Abstract
We describe a label-free imaging method to monitor stem-cell metabolism that discriminates different states of stem cells as they differentiate in living tissues. In this method we use intrinsic fluorescence biomarkers and the phasor approach to fluorescence lifetime imaging microscopy in conjunction with image segmentation, which we use to introduce the concept of the cell phasor. In live tissues we are able to identify intrinsic fluorophores, such as collagen, retinol, retinoic acid, porphyrin, flavins, and free and bound NADH. We have exploited the cell phasor approach to detect a trend in metabolite concentrations along the main axis of the Caenorhabditis elegans germ line. This trend is consistent with known changes in metabolic states during differentiation. The cell phasor approach to lifetime imaging provides a label-free, fit-free, and sensitive method to identify different metabolic states of cells during differentiation, to sense small changes in the redox state of cells, and may identify symmetric and asymmetric divisions and predict cell fate. Our method is a promising noninvasive optical tool for monitoring metabolic pathways during differentiation or disease progression, and for cell sorting in unlabeled tissues.
Keywords: phasor analysis, single cell metabolism
The hallmark of stem cells is their ability to produce a new stem cell by self-renewing, as they generate daughter cells that are committed to differentiation and form specialized tissues (1). The modulation of the balance between self-renewing divisions and differentiation is a central mechanism for stem cells during embryo development, adult tissue regeneration, and homeostasis. Stem-cell differentiation is a complex process mediated both by intrinsic molecular mechanisms and extrinsic signaling. Intrinsic cell polarity, subcellular localization mechanism, asymmetric centrosome and spindles, as well as cell-cycle regulators can establish self-renewing asymmetry during stem-cell division (2, 3). The influence of external chemical and physical stimuli, such as molecular gradients, extracellular matrix remodeling, and niche signaling is crucial for stem-cell plasticity and tissue development (4–6). Extrinsic signals can be propagated through intracellular signal-transduction pathways that converge to genetic networks that control pluripotency. Many different signaling and transcriptional pathways, which are important for development and cell-cycle regulation, converge on regulation of the redox state of stem cells (5, 7, 8). In turn, histone posttranscriptional modifications and epigenetic mechanisms, such as phosphorylation, acetylation, and methylation, sense cellular metabolism and the variation of metabolites levels (9). Hence, redox balance plays an important role in the maintenance and modulation of stem cell self-renewal and differentiation (10–12).
In stem-cell research there is a high demand for noninvasive techniques to investigate self-renewal and differentiation. Methods, such as immunohistochemistry, metabolic assays, and PCR are currently used for cell sorting and to measure stem-cell differentiation both in vitro and in tissues. These techniques require long and invasive procedures that render cells unviable and unrecoverable and that are incompatible with in vivo dynamic observations. Recently, noninvasive optical techniques have been developed to obtain information on cell metabolism and discriminate between different states of cells and tissues (13–20). These methods exploit intrinsic autofluorescence of cells and tissues and multiphoton microscopy.
Multiphoton microscopy is suitable for high-resolution and long-term imaging of living tissues. This type of microscopy allows investigation of the local environment in femtoliter volumes deep in tissues, thanks to its intrinsic 3D resolution, high penetration depth, negligible out-of-focus photobleaching, and reduced photodamage (21). Autofluorescence in live tissues arises from endogenous proteins and physiologically relevant fluorophores, such as collagen, elastin, porphyrin, retinoids, flavins, NADH, hemoglobin, and serotonin (22). NADH and FAD are the main metabolic coenzymes involved in oxidative phosphorylation and glycolysis, and they report on metabolic changes associated with cell carcinogenesis and differentiation (12, 18), and retinoid signaling is involved in differentiation of stem and precursor cells and embryonic development (23). Two-photon excited fluorescence alone cannot assign autofluorescence signal to specific intrinsic molecular sources.
Additional methods have been proposed to assign autofluorescence to specific tissue components, but with limited success. Spectral demixing requires the prior knowledge of the metabolite’s spectra in tissues. Principal component analysis of emission spectra requires identification of the principal components with specific metabolic relevant species. The discrimination between intrinsic fluorescence sources by emission wavelength is also limited by the overlapping of emission spectra of different fluorescent species, such as NADPH and NADH (24). Multiexponential fitting of fluorescence intensity decays is difficult to interpret when multiple fluorescent species are present in the focal volume. Several fluorophores and proteins are characterized by conformational heterogeneity and have complex lifetime distribution with more than one exponential component (25). Moreover processes, such as fluorescence resonance energy transfer, pH variation, scattering, and quenching often occur in tissues. Hence, the choice of a decay model for the intensity decay fitting is often arbitrary and it is difficult to associate specific tissue components to exponential decays (26–28).
Here we use the phasor approach to fluorescence lifetime imaging microscopy (FLIM) (29, 30), which allows a straightforward interpretation of intrinsic fluorescence signal from living tissues directly in terms of physiological relevant fluorophores. We provide images of molecular species by using their phasor fingerprints, without resolving and assigning exponential components to the fluorescence species.
We exploit this property to separate multiple tissue components by cluster analysis of the phasor distribution in FLIM images from seminiferous tubules of a mice testis Oct4 GFP transgene that expresses GFP in undifferentiated germ cells. In our approach each fluorescent molecular source is identified by its specific location in the phasor plot. We identify GFP, collagen, FAD, free and bound NADH, retinol, and retinoic acid within the living tissue by using the pure-species phasor locations. We measure the metabolic state of cells in the Caenorhabditis elegans germ line by image segmentation and calculation of the cell phasor distribution of every cell in the tissue. The C. elegans germ line provides a well-defined model for studying the progression from stem cell self-renewal to differentiation (31–33). The cell phasor analysis shows different metabolic fingerprints within that organ.
Results
Phasor Cluster Analysis Separates Individual Tissue Components.
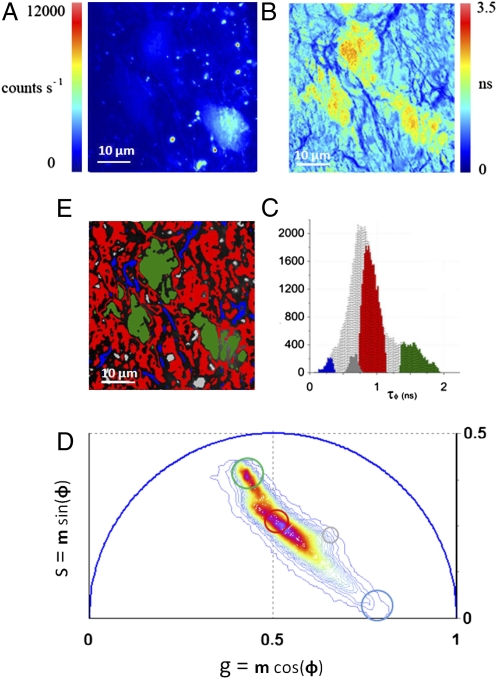
The phasor transformation (described in Materials and Methods and SI Materials and Methods) of FLIM images of a living tissue directly provides maps of individual tissue components without prior assumption on the number of species in the tissues (29, 34–36). The analysis of the FLIM data in the phasor space is performed by detecting clusters of pixel values in specific regions of the phasor plot. Fig. 1A displays a two-photon excited fluorescence image of seminiferous tubules from mice expressing EGFP from an Oct4 transgene. The FLIM image is presented in Fig. 1B in terms of the average lifetime τφ (SI Materials and Methods). The lifetime is relatively homogeneous across the image and the histogram of lifetime values (gray distribution in Fig. 1C) has a range from 0 to 2 ns, with two major peaks at 0.8 and 1.8 ns. We perform the phasor analysis of the FLIM image by a mathematical transformation of the raw data (see Materials and Methods). Fig. 1D displays the 2D phasor plot of the FLIM image. Every pixel of the FLIM image is transformed into a pixel in the phasor plot. All of the pixels are located inside the universal circle of the phasor plot, thus indicating that their decay is multiexponential. The phasor distribution of the living tissue has a complex shape with different clusters. Their positions specifically correspond to different tissue components. In Fig. 1E pixels are highlighted with a color that corresponds to the clusters in the phasor plot in Fig. 1D. Based on morphology, the green-colored cluster selects cells, the blue cluster fibers in the basal membrane, the red cluster the rest of the tissue, and the gray one selects specific bright granules.
Fig. 1.
Phasor analysis of FLIM tissue images. (A) Intensity image of a seminiferous tubule from a mice expressing GFP from an Oct4 transgene. (B) Map of the average lifetime τφ of the FLIM image. (C) τφ Histogram of the FLIM image is represented in gray. Colored areas correspond to τφ of each tissue component identified by the phasor analysis. (D) Phasor histogram of the FLIM image. The line color (from blue to purple  ) corresponds to the 64 levels of the contours that indicate the percent occurrence in the phasor histogram of the pixels of the image. Four clusters corresponding to different tissue components are identified in the phasor distribution with different colors. (E) Phasor color maps of the FLIM image. The colors of pixels correspond to the clusters of tissue components selected in the phasor plot.
) corresponds to the 64 levels of the contours that indicate the percent occurrence in the phasor histogram of the pixels of the image. Four clusters corresponding to different tissue components are identified in the phasor distribution with different colors. (E) Phasor color maps of the FLIM image. The colors of pixels correspond to the clusters of tissue components selected in the phasor plot.
Identifying Tissue Components Using the Phasor Plot.
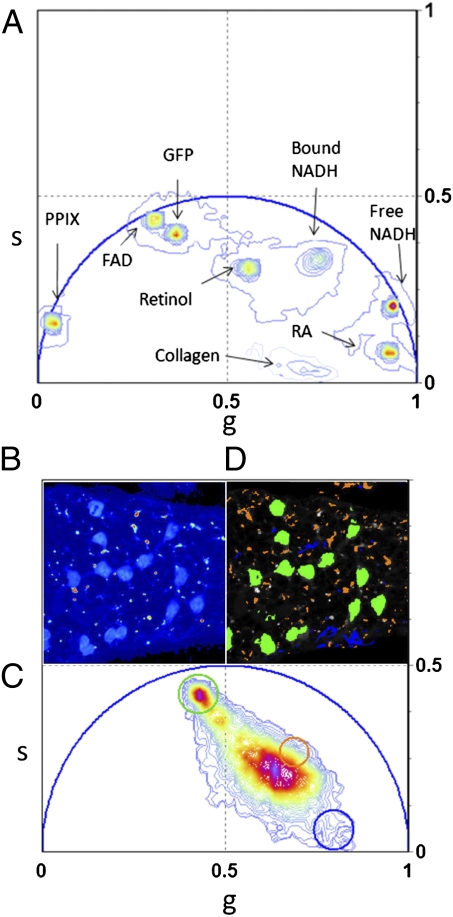
Using the characteristic phasor fingerprint obtained from individual species allows us to identify individual components in a complex system such as a tissue. Fig. 2A shows the phasor location of the most important intrinsic fluorophores. Their positions in the phasor plot are well defined and clearly separated one from the other. The majority of fluorophores have decay with multiple exponential components because of their conformational heterogeneity. The phasor position of pure GFP is near but not exactly on the universal circle (Fig. 2A) because the fluorescence decay of GFP is not single exponential (37). Collagen has a very short lifetime, with a broad distribution of decay components because of the intermolecular cross links (38). Its phasor is located inside the universal circle and closed to the temporal zero. The phasor position of retinol in DMSO has a specific location, which is different from the one of the retinoic acid in DMSO, in agreement with the measured multiexponential decays in ref. 39. FAD and free NADH phasor position are located near the universal circle, but bound NADH is located inside because its lifetime is a combination of more than one exponential (40–42). The phasor position of protoporphyryn IX (in dimethylformamide and methanol) is located on the universal circle because it is characterized by a single lifetime component (43).
Fig. 2.
Phasors of pure chemical species identify tissue components. (A) Phasor location of pure chemical species: GFP, retinol in DMSO, retinoic acid in DMSO, FAD, free NADH, bound NADH with lactate dehydrogenase, and protoporphyrin IX. Color scale defined in Fig. 1D. (B) Intensity image of a seminiferous tubule from a mice expressing GFP from an Oct4 transgene. A chain of spermatogonial stem cells lie on the surface of the seminiferous tubule. (C) Phasor plot of the FLIM image acquired in B. The color scale of the phasor plot histogram was defined in Fig. 1D. The green and the blue cluster are located in the phasor position of pure GFP and pure collagen clusters. (D) Phasor color map. Green and blue pixels contain mostly GFP and collagen.
In Fig. 2 we show that the number of molecular species that can be identified with a unique location in the 2D phasor plot is potentially very large. We note that the separation of many components cannot be obtained when the average lifetime of the species is plotted in a 1D scale, as is usually done in the conventional lifetime analysis approach. All of the components with the same average lifetime but with a different combination of two single exponentials are spread out in a big area of the phasor plot (Fig. S1A). The phasor representation is especially powerful with respect to the classic multiexponential fitting, when resolving many fluorophores or tissue components with multiple exponential lifetimes (Fig. S1B), because it does not require the assignment of the exponentials to the molecular species.
The phasor location of the molecular species in Fig. 2A is used as a reference database to analyze the phasor distribution in Fig. 2C that is acquired from seminiferous tubules from a testis of an Oct-4-GFP mouse. The blue and the green clusters in Fig. 2B are located in the position of the phasor that corresponds to the pure GFP and pure collagen measured in Fig. 2A. In the phasor color map in Fig. 2D, the blue and green pixels represent all of the points in the tissue in which GFP and collagen are in the focal volume, with very small contribution of other fluorescence components. High fractional intensities of GFP with respect to the autofluorescence are located in the germ cells that form a chain on the surface of the tubule. The presence of collagen in the tissue is also confirmed by second harmonic generation microscopy (44). The second harmonic generation signal that is detected in the tissue (Fig. S2) colocalizes with the blue selected collagen pixels in Fig. 2B.
Mapping the Relative Concentrations of Tissue Components in a Seminiferous Tubule.
The phasor representation not only allows identification of molecular species, but also their coexistence in the same pixel of the image. The phasor location is independent from the absolute concentration of a single fluorescent species. However, if two species are coexisting in the same pixel, the phasor location of that pixel lies along the line connecting the phasors of the two pure species (see Materials and Methods and SI Materials and Methods). The phasor position of the bright granule areas that are identified by the orange cluster in the phasor plot in Fig. 2B do not colocalize with any of the pure chemical species of Fig. 2A. However, we identify them as a mixture of retinol and retinoic acid because their position in the phasor plot is located along the straight line between the retinol and the retinoic phasor position. Retinoids are located in Sertoli cells of the seminiferous tubule and regulate germ-cell fate and their transition to meiosis (23). The presence of retinol and retinoic acid was confirmed by spectral imaging (Fig. S2). The average autofluorescence arising from the tissue locates in the central part of the phasor plot (red phasor cluster in Fig. 1A) because it is a mixture of different intrinsic fluorescent components, such as FAD and retinoids.
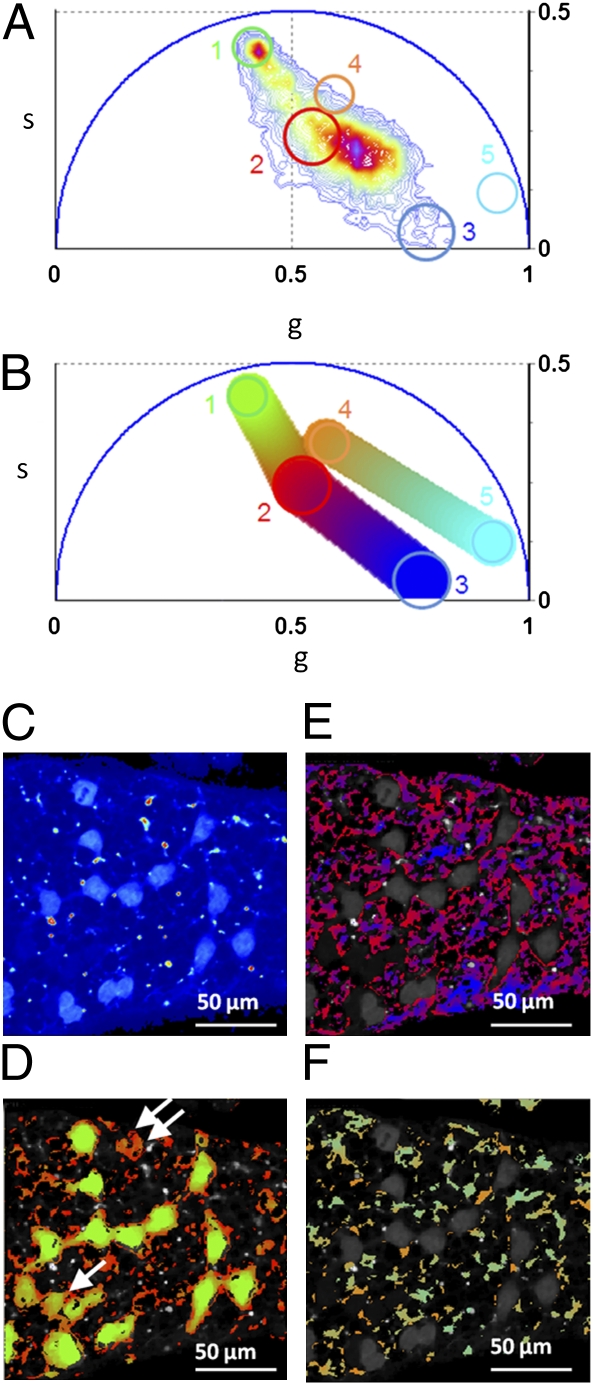
We provide a map of the relative concentration of individual components within the tissues by visualizing their fractional intensities to the signal (Fig. 3). Within the same phasor plot the phasor locations of five different molecular species are selected by different clusters based on the chemical species fingerprint in Fig. 2A. In Fig. 3 A and B the colored clusters are assigned respectively to GFP (green), the average tissue autofluorescence (red), collagen (blue), retinol (orange), and retinoic acid (cyan). We first calculate the relative concentration of GFP with respect to the average autofluorescence, then the average autofluorescence with respect to the collagen, and finally the retinol concentration with respect to the retinoic acid. The relative concentration is calculated in every pixel of the image with a graphic analysis by looking at the position of the pixel in the phasor plot along the line connecting the two molecular species. In Fig. 3B a linear cluster with a color scale from green to red shows all of the possible relative concentrations of GFP and average autofluorescence. Each point along the cluster has a color corresponding to specific fractional intensities. The same color scale is used to map the relative concentration of the two species in Fig. 3D. The phasor position of cells with different expression of GFP lie along the line between the GFP position and the autofluorescence phasor points.
Fig. 3.
Maps of relative concentration of tissue components. (A) Phasor plot of the FLIM image (color scale defined in Fig. 1D). Different clusters are assigned to pure chemical species according to Fig. 2A: GFP, 1; average tissue autofluorescence, 2; collagen, 3; retinol, 4; and retinoic acid, 5. (B) Phasor plot selection using a linear cluster combination that represents all of the possible relative concentrations of GFP and the average autofluorescence, autofluorescence and collagen, and retinol and retinoic acid, respectively. Each point along the line has a color corresponding to specific fractional intensity of the species. (C) Intensity image of a seminiferous tubule from a mice expressing GFP from an Oct4 transgene. (D–F) Maps of relative concentrations of: GFP and autofluorescence (D), autofluorescence and collagen (E), retinol and retinoic acid (F). Pixels in the images are highlighted with the same color scale of the phasor plot. Arrows indicate germ cells with a lower concentration of GFP with respect to autofluorescence.
Using this principle, we directly visualize different states of differentiation of the germ cells within the tissue. In Fig. 3D the last three stem cells of the chain, marked by one arrow, contain a smaller ratio of GFP to autofluorescence with respect to the other cells of the chain; they are differentiating thus decreasing the expression of Oct4-GFP. The germ cell marked with two arrows has lost almost all its Oct4-GFP expression. The relative concentration of autofluorescence and collagen is calculated and mapped in Fig. 3E, with a color scale from red to blue; that of retinol and retinoic acid is shown in Fig. 3F, with a color scale from orange to cyan.
In Vivo Identification of Changes in Metabolic State as Germ Cells Differentiate in C. elegans.
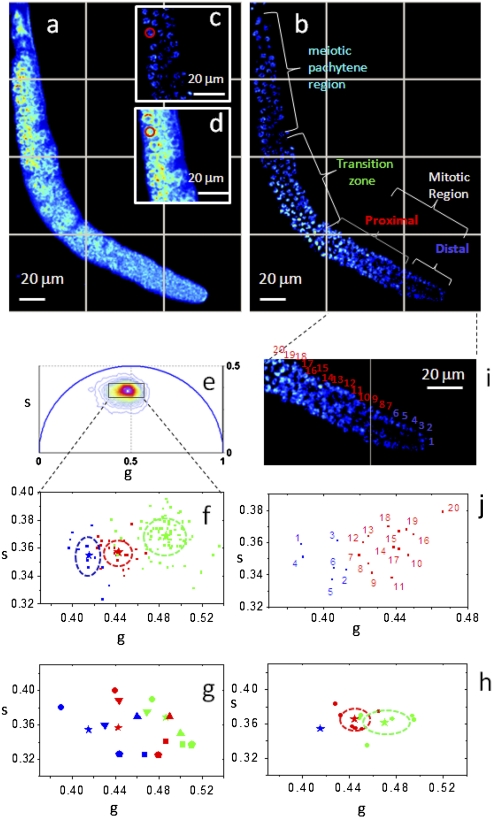
Here we show that the introduction of the cell phasor concept allows identifying gradients of metabolic activity in live tissue. Excitation of intrinsic fluorescent species can be maximized by tuning the excitation wavelength of the Ti:Sapphire laser. Collagen, GFP, and FAD two-photon excitation cross-sections have a peak around 900 nm; NADH is maximal around 740 nm (22, 24). Fig. 4 shows the two-photon excited fluorescence images acquired in the living tissue from the C. elegans germ line excited at 740 nm (Fig. 4A) and at 880 nm (Fig. 4 B and E). The germ line expresses a histone-GFP fusion protein that allows identification of the most obvious steps of differentiation, but does not make it possible to distinguish between stem cells and cells in the earliest stages of differentiation. The distal pool of the mitotic region contains undifferentiated cells maintained in a “stem cell-like state,” but the proximal pool contains cells that are closer to differentiating (32). More proximally, the transition zone contains cells that have differentiated and entered a meiotic prophase (as shown by crescent-shaped DNA), and the meiotic pachytene region contains cells that have further progressed through meiosis.
Fig. 4.
Identification of metabolic states of germ cells during differentiation. Fluorescence intensity image of a C. elegans germ line (A) excited at 740 nm and (B) excited at 880 nm. Histone-GFP fusion protein identifies the position and differentiation state of the germ cells that are indicated with different colors: distal mitotic region (blue), proximal mitotic region (red), transition zone (green), and meiotic pachytene (cyan). A red cursor of 5-μm diameter selects the region of interest of a germ cell in the intensity image at (C) 880 nm and (D) 740 nm. (E) Phasor plot of the FLIM image excited at 740 nm (color scale defined in Fig. 1D). (F) Scatter plot of the cell phasor of all germ cells excited at 740 nm. Every cell phasor (squares) is represented with a color that corresponds to its differentiation state in B. The distribution of distal mitotic cells in blue (blue, n = 14), proximal mitotic region (red, n = 20), transition zone cells (green, n = 83) are clearly separated. The mean values of clusters are represented by the colored stars and the SD by the dotted lines. (G) Scatter plot of the mean values of cell phasor distributions in distal mitotic region (blue), proximal mitotic region (red), and transition zone (green) for n = 6 independent C. elegans germ line. Independent samples are represented with different symbols. (H) Scatter plot of the mean values of the cell phasor distributions for n = 6 independent germ lines. Each sample is translated in the phasor plot as to make all of the distal mitotic region values coincident. The SDs of the proximal mitotic region and transition zone are represented by the dotted lines. (I) Zoomed image of the mitotic region of the C. elegans germ line excited at 880 nm in B. Cells are numbered in a distal to proximal direction. Blue cells belong to the distal mitotic region and red cells belong to the proximal mitotic region. (J) Scatter plot of the phasor average values of the 20 germ cells indicated in I.
Fig. 4E shows the phasor distribution of the FLIM image excited at 740 nm. The intrinsic fluorescence of the tissue is a mixture of FAD and NADH, which are both excited at 740 nm. We confirm the presence of these metabolites by spectral imaging (Fig. S3). We calculate the cell phasors by performing manual image segmentation. We select the regions of interest of germ cells with a circular cursor of 5-μm diameter (red cursor in Fig. 4 C and D). The cell phasor value of a germ cell is calculated within the circular cursor. We plot the cell phasor values of germ cells in the scatter diagram of Fig. 4F (squares). Because of the tight association between the distal-tip cell and germ cells (45), and because the distal-tip cell cross-section on transverse sections decreases with distance to the distal end, we cannot exclude an important contribution of the distal-tip cell to any autofluorescence trend on the distal-proximal axis. This issue will be revisited in future studies. Cell phasors cluster according to their differentiation state (Fig. 4 F and G). The distributions of cell phasors of the distal mitotic region (blue), the proximal mitotic region (red), and the transition zone (green) are significantly different (t test, P < 0.05) (Fig. 4F). The same trend of cell phasor distributions has been found in six independent C. elegans germ lines (Fig. 4G) and their distributions are statistically different (t test, P < 0.05) (Fig. 4H). The phasors of the germ cells along the main axis of the C. elegans germ line (Fig. 4 F and J) show heterogeneity in the concentration of FAD and free and bound NADH (Fig. 2 and Fig. S3). During differentiation, the concentration of FAD decreases, but the concentration of bound NADH increases with respect to free NADH (Fig. S3 A and B). The blue shift in the spectrum during differentiation (Fig. S3 C and D) confirms a decrease in FAD concentration and an increase of the bound/free NADH ratio during differentiation. Fig. 4J shows the cell phasors of germ cells in the mitotic region according to their position from the distal tip to the proximal end (Fig. 4I). The trend in the cell phasors along the mitotic region (Fig. 4 I and J) suggests a gradient in the expression of regulators promoting differentiation and self-renewal, which influence the metabolic states of cells.
Discussion
Here we show that the phasor approach to FLIM can map stem-cell metabolism in label-free living tissues. This method provides a metabolic fingerprint of cells and can identify and classify stem cells and differentiating cells according to their metabolic state. We measure small metabolic changes during differentiation and map metabolic gradients in tissues.
FLIM phasor is a “fit-free” method that requires no assumption or a priori knowledge on the biological system and provides unbiased representation of the FLIM raw data. We separate and identify tissue components by cluster analysis: that is, detecting clusters of pixel values in specific regions in the phasor plot (Fig. 1). This method provides high selectivity in identifying fluorescence components that cannot be separated by a multiexponential fitting or by analyzing the average lifetime. Multiexponential fitting can separate only a limited number of single exponential components (i.e., a few components in a mixture of multiple fluorescence species). The mean lifetime offers a contrast whose physical interpretation is ambiguous and cannot separate tissue components with the same mean lifetime but characterized by different lifetime distributions (Fig. S1A). The phasor approach instead provides excellent discrimination of intrinsic molecular sources in live tissues, where the majority of pixels have a complex multiexponential decay (Fig. 1, and Figs. S1B, S4, and S5). Phasor FLIM allows fast analysis of large datasets and provides a global overview of the decay properties by analyzing all pixels of the image at once.
We determine the phasor location of some relevant endogenous fluorophores, such as collagen, free and bound NADH, FAD, retinol, retinoic acid, and porphyrin. However, the number of fluorescent chemical species that can be identified by their phasor signature is not limited. The phasor location of every molecular species in the histogram is uniquely determined by their fluorescence decay. The phasor fingerprint of chemical species avoids the resolution of lifetime components of fluorophore decay and allows interpreting FLIM images directly in terms of chemical species. The phasor location of endogenous fluorophores is used as a guide to identify them in mice and in C. elegans germ lines (Figs. 2 and 3, and Fig. S3). Phasors allow an easy quantification of the relative concentration of molecular species in living tissues. Phasor coordinates are a linear function of molecular species and a mixture of fluorescent species are identified by a graphic analysis. We measure and map the relative concentration of fluorescent species (Fig. 3 and Fig. S3) directly from the position of the pixel in the phasor plot on the straight line connecting the two chemical species.
FLIM phasor provides metabolic fingerprints of cells through image segmentation and the calculation of cell phasors (Fig. 4). Different positions on the main axis of the C. elegans gonad are statistically attributed to different average phasor values and we can sort them by their metabolic state (Fig. 4 F and H). The evolution of the distal-most cell phasor fingerprint, although moving proximally (Fig. 4F), reflects a decrease in the concentration of FAD and an increase in the ratio of bound/free NADH (Fig. S3), consistent with what would be expected for stem cells and their differentiating descendants (13, 16, 19). Growth factors that promote self-renewal cause stem cells to become more reduced, but signaling molecules that promote differentiation cause progenitors to have a higher oxidation potential (12). The change in the metabolic fingerprint during differentiation (Fig. 4E) may also suggest a change in the binding sites of NADH (Fig. 2 and Fig. S3). The gradient of cell phasors in the mitotic region (Fig. 4J) reflects a progression from undifferentiated stem cells to early differentiation. This finding may also reflect a contribution of the distal dip cell to the signal. Cell phasor heterogeneity among mitotic cells (Fig.4J) could reveal symmetric and asymmetric divisions occurring at the level of the niche. In fact, cell phasor signaling controls the production of stem cells’ daughter and differentiated progenies at the level of individual cells.
FLIM has previously been used to distinguish different states of stem cells only in vitro (13, 16, 19). However, the traditional way of fitting in every pixel of the complex intensity decays with two exponentials assumes that the ratio of the “short” and “long” components is equivalent to the ratio of free/bound NADH. Furthermore, other fluorophores can contribute to the average short or long components and bound NADH is characterized by a multiexponential decay. The 2D phasor representations instead allows us to measure differences in the FLIM signature regardless of the complexity of the lifetime decay (Fig. 4 and Fig. S1).
With phasor FLIM it is possible to easily discriminate undifferentiated stem cells from different stages of differentiation (Fig. 4) and possibly identify asymmetric divisions. Time-lapse phasor FLIM imaging can be performed to obtain information on the dynamics of cell activity, physiological processes thus monitoring tissue development over time. The phasor method of FLIM is capable of measuring small differences in metabolic states among stem cells (Fig. 4I). Relatively small changes in intracellular metabolite levels over a narrow range can modulate cell fate and function with profound difference in outcome (12, 46, 47). Hence, the cell phasor fingerprints of cells could be used to predict stem-cell fate and to characterize stem-cell plasticity and the commitment of stem cells to differentiation. It would be also interesting to monitor the metabolic fingerprint evolution in the phasor plot of different differentiation pathways to cell lineages.
In conclusion, the phasor approach to FLIM provides a more quantitative and straightforward interpretation of physiological processes in living tissues than the classic approach based on average lifetime histograms. Phasor FLIM provides metabolic fingerprints of cells and tissues without any fitting procedures and assumptions that could introduce artifacts in the data interpretation. Hence, phasor FLIM allows an unbiased identification and classification of metabolic states of cells in a similar way to flow cytometry scattergrams.
The phasor approach in tissues is a promising tool in biology, biophotonics, and biomedical research to track in vivo metabolic changes that are associated with stem-cell differentiation, cell carcinogenesis, and apoptosis. This approach could provide important insight into the signaling pathways and regulatory networks, which are involved in cell self-renewal differentiation and oncogenesis in a variety of tissue and organs. The phasor approach to FLIM would also be helpful to monitor cell metabolism and at the same time characterize the 3D microenvironment of tissues by detecting extracellular matrix remodeling and molecular gradients. The ability to observe and isolate cancer cells and stem cells noninvasively based on their metabolic state in living tissues has important implications for early diagnosis and new therapeutic strategies. The detection of malignant transformation of progenitor cells and aberrant differentiation of cancer cells could be performed in vivo. Label-free discrimination between self-renewal and differentiation by a phasor approach to FLIM would be suitable to noninvasively monitor embryonic stem cells and to design new approaches to reprogram somatic cells to a pluripotent stem-cell fate. The phasor approach to FLIM could be of interest to label-free cell sorting and high-throughput screening for drug discovery, cell replacement therapies, and tissue engineering.
Materials and Methods
Mice Seminiferous Tubule Preparation.
One-year-old mice expressing GFP from an Oct4 transgene were scarified. Seminiferous tubules were extracted from the testes and mounted between coverslip in PBS medium. Fresh tissues are imaged within 2 h of the extraction. All animal procedures were performed in strict accordance with the National Institutes of Health and Institutional IACUC guidelines.
Solution preparations are described in SI Materials and Methods.
Worm Maintenance and Germ-Line Preparation.
Strain AZ212 (ruIs32 [pie-1::GFP::H2B + unc-119(+)]) (48), expressing a histone-GFP fusion in germ-line nuclei, was maintained at 20 °C, as previously described (49), with Escherichia coli HB101 as a food source. Worms were staged at L4, 24 h before gonad extrusion in PBS supplemented with 0.2 mM levamisole and rinsing with PBS. Germ lines were imaged within the hour following dissection.
Imaging.
The microscope set-up is described in SI Materials and Methods. The FastFLIM method is described in ref. 35.
Cell Phasor Data Analysis.
Every pixel of the FLIM image is transformed in one pixel in the phasor plot, as previously described, and reported in detail in SI Materials and Methods (29). The coordinates g and s in the phasor plot are calculated from the fluorescence intensity decay of each pixel of the image by using the transformations defined in SI Materials and Methods. The analysis of the phasor distribution is performed by cluster identification. Clusters of pixel values are detected in specific regions of the phasor plot. The cluster assignment is performed by taking into account not only the similar fluorescence properties in the phasor plot but also exploiting the spatial distribution and localization in cellular substructures or tissues; we achieve this by applying a median filter (see SI Materials and Methods) that imposes a correlation between cluster of pixels in the phasor plot and pixels of the image without decreasing the spatial resolution. This process allows better confining of a cluster to a specific phasor value by reducing the statistical error in the phasor associated with each pixel of the image. Regions of the image with different decay profiles and characteristics can be better delineated. To obtain information on the chemical composition of tissues, we compared the size of their phasor distribution with the statistical uncertainty, which depends on the inverse of the square-root of the number of photons collected. If the size of the phasor distribution was comparable to the statistical uncertainty, we selected an independent molecular species using a circular selection cursor. If the phasor distribution size was greater than the statistical uncertainty, we selected a mixture of molecular components using a cursor that joins the two molecular species. Fractional intensities of chemical species in every pixel of the image are evaluated with a graphic analysis in the phasor plot (see SI Materials and Methods). We performed image segmentation on the FLIM data by selecting the region of interest of germ cells within the tissue. The region of interest of cells was selected by using a circular cursor of 5-μm diameter. We calculated the phasor average value within these regions of interest. When measuring the cell phasor, all pixels of the cell (about 1,000) were taken into account and the signal-to-noise ratio of the FLIM signature of cells was higher than in single pixels. All phasor transformation and the data analysis of FLIM data were performed using SimFCS software developed at the Laboratory for Fluorescence Dynamics, University of California at Irvine.
Supplementary Material
Acknowledgments
We thank the Caenorhabditis Genetics Center, funded by the National Institutes of Health National Center for Research Resources, for providing nematode strain AZ212. This work was supported in part by National Institutes of Health Grants P41-RRO3155, P50-GM076516, R01 HD49488, and P01 HD47675 (to P.D.); California Insitute of Regenerative Medicine Grant RC1-00110 (to P.D.); and University of California at Irvine startup funds (to O.C.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108161108/-/DCSupplemental.
References
- 1.Donovan PJ, Gearhart J. The end of the beginning for pluripotent stem cells. Nature. 2001;414(6859):92–97. doi: 10.1038/35102154. [DOI] [PubMed] [Google Scholar]
- 2.Knoblich JA. Asymmetric cell division during animal development. Nat Rev Mol Cell Biol. 2001;2(1):11–20. doi: 10.1038/35048085. [DOI] [PubMed] [Google Scholar]
- 3.Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- 4.Morrison SJ, Spradling AC. Stem cells and niches: Mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pera MF, Tam PP. Extrinsic regulation of pluripotent stem cells. Nature. 2010;465:713–720. doi: 10.1038/nature09228. [DOI] [PubMed] [Google Scholar]
- 6.Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441:1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 7.Fjeld CC, Birdsong WT, Goodman RH. Differential binding of NAD+ and NADH allows the transcriptional corepressor carboxyl-terminal binding protein to serve as a metabolic sensor. Proc Natl Acad Sci USA. 2003;100:9202–9207. doi: 10.1073/pnas.1633591100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Q, Piston DW, Goodman RH. Regulation of corepressor function by nuclear NADH. Science. 2002;295:1895–1897. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
- 9.Sahar S, Sassone-Corsi P. Metabolism and cancer: The circadian clock connection. Nat Rev Cancer. 2009;9:886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 10.Ito K, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- 11.Ogasawara MA, Zhang H. Redox regulation and its emerging roles in stem cells and stem-like cancer cells. Antioxid Redox Signal. 2009;11:1107–1122. doi: 10.1089/ars.2008.2308. [DOI] [PubMed] [Google Scholar]
- 12.Smith J, Ladi E, Mayer-Proschel M, Noble M. Redox state is a central modulator of the balance between self-renewal and differentiation in a dividing glial precursor cell. Proc Natl Acad Sci USA. 2000;97:10032–10037. doi: 10.1073/pnas.170209797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo HW, et al. Reduced nicotinamide adenine dinucleotide fluorescence lifetime separates human mesenchymal stem cells from differentiated progenies. J Biomed Opt. 2008;13:050505. doi: 10.1117/1.2990752. [DOI] [PubMed] [Google Scholar]
- 14.Kantelhardt SR, et al. Imaging of brain and brain tumor specimens by time-resolved multiphoton excitation microscopy ex vivo. Neuro-oncol. 2007;9(2):103–112. doi: 10.1215/15228517-2006-034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasischke KA, Vishwasrao HD, Fisher PJ, Zipfel WR, Webb WW. Neural activity triggers neuronal oxidative metabolism followed by astrocytic glycolysis. Science. 2004;305(5680):99–103. doi: 10.1126/science.1096485. [DOI] [PubMed] [Google Scholar]
- 16.König K, Uchugonova A, Gorjup E. Multiphoton fluorescence lifetime imaging of 3D-stem cell spheroids during differentiation. Microsc Res Tech. 2011;74(1):9–17. doi: 10.1002/jemt.20866. [DOI] [PubMed] [Google Scholar]
- 17.Rice WL, Kaplan DL, Georgakoudi I. Two-photon microscopy for non-invasive, quantitative monitoring of stem cell differentiation. PLoS ONE. 2010;5:e10075. doi: 10.1371/journal.pone.0010075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skala MC, et al. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc Natl Acad Sci USA. 2007;104:19494–19499. doi: 10.1073/pnas.0708425104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uchugonova A, König K. Two-photon autofluorescence and second-harmonic imaging of adult stem cells. J Biomed Opt. 2008;13:054068. doi: 10.1117/1.3002370. [DOI] [PubMed] [Google Scholar]
- 20.Bird DK, et al. Metabolic mapping of MCF10A human breast cells via multiphoton fluorescence lifetime imaging of the coenzyme NADH. Cancer Res. 2005;65:8766–8773. doi: 10.1158/0008-5472.CAN-04-3922. [DOI] [PubMed] [Google Scholar]
- 21.Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat Methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- 22.Zipfel WR, et al. Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation. Proc Natl Acad Sci USA. 2003;100:7075–7080. doi: 10.1073/pnas.0832308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowles J, et al. Retinoid signaling determines germ cell fate in mice. Science. 2006;312:596–600. doi: 10.1126/science.1125691. [DOI] [PubMed] [Google Scholar]
- 24.Huang S, Heikal AA, Webb WW. Two-photon fluorescence spectroscopy and microscopy of NAD(P)H and flavoprotein. Biophys J. 2002;82:2811–2825. doi: 10.1016/S0006-3495(02)75621-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alcala JR, Gratton E, Prendergast FG. Fluorescence lifetime distributions in proteins. Biophys J. 1987;51:597–604. doi: 10.1016/S0006-3495(87)83384-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medine CN, McDonald A, Bergmann A, Duncan RR. Time-correlated single photon counting FLIM: some considerations for physiologists. Microsc Res Tech. 2007;70:420–425. doi: 10.1002/jemt.20425. [DOI] [PubMed] [Google Scholar]
- 27.Pelet S, Previte MJ, Laiho LH, So PT. A fast global fitting algorithm for fluorescence lifetime imaging microscopy based on image segmentation. Biophys J. 2004;87:2807–2817. doi: 10.1529/biophysj.104.045492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verveer PJ, Squire A, Bastiaens PI. Global analysis of fluorescence lifetime imaging microscopy data. Biophys J. 2000;78:2127–2137. doi: 10.1016/S0006-3495(00)76759-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Digman MA, Caiolfa VR, Zamai M, Gratton E. The phasor approach to fluorescence lifetime imaging analysis. Biophys J. 2008;94(2):L14–L16. doi: 10.1529/biophysj.107.120154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jameson DM, Gratton E, Hall RD. The measurement and analysis of heterogeneous emissions by multifrequency phase and modulation fluorometry. Appl Spectrosc Rev. 1984;20(1):55–106. doi: 10.1146/annurev.bb.13.060184.000541. [DOI] [PubMed] [Google Scholar]
- 31.Cinquin O. Purpose and regulation of stem cells: A systems-biology view from the Caenorhabditis elegans germ line. J Pathol. 2009;217(2):186–198. doi: 10.1002/path.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cinquin O, Crittenden SL, Morgan DE, Kimble J. Progression from a stem cell-like state to early differentiation in the C. elegans germ line. Proc Natl Acad Sci USA. 2010;107:2048–2053. doi: 10.1073/pnas.0912704107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hubbard EJ. Caenorhabditis elegans germ line: A model for stem cell biology. Dev Dyn. 2007;236:3343–3357. doi: 10.1002/dvdy.21335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clayton AH, Hanley QS, Verveer PJ. Graphical representation and multicomponent analysis of single-frequency fluorescence lifetime imaging microscopy data. J Microsc. 2004;213(Pt 1):1–5. doi: 10.1111/j.1365-2818.2004.01265.x. [DOI] [PubMed] [Google Scholar]
- 35.Colyer RA, Lee C, Gratton E. A novel fluorescence lifetime imaging system that optimizes photon efficiency. Microsc Res Tech. 2008;71:201–213. doi: 10.1002/jemt.20540. [DOI] [PubMed] [Google Scholar]
- 36.Redford GI, Clegg RM. Polar plot representation for frequency-domain analysis of fluorescence lifetimes. J Fluoresc. 2005;15:805–815. doi: 10.1007/s10895-005-2990-8. [DOI] [PubMed] [Google Scholar]
- 37.Hess ST, Sheets ED, Wagenknecht-Wiesner A, Heikal AA. Quantitative analysis of the fluorescence properties of intrinsically fluorescent proteins in living cells. Biophys J. 2003;85:2566–2580. doi: 10.1016/s0006-3495(03)74679-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bornstein P, Kang AH, Piez KA. The nature and location of intramolecular cross-links in collagen. Proc Natl Acad Sci USA. 1966;55:417–424. doi: 10.1073/pnas.55.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bel'Kov MV, Bondarev SL. Fluorescence spectra and kinetics of isomers and dimers of retinoic acid. J Appl Spectrosc. 1990;53:1271–1275. [Google Scholar]
- 40.Chia TH, Williamson A, Spencer DD, Levene MJ. Multiphoton fluorescence lifetime imaging of intrinsic fluorescence in human and rat brain tissue reveals spatially distinct NADH binding. Opt Express. 2008;16:4237–4249. doi: 10.1364/oe.16.004237. [DOI] [PubMed] [Google Scholar]
- 41.König K, Riemann I. High-resolution multiphoton tomography of human skin with subcellular spatial resolution and picosecond time resolution. J Biomed Opt. 2003;8:432–439. doi: 10.1117/1.1577349. [DOI] [PubMed] [Google Scholar]
- 42.Lakowicz JR, Szmacinski H, Nowaczyk K, Johnson ML. Fluorescence lifetime imaging of free and protein-bound NADH. Proc Natl Acad Sci USA. 1992;89:1271–1275. doi: 10.1073/pnas.89.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brancaleon L, et al. Characterization of the photoproducts of protoporphyrin IX bound to human serum albumin and immunoglobulin G. Biophys Chem. 2004;109:351–360. doi: 10.1016/j.bpc.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 44.Campagnola PJ, Loew LM. Second-harmonic imaging microscopy for visualizing biomolecular arrays in cells, tissues and organisms. Nat Biotechnol. 2003;21:1356–1360. doi: 10.1038/nbt894. [DOI] [PubMed] [Google Scholar]
- 45.Crittenden SL, Leonhard KA, Byrd DT, Kimble J. Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Mol Biol Cell. 2006;17:3051–3061. doi: 10.1091/mbc.E06-03-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lonergan T, Brenner C, Bavister B. Differentiation-related changes in mitochondrial properties as indicators of stem cell competence. J Cell Physiol. 2006;208(1):149–153. doi: 10.1002/jcp.20641. [DOI] [PubMed] [Google Scholar]
- 47.Parker GC, Acsadi G, Brenner CA. Mitochondria: Determinants of stem cell fate? Stem Cells Dev. 2009;18:803–806. doi: 10.1089/scd.2009.1806.edi. [DOI] [PubMed] [Google Scholar]
- 48.Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.