Abstract
Mammalian mtDNA is packaged in DNA-protein complexes denoted mitochondrial nucleoids. The organization of the nucleoid is a very fundamental question in mitochondrial biology and will determine tissue segregation and transmission of mtDNA. We have used a combination of stimulated emission depletion microscopy, enabling a resolution well below the diffraction barrier, and molecular biology to study nucleoids in a panel of mammalian tissue culture cells. We report that the nucleoids labeled with antibodies against DNA, mitochondrial transcription factor A (TFAM), or incorporated BrdU, have a defined, uniform mean size of ∼100 nm in mammals. Interestingly, the nucleoid frequently contains only a single copy of mtDNA (average ∼1.4 mtDNA molecules per nucleoid). Furthermore, we show by molecular modeling and volume calculations that TFAM is a main constituent of the nucleoid, besides mtDNA. These fundamental insights into the organization of mtDNA have broad implications for understanding mitochondrial dysfunction in disease and aging.
Keywords: mitochondria, nanoscopy
Mitochondria have evolved from eubacteria-like precursors and the mitochondrial DNA (mtDNA) represents a vestige of the ancestral genome of the endosymbiont (1). The mtDNA was discovered in the 1960s by electron microscopy analysis of chicken tissue sections (2) and has a contour length of 5 μm (3). The mammalian mtDNA is a highly compacted genome of ∼16.5 kb and is very gene-dense, despite its small size, encoding 13 proteins, 2 ribosomal RNAs, and 22 tRNAs (4, 5). The mtDNA was initially considered to be naked, unprotected, and vulnerable to damage. However, work during the last decades has clarified that mtDNA is protein-coated and packaged into aggregates denoted nucleoids (6). In budding yeast, the high-mobility-group (HMG) box domain protein ABF2 binds and fully coats mtDNA and is essential for mtDNA maintenance (7). The mammalian ABF2 homolog, mitochondrial transcription factor A (TFAM), was first identified as a mitochondrial transcriptional activator by a biochemical approach (8) and subsequent work has confirmed that it is an indispensable component of the basal mtDNA transcription machinery (9). Similar to other HMG box proteins, TFAM is able to bind, wrap, bend, and unwind DNA without sequence specificity (10). Furthermore, TFAM is quite abundant and coats mtDNA in Xenopus laevis (11), chicken (12), mouse (13, 14), and human cells (15, 16). In vivo data from mouse models has demonstrated that disruption of the Tfam gene leads to loss of mtDNA and embryonic lethality (17), whereas increase of TFAM protein levels leads to increase of mtDNA copy number (13). Confocal microscopy has shown that mtDNA and TFAM colocalize in mammalian cells and are present in punctuate aggregates corresponding to nucleoids (18, 19). A large number of putative nucleoid proteins have been identified by using biochemical approaches to identify proteins that can be cross-linked to or copurified with mtDNA (20) or that colocalize with mtDNA on confocal microscopy (18, 19). Association of a protein that is essential for mtDNA maintenance with mtDNA does not necessarily mean that it has a role in structural organization of the nucleoid. Currently, TFAM is the only protein that fulfils a more stringent definition of a structural component of the mammalian nucleoid (21). It has been reported that up-regulation of mtDNA copy number can result in nucleoid size variability and the formation of larger nucleoids (22). Confocal microscopy studies of nucleoids have reported sizes of these structures that are close to or even substantially below the diffraction limit of ∼260 nm. Hence, conventional light microscopy is not suited to determine the size of nucleoids. Furthermore, an understanding of principles governing the replication and segregation of mtDNA (21) will require a definition of the mtDNA copy number per nucleoid. The organization of the nucleoid is thus a very fundamental question in mammalian mitochondrial genetics. We have used stimulated emission depletion (STED) microscopy (23, 24), enabling a resolution well below the diffraction barrier, to study mitochondrial nucleoids and report here that they have a very uniform mean size in a variety of mammalian species. In addition, by combining molecular biology and STED microscopy, we report that many nucleoids contain just a single mtDNA molecule and that TFAM is the main protein component.
Results
Mitochondrial Nucleoids Have a Uniform Mean Size in Mammalian Cells.
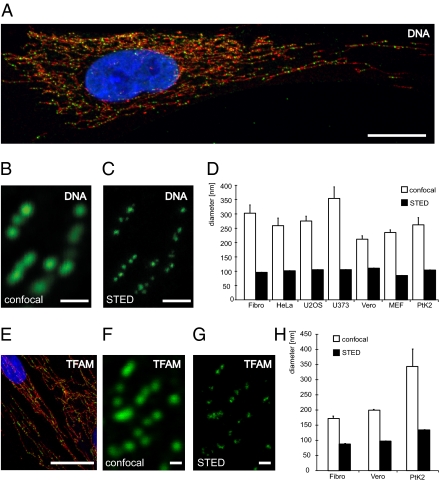
Confocal imaging of mitochondrial nucleoids visualized by DNA antibodies results in a punctuate pattern within the mitochondrial network of human fibroblasts (Fig. 1A). Image analysis revealed that the nucleoids have an apparent average diameter of ∼300 nm (Fig. 1 B and D), which is close to the lateral resolution limit of light microscopy. Confocal microscopy can thus not be used to assign a size to mammalian nucleoids because their actual sizes may be obscured by the diffraction-limited resolution. To accurately determine nucleoid size and size variability, we used STED microscopy. We found that the nucleoids labeled with DNA-specific antiserum had a considerably smaller size than the one suggested by confocal microscopy (Fig. 1 B and C). We quantitatively analyzed super-resolution micrographs and found that the mean apparent diameter of nucleoids was 99 nm in human fibroblasts (Fig. 1 C and D). The STED microscopy set-up we used enables a resolution of 40 to 50 nm (25) and, hence, the observed nucleoid diameter of ∼100 nm represents the true size of antibody-labeled nucleoids. We proceeded to study a variety of mammalian cell lines, such as human primary culture fibroblasts (Fibro), human cervix adenocarcinoma cells (HeLa), human osteosarcoma cells (U2OS), human glioblastoma cells (U373), culture mouse embryonic fibroblasts (MEF), African green monkey (Cercopithecus aethiops) kidney epithelial cells (Vero), and potoroo (Potorous tridactylus) kidney cells (PtK2), by using DNA antibodies and found an average nucleoid size of 271 ± 23 nm (n = 22,918) by confocal microscopy (Fig. 1D). Unexpectedly, STED microscopy revealed a very uniform nucleoid size of 85 to 111 nm (mean 102 ± 0.3 nm, n = 38,777) in all studied mammalian species (Fig. 1D). We proceeded to use TFAM as an independent marker of nucleoid size to confirm the results obtained by using DNA antibodies. Confocal microscopy with antibodies detecting TFAM in human fibroblasts revealed a punctuate pattern (Fig. 1E) very similar to the one observed with DNA antibody staining (Fig. 1A). Double staining with TFAM antibodies and the DNA-binding dye DAPI resulted in the expected colocalization of both markers in all studied nucleoids (Fig. S1). We proceeded to analyze human fibroblasts, Vero cells, and PtK2 cells with TFAM antibodies. We found a mean nucleoid size of 238 ± 21 nm (n = 7,414) by confocal microscopy (Fig. 1 F and H) and a uniform nucleoid size of 88 to 134 nm (mean 107 ± 0.6 nm, n = 11,009) by STED microscopy (Fig. 1 G and H). Our findings thus show that the antibody-decorated nucleoid has a very uniform mean size of ∼100 nm in all mammals, and by subtracting the estimated size of the antibodies (26, 27), the nucleoid can be predicted to have a diameter of ∼70 nm. The uniform mean nucleoid size in mammals suggests a strong evolutionary conservation in the way mtDNA is organized and maintained.
Fig. 1.
The nucleoid has a uniform mean size in mammalian cells. (A) mtDNA (green, DNA antibodies) is localized in nucleoids in the tubular mitochondria (red, anti-TOM20) of human fibroblasts. DAPI-staining of nucleus in blue. (B) Using confocal microscopy, nucleoids labeled with an antiserum against DNA appear to be solid structures with an average diameter of ∼300 nm in human fibroblasts. (C) Applying super-resolution STED microscopy, we found that the apparent nucleoids, as detected by confocal microscopy, frequently are agglomerates of several smaller structures, each with an average diameter of ∼99 nm. The section shown is the same as the one in B. (D) Sizes of nucleoids labeled with a DNA antibody determined by confocal and STED imaging in mammalian cell lines, such as, human primary culture fibroblasts (Fibro), human cervix adenocarcinoma cells (HeLa), human osteosarcoma cells (U2OS), human glioblastoma cells (U373), mouse embryonic fibroblasts (MEF), African green monkey kidney epithelial cells (Vero), and potoroo kidney cells (PtK2). The indicated sizes denote the antibody labeled nucleoids. (E) TFAM (green, anti-TFAM) is located in nucleoids in the mitochondrial network (red, anti-TOM20) of human fibroblasts. Nuclear DAPI-staining in blue. (F) Using confocal microscopy, the nucleoids labeled with an antiserum against TFAM appear in a punctuate pattern. (G) STED microscopy reveals a mean diameter of ∼88 nm for nucleoids labeled with a TFAM antibody in human fibroblasts. The section shown is the same as the one in F. (H) Quantification of the sizes of nucleoids labeled with TFAM antibodies determined by confocal and STED imaging in three mammalian cell lines. Error bars indicate SEM. (Scale bars: 20 μm in A and E, 0.5 μm in B, C, F, and G.)
More Nucleoids per Cell Are Visible with STED than with Confocal Microscopy.
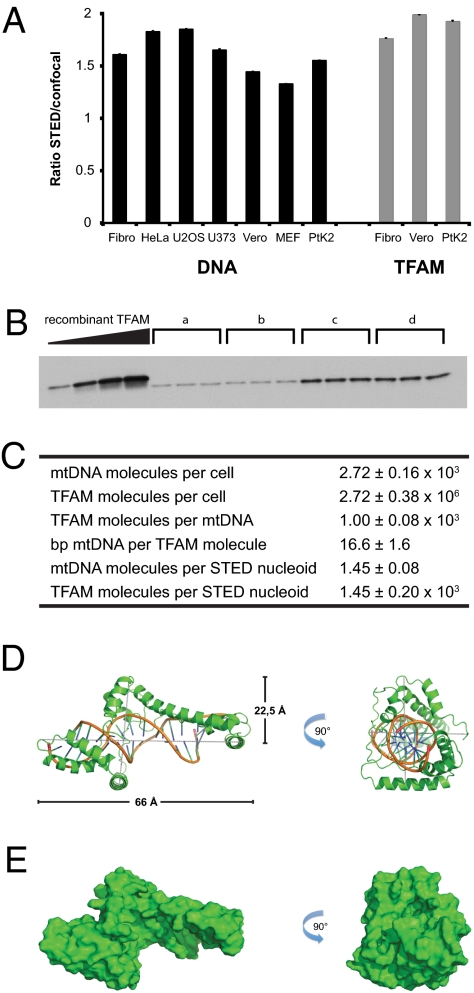
Using confocal microscopy we found on average 1,184 ± 59 nucleoids per cell with DNA antibodies (n = 486 cells) and 964 ± 50 nucleoids per cell with TFAM antibodies (n = 325 cells) in human fibroblasts. Our initial analyses suggested that the number of nucleoids observed by STED microscopy were larger than the number observed by confocal microscopy (Fig. 1 B, C, F, and G). When we compared images of different mammalian cell lines taken with a STED and confocal microscope, we observed on average ∼1.6 times more nucleoids when using antibodies against DNA and ∼1.9 times more nucleoids when using an antiserum against TFAM (Fig. 2A). Using these factors one can use confocal image data to calculate the accurate number of nucleoids.
Fig. 2.
Combination of molecular biology methods and super-resolution microscopy. (A) Ratio of the number of nucleoids detected by STED microscopy versus the number of nucleoids detected by confocal microscopy. Black bars: labeling with an antiserum against DNA; gray bars: labeling with an antiserum against TFAM. (B) Representative quantitative Western blot measurement of endogenous TFAM protein levels in human fibroblasts. “a” to “d” indicate protein extracts in triplicates from a determined number of cells. His-tagged, purified recombinant TFAM was used as standard in known concentrations (16.5, 33, 49.5, and 66 ng). (C) Table displaying the results of the quantitative real-time PCR, the quantitative Western blots and the resulting ratios of combining molecular biology methods with super-resolution microscopy. (D) Overall predicted structure of human TFAM bound to DNA. A cartoon representation shows TFAM in green and DNA by element with backbone in orange. (E) Surface representation of human TFAM bound to DNA. The solvent accessible surface is shown in green and has the same orientations as in D. Errors and error bars indicate SEM.
Many Nucleoids Contain a Single Copy of mtDNA.
To quantify the number of mtDNA and TFAM molecules per nucleoid, we combined STED microscopy with molecular biology approaches. We used DNA antibodies and confocal microscopy in combination with the factor described above and found 1,883 ± 106 nucleoids per human fibroblast. Next, we used quantitative real-time PCR (qRT-PCR) to calculate the mtDNA copy number per cell by using cloned mtDNA fragments as a standard. We found on average 2,721 ± 156 copies of mtDNA per human fibroblast when using three different Taqman probes (Fig. 2C). There are thus on average ∼1.4 mtDNA molecules per nucleoid, which indicates that many nucleoids just contain a single copy of mtDNA.
TFAM Is a Main Component of the Nucleoid.
We proceeded to calculate the number of TFAM molecules per cell. We expressed a His-tagged form of TFAM lacking the mitochondrial targeting signal and analyzed purified protein with MALDI mass fingerprinting to confirm identity. Further mass spectrometry analyses showed that the recombinant protein was free from major contamination and had a molecular mass of 25.3 kDa (Fig. S2A), which is very close to the predicted molecular mass of 25.4 kDa (8). With the recombinant TFAM protein as standard, we found on average 0.11 pg TFAM per cell by Western blot analyses of total protein extracts from human fibroblasts (Fig. 2B and Fig. S2B), which corresponds to 2.7 × 106 TFAM molecules per cell or 1.4 × 103 TFAM molecules per nucleoid (Fig. 2C). There are thus ∼1,000 TFAM molecules per mtDNA molecule or one TFAM molecule per 16.6 bp of mtDNA in human fibroblasts, in good agreement with our previous estimates from mouse (13, 14). There is no available atomic structure of the complete intramitochondrial form of TFAM bound to mtDNA, and we therefore predicted the 3D structure by Phyre and I-Tasser (Fig. 2 D and E). The calculated volume of TFAM bound to mtDNA is 1.1 × 105 Å3 if an approximate cylindrical volume is used (Fig. 2D) or 3.0 × 104 Å3 if a more detailed solvent excluded volume is calculated (Fig. 2E). These predicted sizes argue that a single spherical nucleoid of an apparent diameter of ∼100 nm maximally can contain 5.0 × 103 (cylindrical volume) or 17.5 × 103 (solvent excluded volume) TFAM/mtDNA complexes. However, the real size of the nucleoid is predicted to be ∼70 nm if the volumes of the bound antibodies are deducted (26, 27), meaning that it could contain a maximally 1.7 × 103 (cylindrical volume) or 6.0 × 103 (solvent excluded volume) TFAM/mtDNA complexes, which are in reasonably good agreement with the experimentally determined number of 1.4 × 103 TFAM molecules per nucleoid. TFAM is thus likely a main protein component of the nucleoid, but this estimate does not exclude that other proteins also are important structural elements.
Nucleoids Are Frequently Observed in Clusters.
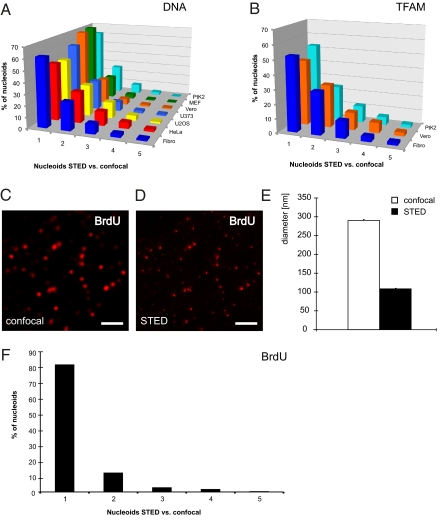
A subset of single nucleoids that were observable by confocal microscopy could be resolved into clusters of multiple nucleoids when analyzed by STED microscopy (Fig. 1 B, C, F, and G). We found that a surprisingly large fraction (26–54%) of all nucleoids identified by DNA antibodies and confocal microscopy could be resolved into clusters containing multiple nucleoids when analyzed by STED microscopy in all studied mammalian cells (Fig. 3A; for details on the algorithms used see SI Materials and Methods). Similarly, a substantial fraction of nucleoids identified with TFAM antibodies could be resolved into multiple nucleoids by STED microscopy (Fig. 3B).
Fig. 3.
Nucleoids are frequently distributed in clusters. (A) Ratio of the number of DNA antibody-labeled nucleoids that were detected with STED microscopy versus those detected by confocal microscopy. The numbers denote the quantity of nucleoids detected by STED microscopy in a confocally assigned nucleoid. (B) Cells labeled with an antiserum against TFAM analyzed as in A. (C) Confocal imaging of BrdU-stained nucleoids in human fibroblasts after 60 min of BrdU incubation. (Scale bar, 2 μm.) (D) STED micrograph of the same section as in C. (Scale bar, 2 μm.) (E) Sizes of nucleoids in human fibroblasts labeled with BrdU antibodies and imaged by confocal and STED microscopy. (F) Cells treated for 60 min with BrdU and labeled with an antiserum against BrdU analyzed as in A. Error bars indicate SEM.
Replication of mtDNA Only Occurs in a Subset of Nucleoids.
To gain further insights in the structure of mitochondrial nucleoids we used bromodeoxyuridine (BrdU), which is a synthetic nucleoside that is incorporated into newly synthesized DNA and is detectable by antibodies. The mean diameter of BrdU-stained nucleoids in human fibroblasts was 290 nm with confocal microscopy and 109 nm with STED microscopy (Fig. 3 C–E), which is in very good agreement with the results obtained above (Fig. 1 D and H). Only 18% of the nucleoids identified by confocal microscopy contained more than one BrdU-containing nucleoid when analyzed by STED microscopy after 60 min of BrdU incubation (Fig. 3F). On average, we observed only ∼1.3 times more nucleoids with antibodies against BrdU when we compared results from STED and confocal microscopy (Fig. 3F). These results indicate that most frequently only one nucleoid in a cluster has ongoing mtDNA replication.
Nucleoid Distribution.
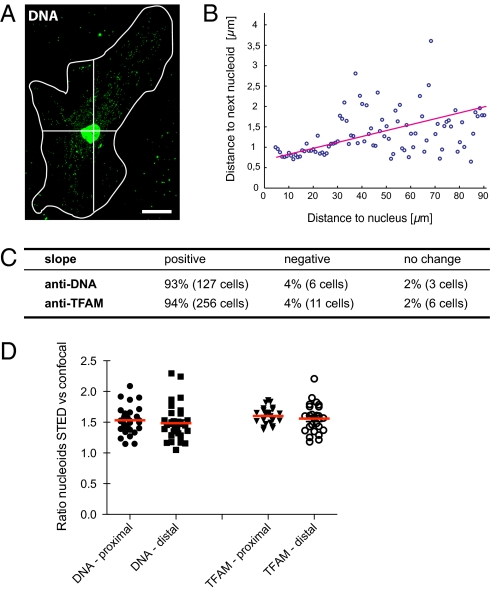
To clarify if the distribution of nucleoids changes with the distance to the nucleus, we determined for each nucleoid the distance to its nearest neighbor as a function of the distance to the nucleus (Fig. 4 A and B). We observed an increase in the nearest neighbor distances when comparing perinuclear nucleoids to those in the cell periphery in 93% of the DNA antibody-labeled cells (n = 127 cells) and 94% of TFAM antibody-labeled cells (n = 256 cells) (Fig. 4C). We proceeded to use STED microscopy to analyze if there was any difference in clustering of nucleoids in relation to the distance from the nucleus. We compared the ratio of nucleoids detected by STED versus confocal microscopy and found no relevant difference between nucleoids with a location proximal (1.53 ± 0.04; n = 32 cells) or distal (1.48 ± 0.05) to the nucleus (Fig. 4D). In addition, with TFAM antibodies there was no relevant difference between nucleoids proximal (1.61 ± 0.03; n = 27 cells) and distal (1.58 ± 0.05) to the nucleus (Fig. 4D). In summary, whereas the average nucleoid-to-nucleoid distance is smaller in proximity to the nucleus, the clustering is independent of the distance from the nucleus. These findings indicate that although there are more proximal nucleoids, there is a very efficient distribution of nucleoids, which may ensure good access to mtDNA-encoded gene products throughout the mitochondrial network.
Fig. 4.
Nucleoid distribution as a function of the distance to the nucleus. (A) Representative cell labeled with an antiserum against DNA used for the analysis presented in B. The white line denotes the cell border; the cross indicates the center (nucleus) of the cell. (Scale bar, 20 μm.) (B) Plot of the nearest neighbor distance of individual nucleoids as depending of the distance from the nucleus. The circles represent 100 equally spaced bins, all together representing 986 nucleoids. (C) Table summarizing the analysis of altogether 409 cells as exemplified in A and B. (D) Ratio of the number of nucleoids labeled with DNA or TFAM antibodies detected by STED microscopy versus the number of nucleoids detected by confocal microscopy in dependence of the relative cellular location.
Discussion
We have performed STED super-resolution imaging of nucleoids and report a surprisingly uniform apparent mean nucleoid size of ∼100 nm in a variety of mammalian cells. There are just a few reports published about the size of mitochondrial nucleoids in mammals and a size of 0.1 to 0.3 μm was reported in cells transfected with Twinkle-GFP or cells imaged with antibodies against TFAM or mtSSB (18). A nucleoid size of 0.4 to 0.9 μm was reported with PicoGreen and antibodies against DNA in mammalian cells imaged by confocal microscopy (28). We did an extensive confocal microscopy image analysis of a panel of mammalian cell lines and found a mean nucleoid size of ∼270 nm, which is close to the lateral optical resolution limit of light microscopy. We used STED microscopy in conjunction with three different markers (i.e., DNA antibodies, TFAM antibodies, and immunodetection of incorporated BrdU) to study mammalian cell lines and all of these markers independently gave a very similar apparent mean nucleoid size of ∼100 nm. Because the antibody labeling inescapably increases the volume of the nucleoid, the determined sizes moderately overemphasize the actual nucleoid sizes. Taking this into account (26, 27), the true nucleoid size may be estimated as ∼70 nm. Our results are consistent with transmission electron microscopy (TEM) studies that have reported sizes of ∼70 nm of immunogold-labeled single nucleoids (29). TEM has also been used to visualize nucleoids after heavy metal labeling of mtDNA and the nucleoid sizes on published images are consistent with our results, although no detailed quantification was performed (3, 30). Determination of nucleoid size with electron microscopy is difficult as the analyzed ultrathin sections often only contain part of the nucleoid (30), which may explain the large range (32–132 nm) of nucleoid sizes in some reports (29). An additional issue with TEM is that reconstruction of serial sections is difficult, meaning that typically only a subset of the nucleoids in a cell are visualized. The STED microscopy imaging approach we used has the advantage that it involves no tissue sectioning and allows an automated unbiased imaging of all nucleoids of a cell.
We also report that the TFAM protein is a main component of the mammalian nucleoid, consistent with its conserved role in mtDNA maintenance in mammals and other vertebrates (9). There are some reports that there is only one TFAM protein molecule per several hundred base pairs of mtDNA (31). However, we report here a much higher abundance and found one TFAM protein molecule per ∼16.6 bp of mtDNA, which is in good agreement with the molecular modeling results we present. The finding that TFAM is very abundant is consistent with several other reports, demonstrating one TFAM molecule per 10 to 20 bp of mtDNA (13–15). In addition, biochemical characterization of purified nucleoids has shown that TFAM is the main protein component (20). TFAM is thus likely the main factor packaging and organizing mtDNA into nucleoids, but this finding does not exclude that other proteins may also be of importance.
The copy number of mtDNA per nucleoid is of fundamental importance, as this arrangement will influence germ-line transmission (32) and segregation of mtDNA in somatic tissues in disease and aging (21, 33). It has been reported that each nucleoid contains on average 6 to 10 copies (29), 2 to 8 copies (19), or ∼5 copies of mtDNA (34). We used confocal microscopy analyses and qPCR determination of mtDNA copy number and found on average ∼2.3 mtDNA molecules per nucleoid in human fibroblasts. However, with STED microscopy we detected ∼60% more nucleoids per cell than with confocal microscopy, and each nucleoid therefore only contains on average ∼1.4 mtDNA molecules. This finding means that the majority of the nucleoids just contain a single copy of mtDNA. This arrangement is consistent with the so-called “faithful” nucleoid model (34), whereby nucleoids as a general rule do not exchange genomes with each other.
Interestingly, a large proportion of the nucleoids detected on confocal microscopy contained two or more nucleoids when imaged by STED microscopy. The existence of clusters of nucleoids could, at least partly, be the result of a random distribution. We found that cluster of nucleoids were occurring both proximally and distally to the nucleus, which argues that the cluster may not be explained by juxtaposition of newly replicated mtDNA molecules, as others have reported that mtDNA replication preferentially occurs close to the nucleus (35). Another possibility is that the clusters represent some type of higher order structural/functional arrangement, consistent with the idea that there is a layered structure of protein complexes interacting with nucleoids (20).
In summary, we present here an improved definition of the mammalian mitochondrial nucleoid and report that it is a defined structure with a mean size of ∼100 nm (antibody-decorated) containing on average ∼1.4 mtDNA molecules. This means that the smallest inheritable or segregating unit of mtDNA is a single copy. These findings have broad implications for understanding segregation and transmission of mtDNA in disease and aging.
Materials and Methods
Cell Culture.
In this study seven different cell lines were analyzed: HeLa (human cervix carcinoma), U2OS (human osteosarcoma), U373 (human glioblastoma astrocytoma), Vero (Cercopithecus aethiops kidney), PtK2 (Potorous tridactylus kidney), MEF, and primary human skin fibroblasts. The cells were cultivated in DMEM with Glutamax and 4.5 g/L glucose (Invitrogen) supplemented with 50 U/mL penicillin, 50 μg/mL streptomycin, 1 mM Na-pyruvate, and 10% (vol/vol) FCS (Invitrogen) at 37 °C, 5% CO2.
Cell numbers were determined by a Vi-CELL XR system (Beckman-Coulter).
Sample Preparation for Fluorescence Microscopy.
For immunolabeling, cells were grown on coverslips over night, fixed with 1% to 8% (wt/vol) formaldehyde in PBS (137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, pH7) for 5 min at 37 °C (for TFAM labeling) or methanol (abs.) for 5 min at −20 °C (for DNA labeling), extracted with 0.5% (vol/vol) Triton X-100 in PBS, blocked with 5% (wt/vol) BSA in PBS, and incubated with monoclonal mouse antibodies against TFAM (Abnova), DNA (PROGEN), or TOM20 (Santa Cruz). The primary antibodies were detected with secondary antibodies (sheep anti-mouse and goat anti-rabbit; Jackson Immuno Research Laboratories) custom-labeled with ATTO532 (AttoTec) or KK114 (36). After immunolabeling, the samples were mounted in MOWIOL with 0.1% (wt/vol) 1,4-Diazabicyclo[2.2.2]octane (DABCO) and 2.5 μg/mL DAPI (Sigma Aldrich).
BrdU labeling was performed using the 5-Bromo-2′-deoxy-uridine Labeling and Detection Kit I (Roche Diagnostics) according to the manufacturer’s protocol. In brief, the cells were incubated for 60 min in BrdU containing cell-culture medium. After washing in cell-culture medium without BrdU and fixation in ethanol fixative (15 mM glycine; ∼70% ethanol, pH2), the BrdU labels in the DNA were detected using a BrdU-specific antibody contained in the kit. The primary antibodies were detected as indicated above. Mounting of the samples was performed in Mowiol.
Quantitative Western Analysis.
Recombinant human TFAM was purified as described previously (37) and analyzed by mass spectrometry (see below). The concentration was determined by OD280 measurements and Criterion stain free gel PAGE (Bio-Rad) with alcohol dehydrogenase from Saccharomyces cerevisiae (Sigma-Aldrich) as standard. For PAGE analyses with Criterion Tris-HCl Gels (Bio-Rad), cell pellets were resuspended in a buffer containing 100 mM Tris-Cl, pH6.8, 4% SDS, 20% glycerol (Sigma-Aldrich) and Complete Protease Inhibitor Mixture (Roche Diagnostics), vortexed, incubated at 95 °C for 5 min, and sonicated for 5 cycles at 30 s. Before PAGE, 200 mM DTT and 0.2% bromophenolblue (Sigma-Aldrich) were added.
Western Blot analyses were performed with monoclonal mouse antibodies against human TFAM and secondary anti-mouse IgG horseradish peroxidase linked antibodies visualized by Amersham ECL Western Blotting Detection Reagents (GE Healthcare.). We quantified human TFAM protein levels by using a standard of recombinant human TFAM protein.
Mass Spectrometry.
The sample was desalted using ZipTip C4 (Millipore) according to the manufacturer’s instructions and eluted with 50% acetonitrile containing 1% formic acid directly into an off-line nano flow needle (EconoTipTM; New Objective). Electrospray ionization (ESI) experiments in positive ESI-MS mode were performed with a hybrid quadrupole-orthogonal acceleration time-of-flight mass spectrometer (XevoTM Q-Tof; Waters). The ESI-source was operated at a temperature of 100 °C with cone gas flow of 20 L/h. To the off-line nano flow needle, a potential of 3,500 V was applied. The cone voltage was 24 V and the collision energy was set to 6 V. Full-scan mass spectra were acquired in the continuous data acquisition mode in the range m/z 600 to 2,300 at a scan rate of 1 s and an interscan delay of 0.025 s per scan. MaxEnt 1 in the MassLynx 4.1 softare package (Waters) was used for deconvolution of the multiple charged ion series with following parameters: resolution 1, width at half height 0.6, range 957:1,435, minimum intensity ratios left/right 50%.
Structure Prediction and Volume Calculation of hTFAM Bound to DNA.
The sequence of human TFAM (hTFAM) without the mitochondrial targeting peptide, comprising residues 43 to 246 was used for structure prediction by Phyre (38) and I-Tasser (39). Structural similarity was found to the NMR structure of the hybrid protein SRY.B derived from human SRY and rat HMGB1 (PDB accession code 2GZK; E-value 1.9 × 10−17), which contains two tandem HMG boxes bound to a double stranded DNA sequence of 16 bp (40). In addition, the sequence of the intramitochondrial form of human TFAM was sent to the structure prediction server I-Tasser (39), which calculated a complete model with a confidence score of −0.99. The DNA prediction by Phyre was modeled into the TFAM structure built by I-Tasser by aligning the two models using the SSM method followed by manual adjustments in COOT (41), resulting in a final model with the dimensions of x = 66 Å, y = 45 Å, and z = 39 Å. Dimension calculations of the hTFAM-DNA model and figures were prepared with Pymol (http://www.pymol.org/). Volume calculation of the model was performed either from the dimensions calculated by Pymol or from the coordinate file by the program VOIDOO (42).
Quantification of mtDNA Content.
Cell pellets were lysed in QuantiLyse buffer by incubation at 50 °C for 30 min and then 95 °C for 10 min (43) and used for the subsequent qPCR reaction with TaqMan probes for human MT-ND1, MT-ND2 and MT-7S (Applied Biosystems) on a 7900HT Fast Real-Time PCR System (Applied Biosystems) according to the manufacturer’s protocols. Subcloned fragments of human mtDNA (48–743 bp, 2,573–3,423 bp, 4,121–5,030 bp of human mtDNA according to GenBank NC_012920) in plasmids were used as standards for the absolute quantification of mtDNA.
Microscopy and Image Analysis.
Detailed description can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Stefan W. Hell for continuous support and Benjamin Harke for help with the stimulated emission depletion microscope; Thomas Franz and Xinping Li of the BIO-MS facility for support with the mass spectometry analysis; and Bianca Habermann of the bioinformatics group of the Max Planck Institute for Biology of Ageing for help calculating the mitochondrial transcription factor A volumes. This work was supported by the Deutsche Forschungsgemeinschaft, SFB 829 (N.-G.L.); and the Deutsche Forschungsgemeinschaft Research Center for Molecular Physiology of the Brain (S.J.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109263108/-/DCSupplemental.
References
- 1.Lang BF, et al. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387:493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- 2.Nass S, Nass MM. Intramitochondrial fibers with DNA characteristics. Ii. Enzymatic and other hydrolytic treatments. J Cell Biol. 1963;19:613–629. doi: 10.1083/jcb.19.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nass MM. Mitochondrial DNA. I. Intramitochondrial distribution and structural relations of single- and double-length circular DNA. J Mol Biol. 1969;42:521–528. doi: 10.1016/0022-2836(69)90240-x. [DOI] [PubMed] [Google Scholar]
- 4.Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 5.Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26(2 Pt 2):167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 6.Chen XJ, Butow RA. The organization and inheritance of the mitochondrial genome. Nat Rev Genet. 2005;6:815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- 7.MacAlpine DM, Perlman PS, Butow RA. The high mobility group protein Abf2p influences the level of yeast mitochondrial DNA recombination intermediates in vivo. Proc Natl Acad Sci USA. 1998;95:6739–6743. doi: 10.1073/pnas.95.12.6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parisi MA, Clayton DA. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252:965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- 9.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 10.Fisher RP, Lisowsky T, Parisi MA, Clayton DA. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J Biol Chem. 1992;267:3358–3367. [PubMed] [Google Scholar]
- 11.Shen EL, Bogenhagen DF. Developmentally-regulated packaging of mitochondrial DNA by the HMG-box protein mtTFA during Xenopus oogenesis. Nucleic Acids Res. 2001;29:2822–2828. doi: 10.1093/nar/29.13.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsushima Y, et al. Functional domains of chicken mitochondrial transcription factor A for the maintenance of mitochondrial DNA copy number in lymphoma cell line DT40. J Biol Chem. 2003;278:31149–31158. doi: 10.1074/jbc.M303842200. [DOI] [PubMed] [Google Scholar]
- 13.Ekstrand MI, et al. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 14.Pellegrini M, et al. MTERF2 is a nucleoid component in mammalian mitochondria. Biochim Biophys Acta. 2009;1787:296–302. doi: 10.1016/j.bbabio.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 15.Alam TI, et al. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 2003;31:1640–1645. doi: 10.1093/nar/gkg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaufman BA, et al. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol Biol Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsson NG, et al. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 18.Garrido N, et al. Composition and dynamics of human mitochondrial nucleoids. Mol Biol Cell. 2003;14:1583–1596. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Legros F, Malka F, Frachon P, Lombès A, Rojo M. Organization and dynamics of human mitochondrial DNA. J Cell Sci. 2004;117:2653–2662. doi: 10.1242/jcs.01134. [DOI] [PubMed] [Google Scholar]
- 20.Bogenhagen DF, Rousseau D, Burke S. The layered structure of human mitochondrial DNA nucleoids. J Biol Chem. 2008;283:3665–3675. doi: 10.1074/jbc.M708444200. [DOI] [PubMed] [Google Scholar]
- 21.Park CB, Larsson N-G. Mitochondrial DNA mutations in disease and aging. J Cell Biol. 2011;193:809–818. doi: 10.1083/jcb.201010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ylikallio E, Tyynismaa H, Tsutsui H, Ide T, Suomalainen A. High mitochondrial DNA copy number has detrimental effects in mice. Hum Mol Genet. 2010;19:2695–2705. doi: 10.1093/hmg/ddq163. [DOI] [PubMed] [Google Scholar]
- 23.Hell SW. Single Molecule Spectroscopy in Chemistry, Physics and Biology. Berlin: Springer; 2009. Far-field optical nanoscopy; pp. 365–398. [Google Scholar]
- 24.Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: Stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 25.Harke B, et al. Resolution scaling in STED microscopy. Opt Express. 2008;16:4154–4162. doi: 10.1364/oe.16.004154. [DOI] [PubMed] [Google Scholar]
- 26.Dyba M, Jakobs S, Hell SW. Immunofluorescence stimulated emission depletion microscopy. Nat Biotechnol. 2003;21:1303–1304. doi: 10.1038/nbt897. [DOI] [PubMed] [Google Scholar]
- 27.Weber K, Rathke PC, Osborn M. Cytoplasmic microtubular images in glutaraldehyde-fixed tissue culture cells by electron microscopy and by immunofluorescence microscopy. Proc Natl Acad Sci USA. 1978;75:1820–1824. doi: 10.1073/pnas.75.4.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holt IJ, et al. Mammalian mitochondrial nucleoids: Organizing an independently minded genome. Mitochondrion. 2007;7:311–321. doi: 10.1016/j.mito.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 29.Iborra FJ, Kimura H, Cook PR. The functional organization of mitochondrial genomes in human cells. BMC Biol. 2004;2:9. doi: 10.1186/1741-7007-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prachar J. Mouse and human mitochondrial nucleoid—Detailed structure in relation to function. Gen Physiol Biophys. 2010;29(2):160–174. [PubMed] [Google Scholar]
- 31.Cotney J, Wang Z, Shadel GS. Relative abundance of the human mitochondrial transcription system and distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression. Nucleic Acids Res. 2007;35:4042–4054. doi: 10.1093/nar/gkm424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart JB, Freyer C, Elson JL, Larsson NG. Purifying selection of mtDNA and its implications for understanding evolution and mitochondrial disease. Nat Rev Genet. 2008;9:657–662. doi: 10.1038/nrg2396. [DOI] [PubMed] [Google Scholar]
- 33.Larsson NG. Somatic mitochondrial DNA mutations in mammalian aging. Annu Rev Biochem. 2010;79:683–706. doi: 10.1146/annurev-biochem-060408-093701. [DOI] [PubMed] [Google Scholar]
- 34.Gilkerson RW, Schon EA, Hernandez E, Davidson MM. Mitochondrial nucleoids maintain genetic autonomy but allow for functional complementation. J Cell Biol. 2008;181:1117–1128. doi: 10.1083/jcb.200712101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis AF, Clayton DA. In situ localization of mitochondrial DNA replication in intact mammalian cells. J Cell Biol. 1996;135:883–893. doi: 10.1083/jcb.135.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolmakov K, et al. Red-emitting rhodamine dyes for fluorescence microscopy and nanoscopy. Chem Eur J. 2010;16(1):158–166. doi: 10.1002/chem.200902309. [DOI] [PubMed] [Google Scholar]
- 37.Falkenberg M, et al. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 38.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 39.Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stott K, Tang GS, Lee KB, Thomas JO. Structure of a complex of tandem HMG boxes and DNA. J Mol Biol. 2006;360(1):90–104. doi: 10.1016/j.jmb.2006.04.059. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Kleywegt GJ, Jones TA. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 2):178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- 43.Pierce KE, Rice JE, Sanchez JA, Wangh LJ. QuantiLyse: Reliable DNA amplification from single cells. Biotechniques. 2002;32:1106–1111. doi: 10.2144/02325pf01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.