the att sites necessary for multisite cloning. Maize plants were transformed as described (69). Lateral buds were dissected from transgenic plants, and GT1-YFP fluorescence was imaged using a Zeiss 710 confocal microscope.
Plant Material and Growing Conditions for Supplemental FR Experiments.
Seeds were sown in flats containing cells 6 × 6 × 5.5 cm−3 in volume filled with a soil mix containing 35% (vol/vol) peat moss, 10% (vol/vol) vermiculite, 35% (vol/vol) baked clay, 10% (vol/vol) sand and 10% (vol/vol) topsoil. Seedlings were grown in a high-light-intensity growth chamber illuminated with incandescent and metal halide lamps (500–600 micromol per meter squared per second (umol m−2 sec−1)). Plants were grown under a 12-h light/12-h dark photoperiod at 31 °C light/22 °C dark. Supplemental FR was applied laterally with FR-emitting diodes. FR treatment was started after the buds in the first or second leaf axils were well formed, as determined by examining their developmental progression, and continued for 2 d during the light period. Buds were harvested under a dissecting microscope, and their length was measured using a micrometer if it was less than 3 mm. Buds longer than 3 mm were measured using a ruler.
Gene Expression Analysis by Quantitative Real-Time PCR.
Expression levels of gt1 and tb1 in axillary buds were quantified by quantitative real-time–PCR (70). Buds from the first or second leaf axils were dissected and immersed in a lysis-binding solution, and RNA was extracted using TRIzol (Invitrogen). RNAs were quantified using a NanoDrop 1000 (Thermo Scientific), and 1.5 μg of RNA from each sample was treated with DNase I (Invitrogen). Half of the DNase-treated 1.5 μg of RNA was reverse-transcribed using SuperScript III according to the manufacturer's protocol (Invitrogen), whereas the remaining half was used as a negative RT control. Quantitative PCR was performed using SYBR green (Sigma) on an ABI 7900HT (Applied Biosystems). The target cycle threshold values were normalized using 18S rRNA. The relative expression level was analyzed using the mean normalized threshold value as a reference for all the samples in each biological replicate. At least three biological replicates were used for the expression levels of gt1 in teosinte and tb1 in gt1-1 and WT (A619), and two biological replicates were used for the expression of Sorghum Gt1 (SbGt1). The expression of gt1 in tb1 mutants was analyzed in single buds. Both DNA and RNA were extracted from single buds. The DNA was used for genotyping, whereas the RNA was used to measure the expression level of gt1. Therefore, the expression level of gt1 is the mean of the level in eight heterozygous and seven homozygous axillary buds. The forward and reverse primer sequences were (TTCCCTCAACGTGAGCTTCT/ TTCATCGTCACACAGCCAAT) for Tb1, (GCTGCGAGGAGGAAGAGAG/CTGCCGAGCCTTCTTCTG) for Gt1, (GCAGCAGCTCGATCTCTTCT/AGCCCATGGTTCTTCAGCTA) for SbGt1, and (ATTCTATGGGTGGTGGTGCAT/TCAAACTTCGCGGCCTAAA) for18S rRNA.
Supplementary Material
Acknowledgments
The gt1-ref seed stock was provided by the Maize Genetics Coop Stock Center. Maize transgenics were produced by the Iowa State University transformation facility. Keoni Kauwe provided assistance in formatting Figs. 2 and 4. Funding for this work was provided by the National Science Foundation (Grants DBI-0820619, DBI-0604923, and DBI-0501862) and US Department of Agriculture (Hatch Grant MSN101593).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JF420894–JF421123).
This article is a PNAS Direct Submission.
See Author Summary on page 13375.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102819108/-/DCSupplemental.
References
- 1.McSteen P. Hormonal regulation of branching in grasses. Plant Physiol. 2009;149:46–55. doi: 10.1104/pp.108.129056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sachs T, Thimann KV. The role of auxins and cytokinins in the release of buds from dominance. Am J Bot. 1967;54:136–144. [Google Scholar]
- 3.Thimann KV, Skoog F. Studies on the Growth Hormone of plants: III. The Inhibiting Action of the Growth Substance on Bud Development. Proc Natl Acad Sci USA. 1933;19:714–716. doi: 10.1073/pnas.19.7.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li C-J, Guevara E, Herrerea J, Bangerth F. Effect of apex excision and replacement by 1-naphthylacetic acid on cytokinin concentration and apical dominance in pea plants. Physiol Plant. 1995;94:465–469. [Google Scholar]
- 5.Bangerth F. Response of cytokinin concentration in the xylem exudate of bean (Phaseolus vulgaris L) plants to decapitation and auxin treatment, and relationship to apical dominance. Planta. 1994;194:439–442. [Google Scholar]
- 6.Gomez-Roldan V, et al. Strigolactone inhibition of shoot branching. Nature. 2008;455:189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- 7.Umehara M, et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature. 2008;455:195–200. doi: 10.1038/nature07272. [DOI] [PubMed] [Google Scholar]
- 8.Lincoln C, Britton JH, Estelle M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell. 1990;2:1071–1080. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu M, Zhu L, Shou HX, Wu P. A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 2005;46:1674–1681. doi: 10.1093/pcp/pci183. [DOI] [PubMed] [Google Scholar]
- 10.Li PJ, et al. LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res. 2007;17:402–410. doi: 10.1038/cr.2007.38. [DOI] [PubMed] [Google Scholar]
- 11.Helliwell CA, et al. The Arabidopsis AMP1 gene encodes a putative glutamate carboxypeptidase. Plant Cell. 2001;13:2115–2125. doi: 10.1105/TPC.010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tantikanjana T, et al. Control of axillary bud initiation and shoot architecture in Arabidopsis through the SUPERSHOOT gene. Genes Dev. 2001;15:1577–1588. doi: 10.1101/gad.887301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stirnberg P, van De Sande K, Leyser HMO. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development. 2002;129:1131–1141. doi: 10.1242/dev.129.5.1131. [DOI] [PubMed] [Google Scholar]
- 14.Sorefan K, et al. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 2003;17:1469–1474. doi: 10.1101/gad.256603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Snowden KC, et al. The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell. 2005;17:746–759. doi: 10.1105/tpc.104.027714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Booker J, et al. MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev Cell. 2005;8:443–449. doi: 10.1016/j.devcel.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 17.Beveridge CA, Dun EA, Rameau C. Pea has its tendrils in branching discoveries spanning a century from auxin to strigolactones. Plant Physiol. 2009;151:985–990. doi: 10.1104/pp.109.143909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- 19.Kosugi S, Ohashi Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell. 1997;9:1607–1619. doi: 10.1105/tpc.9.9.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kosugi S, Ohashi Y. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 2002;30:337–348. doi: 10.1046/j.1365-313x.2002.01294.x. [DOI] [PubMed] [Google Scholar]
- 21.Li CX, Potuschak T, Colón-Carmona A, Gutiérrez RA, Doerner P. Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci USA. 2005;102:12978–12983. doi: 10.1073/pnas.0504039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaudin V, et al. The expression of D-cyclin genes defines distinct developmental zones in snapdragon apical meristems and is locally regulated by the Cycloidea gene. Plant Physiol. 2000;122:1137–1148. doi: 10.1104/pp.122.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trémousaygue D, et al. Internal telomeric repeats and ‘TCP domain’ protein-binding sites co-operate to regulate gene expression in Arabidopsis thaliana cycling cells. Plant J. 2003;33:957–966. doi: 10.1046/j.1365-313x.2003.01682.x. [DOI] [PubMed] [Google Scholar]
- 24.Dubois PG, Brutnell TP. Light signal transduction networks in maize. In: Hake S, Bennetzen JL, editors. Handbook of Maize: Its Biology. Vol. 1. New York: Springer; 2009. pp. 205–228. [Google Scholar]
- 25.Casal JJ, Sanchez RA, Deregibus VA. The effect of plant-density on tillering: The involvement of R/FR ratio and the proportion of radiation intercepted per plant. Environ Exp Bot. 1986;26:365–371. [Google Scholar]
- 26.Casal JJ. Light quality effects on the appearance of tillers of different order in wheat (Triticum aestivum) Ann Appl Biol. 1988;112:167–173. [Google Scholar]
- 27.Power JF, Alessi J. Tiller development and yield of standard and semidwarf spring wheat varieties as affected by nitrogen fertilizer. The Journal of Agricultural Science. 1978;90:97–108. [Google Scholar]
- 28.Kebrom TH, Brutnell TP. The molecular analysis of the shade avoidance syndrome in the grasses has begun. J Exp Bot. 2007;58:3079–3089. doi: 10.1093/jxb/erm205. [DOI] [PubMed] [Google Scholar]
- 29.Doust AN. Grass architecture: Genetic and environmental control of branching. Curr Opin Plant Biol. 2007;10:21–25. doi: 10.1016/j.pbi.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 30.Doust A. Architectural evolution and its implications for domestication in grasses. Ann Bot (Lond) 2007;100:941–950. doi: 10.1093/aob/mcm040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doebley J, Stec A. Inheritance of the morphological differences between maize and teosinte: Comparison of results for two F2 populations. Genetics. 1993;134:559–570. doi: 10.1093/genetics/134.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Briggs WH, McMullen MD, Gaut BS, Doebley J. Linkage mapping of domestication loci in a large maize teosinte backcross resource. Genetics. 2007;177:1915–1928. doi: 10.1534/genetics.107.076497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doust AN, Devos KM, Gadberry MD, Gale MD, Kellogg EA. Genetic control of branching in foxtail millet. Proc Natl Acad Sci USA. 2004;101:9045–9050. doi: 10.1073/pnas.0402892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiao Y, et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet. 2010;42:541–544. doi: 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- 35.Miura K, et al. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet. 2010;42:545–549. doi: 10.1038/ng.592. [DOI] [PubMed] [Google Scholar]
- 36.Wang RL, Stec A, Hey J, Lukens L, Doebley J. The limits of selection during maize domestication. Nature. 1999;398:236–239. doi: 10.1038/18435. [DOI] [PubMed] [Google Scholar]
- 37.Aguilar-Martínez JA, Poza-Carrión C, Cubas P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell. 2007;19:458–472. doi: 10.1105/tpc.106.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finlayson SA, Krishnareddy SR, Kebrom TH, Casal JJ. Phytochrome regulation of branching in Arabidopsis. Plant Physiol. 2010;152:1914–1927. doi: 10.1104/pp.109.148833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kebrom TH, Burson BL, Finlayson SA. Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol. 2006;140:1109–1117. doi: 10.1104/pp.105.074856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng PC, Greyson RI, Walden DB. Organ initiation and the development of unisexual flowers in the tassel and ear of Zea mays. Am J Bot. 1983;70:450–462. [Google Scholar]
- 41.Shaver DL. Perennial maize. J Hered. 1967;58:271–273. [Google Scholar]
- 42.Tracy WF, Everett HL. Variable penetrance and expressivity of grassy tillers, gt. Maize Genetics Cooperation News Letter. 1982;56:77–78. [Google Scholar]
- 43.Michelmore RW, Paran I, Kesseli RV. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA. 1991;88:9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pereira A, Cuypers H, Gierl A, Schwarz-Sommer Z, Saedler H. Molecular analysis of the En/Spm transposable element system of Zea mays. EMBO J. 1986;5:835–841. doi: 10.1002/j.1460-2075.1986.tb04292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pereira A, et al. Genetic and molecular analysis of the Enhancer (En) transposable element system of Zea mays. EMBO J. 1985;4:17–23. doi: 10.1002/j.1460-2075.1985.tb02311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bennetzen JL, Swanson J, Taylor WC, Freeling M. DNA insertion in the first intron of maize Adh1 affects message levels: Cloning of progenitor and mutant Adh1 alleles. Proc Natl Acad Sci USA. 1984;81:4125–4128. doi: 10.1073/pnas.81.13.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCarty DR, Meeley RB. Transposon resources for forward and reverse genetics in maize. In: Hake S, Bennetzen JL, editors. Handbook of Maize. Vol. 2. New York: Springer; 2009. pp. 561–584. [Google Scholar]
- 48.Henriksson E, et al. Homeodomain leucine zipper class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant Physiol. 2005;139:509–518. doi: 10.1104/pp.105.063461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Son O, et al. Induction of a homeodomain-leucine zipper gene by auxin is inhibited by cytokinin in Arabidopsis roots. Biochem Biophys Res Commun. 2005;326:203–209. doi: 10.1016/j.bbrc.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 50.Komatsuda T, et al. Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Natl Acad Sci USA. 2007;104:1424–1429. doi: 10.1073/pnas.0608580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu XM, Jackson D. Lights at the end of the tunnel: New views of plasmodesmal structure and function. Curr Opin Plant Biol. 2010;13:684–692. doi: 10.1016/j.pbi.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 52.Doebley J, Stec A, Gustus C. teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics. 1995;141:333–346. doi: 10.1093/genetics/141.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Childs KL, et al. The sorghum photoperiod sensitivity gene, Ma3, encodes a phytochrome B. Plant Physiol. 1997;113:611–619. doi: 10.1104/pp.113.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quijada P, Shannon LM, Glaubitz JC, Studer AJ, Doebley J. Characterization of a major maize domestication Qtl on the short arm of chromosome 1. Maydica. 2009;54:401–408. [Google Scholar]
- 55.Sessa G, et al. Identification of distinct families of HD-Zip proteins in Arabidopsis thaliana. In: Puidgomenech P, Coruzzi G, editors. Plant Molecular Biology: Molecular-Genetic Analysis of Plant Development and Metabolism. Berlin: Springer; 1994. pp. 411–426. [Google Scholar]
- 56.Saddic LA, et al. The LEAFY target LMI1 is a meristem identity regulator and acts together with LEAFY to regulate expression of CAULIFLOWER. Development. 2006;133:1673–1682. doi: 10.1242/dev.02331. [DOI] [PubMed] [Google Scholar]
- 57.Ariel F, et al. Environmental regulation of lateral root emergence in Medicago truncatula requires the HD-Zip I transcription factor HB1. Plant Cell. 2010;22:2171–2183. doi: 10.1105/tpc.110.074823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manavella PA, Dezar CA, Ariel FD, Drincovich MF, Chan RL. The sunflower HD-Zip transcription factor HAHB4 is up-regulated in darkness, reducing the transcription of photosynthesis-related genes. J Exp Bot. 2008;59:3143–3155. doi: 10.1093/jxb/ern170. [DOI] [PubMed] [Google Scholar]
- 59.Tanaka S, Nakamura S, Mochizuki N, Nagatani A. Phytochrome in cotyledons regulates the expression of genes in the hypocotyl through auxin-dependent and -independent pathways. Plant Cell Physiol. 2002;43:1171–1181. doi: 10.1093/pcp/pcf133. [DOI] [PubMed] [Google Scholar]
- 60.Calderon-Urrea A, Dellaporta SL. Cell death and cell protection genes determine the fate of pistils in maize. Development. 1999;126:435–441. doi: 10.1242/dev.126.3.435. [DOI] [PubMed] [Google Scholar]
- 61.Smith H. Physiological and ecological function within the phytochrome family. Annu Rev Plant Physiol. 1995;46:289–315. [Google Scholar]
- 62.McLaughlin SB, Kszos LA. Development of switchgrass (Panicum virgatum) as a bioenergy feedstock in the United States. Biomass Bioenergy. 2005;28:515–535. [Google Scholar]
- 63.Neuffer MG. Mutagenesis. In: Freeling M, Walbot V, editors. The Maize Handbook. New York: Springer; 1994. pp. 212–218. [Google Scholar]
- 64.Whipple CJ, et al. A conserved mechanism of bract suppression in the grass family. Plant Cell. 2010;22:565–578. doi: 10.1105/tpc.109.073536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- 66.Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:430–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 67.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 68.Jackson D, Veit B, Hake S. Expression of maize knotted1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development. 1994;120:405–413. [Google Scholar]
- 69.Mohanty A, et al. Advancing cell biology and functional genomics in maize using fluorescent protein-tagged lines. Plant Physiol. 2009;149:601–605. doi: 10.1104/pp.108.130146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kebrom TH, Brutnell TP, Finlayson SA. Suppression of sorghum axillary bud outgrowth by shade, phyB and defoliation signalling pathways. Plant Cell Environ. 2010;33:48–58. doi: 10.1111/j.1365-3040.2009.02050.x. [DOI] [PubMed] [Google Scholar]