Abstract
EspR is a transcriptional regulator that activates the ESX-1 secretion system during Mycobacterium tuberculosis infection and is critical for pathogenesis. It is unique among DNA-binding proteins as it is secreted as part of a feedback regulatory loop that serves to mitigate transcriptional activity. Here we report the crystal structure of a functional EspR dimer at 2.5-Å resolution. The amino-terminal half of EspR is a helix-turn-helix (HTH) DNA-binding domain and the carboxy terminus consists of a dimerization domain with similarity to the SinR:SinI sporulation regulator of Bacillus subtilis. Surprisingly, the HTH domains of EspR are arranged in an unusual conformation in which they are splayed at an oblique angle to each other, suggesting that EspR binds DNA in a profoundly different way than most other known HTH regulators. By mapping the EspR binding sites in the espACD promoter, using both in vivo and in vitro binding assays, we show that the EspR operators are located unusually far from the promoter. The EspR dimer binds to these sites cooperatively, but the two “half-sites” contacted by each DNA recognition motif are separated by 177 base pairs. The distinctive structure of EspR and the exceptional arrangement of its operator contacts suggest that it could promote DNA looping in its target promoter. We hypothesize that direct DNA looping mediated by single-site binding of each EspR monomer may facilitate transcriptional control of this important virulence system.
Keywords: transcription, type VII secretion
Mycobacterium tuberculosis, an intracellular pathogen that exerts an enormous toll on global human health, has evolved to respond to changing conditions within the infected host (1). In particular, the ESX-1 secretion system, which delivers effector proteins into host cells and is essential for M. tuberculosis virulence (2), is under positive transcriptional control by the helix-turn-helix (HTH) transcription factor EspR (3). Because the major secreted substrates ESAT-6 (early secretory antigenic target, 6 kDa) and CFP-10 (culture filtrate protein, 10 kDa) are not only required for virulence but are also highly immunogenic T-cell antigens, dynamic regulation during infection of macrophages may function to balance virulence and immunogenicity. Thus, precise regulation of the delivery of ESX-1 substrates during infection may be essential to M. tuberculosis pathogenesis (3).
EspR promotes ESX-1 secretion by activating transcription of the espACD (Rv3616c-Rv3614c) operon immediately upon macrophage infection (3). EspA, EspC, and EspD are required for ESX-1 secretion, and both EspA and EspC are ESX-1 secreted substrates (4, 5). Interestingly, not only is EspR an activator of the system, it is itself a substrate of ESX-1 and its secretion provides a simple negative feedback loop that serves to limit transcription by lowering the concentration of EspR in the cell. Thus, EspR activates ESX-1 secretion early after phagocytosis, but its activity is titrated away from the cell later during macrophage infection. Therefore, EspR is not only a key regulator of M. tuberculosis virulence, but it is controlled by a unique mechanism.
In this work, we have determined the crystal structure of EspR to a maximum resolution of 2.5 Å and have identified its operator binding sites in the espACD regulatory region. Our studies reveal that EspR has both an unusual arrangement of its DNA-interaction domains as well as complex binding characteristics, distinguishing it from other known HTH regulators.
Results
EspR Crystallization and Structure Determination.
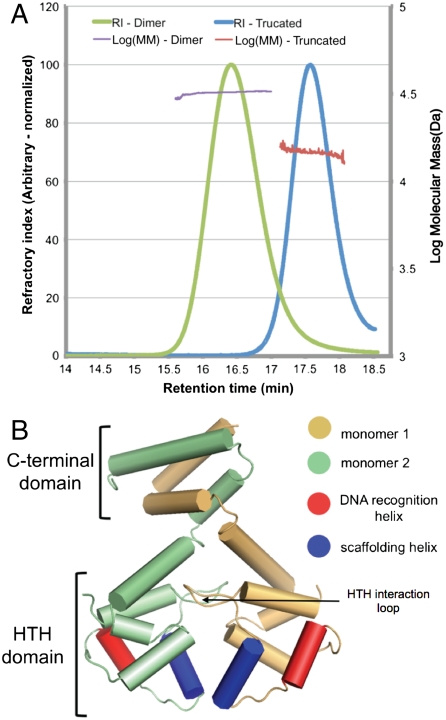
We overexpressed recombinant EspR in Escherichia coli, purified it via a combination of affinity and size-exclusion chromatography, and removed the tag to produce full-length EspR with three additional residues from the affinity tag on the amino terminus. Gel filtration revealed that the protein migrated as a single species and with an estimated molecular mass of approximately 30 kDa, suggesting that the protein consisted of a dimer of two 15.2-kDa monomers. Inline light-scattering and refractivity analysis indicated that the protein had an estimated, shape-independent molecular mass of approximately 33 kDa, within experimental error of the predicted molecular mass of the dimer (Fig. 1A). The protein was uniformly monodisperse and there was no evidence for other molecular mass species. Thus, EspR likely exists as a dimer in solution.
Fig. 1.
The dimeric EspR crystal structure. (A) Full-length (amino acids 1–132) and a truncated form of EspR (amino acids 1–107) from M. tuberculosis were expressed in E. coli, purified, and injected on a Superose 200 gel filtration column. The refractory index (RI) and light scattering (see Materials and Methods) were used to calculate a molecular mass (MM) of 31.8 kDa for the full-length and 15.1 kDa for the truncated EspR. The predicted masses of the full-length EspR monomer and of the truncated form are 14.7 and 12.5 kDa. (B) Schematic of the EspR dimer demonstrating that the two monomers, colored in gold and green, interact at both the C-terminal domain and in the HTH domain. The DNA recognition helix (red) is predicted to fit into the major groove of DNA and the scaffolding helix (blue) is predicted to make additional contacts with the DNA.
EspR crystallized in the spacegroup P32 and we determined its structure at a resolution of 2.5 Å using a selenomethionine derivative and multiwavelength anomalous phasing (Table S1 lists crystallographic information and statistics). Although the solution was complicated by pseudotranslation and perfect merohedral twinning, we were able to refine to an Rfree of 23% using least-squares, twinned refinement (6). As was the case for another group with a related crystal form, we were unable to solve the structure with molecular replacement (7). Attempts to fit our data with models of SinR, the closest structural homolog discovered by tertiary structure prediction (3, 8), and with models of other classic HTH proteins including lambda (9) and phage 434 repressors (10), failed to produce a solution, likely because these were monomeric domains that were only distantly related to EspR sequence (< 25% identical).
Structural Overview.
Although there are 18 monomers in the asymmetric unit (molecules A–R), they are arranged into a set of nine closely interacting dimers. An analysis with the PISA suite of programs (11) suggested that only the intradimer interfaces (between molecules A–B, C–D, etc.) have a significant role in complex formation. These interfaces were all given a “complexation significance score” of 1.0 (on a scale of 0–1.0), implying that the interface plays an essential role in complex formation. All other monomer–monomer contacts in the crystal are scored as incidental artifacts of crystallization. Thus, the rest of this report will discuss only the dimer formed between molecule A and B, which we believe represents the biological assembly. We have also solved a second crystal form (P212121 spacegroup) with molecular replacement of our original, monomeric model and, though it was of lower resolution (ca. 3.2 Å), the initial electron density maps clearly revealed the same dimeric structure but with extensive disorder that precluded further refinement. However, it was clear from rough electron density maps that the dimeric structure found in the trigonal crystal form was retained in the low-resolution, orthorhombic form.
Overall, the two EspR monomers in the dimer form an extensive interface burying 3,017 Å2 of surface area (as calculated in ref. 12) for each molecule at the interface (Fig. 1B). The monomer is an all-helical structure with distinct amino- and carboxyl-terminal domains. The N terminus (amino acids 1–80) is a classical HTH domain, and the two HTH domains in the dimer are packed closely against one another. The carboxyl-terminal domains (CTDs; amino acids 81–132) of each monomer create a series of four crisscrossing helices forming a hydrophobic interaction, suggesting that this domain is necessary for EspR constitutive dimerization.
Individual HTH Domain.
The HTH domain is perhaps the most widely used DNA-binding motif in prokaryotes and a large number of HTH-containing proteins have been extensively studied in structural detail (13). A search using the DALI server (14) revealed that the most closely related HTH domain where a DNA-bound structure is available is the phage 434 repressor (10), and the domain’s structure is also extremely similar to CI and Cro repressors of lambda (9, 15). The hydrophobic folding of the EspR HTH domain is essentially conserved with the 434 repressor, centered around the buried residues L9, L12, V29, L64, and Y74. Although individual residues shown to be important for binding DNA are not conserved with the 434 repressor, the network of hydrogen donors required for DNA binding is available on the recognition and scaffolding helices of EspR and there are no steric hindrances to DNA binding.
The overall position of canonical DNA interacting elements, the DNA recognition and scaffolding helices (13), are also well conserved with 434 repressor although there are some notable differences. First, the scaffolding helix is rotated approximately 20° relative to the scaffolding helices of the DNA-bound form of the 434 repressor, as well as compared to the CI and Cro repressors from lambda phage. There is also an extensive loop N-terminal to the scaffolding helix (amino acids 16–25), which is involved in the dimerization interface (Fig. 1B, “HTH interaction loop”).
Dimer Interface.
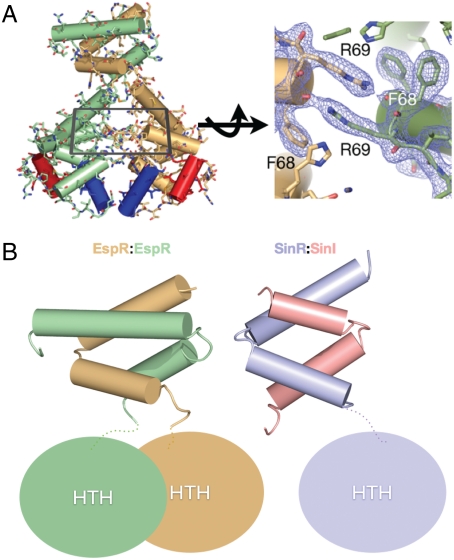
The two monomers (chain A and B) are related to one another by pseudo-twofold symmetry (Fig. 1B). Twofold symmetry is not exact, due in part to the fact that the full C terminus is seen in the B monomer but the final residues of the A chain are disordered. The dimer interface consists of two distinct regions in the N- and C-terminal domains. In the N-terminal HTH domain, the two monomers form an extensive electrostatic and hydrogen bonding network (Fig. 2A). This interaction also includes symmetrical arginines (R69) that stack hydrophobically with each other, as well as symmetrical phenylalanines (F67 and F68) that further stabilize the dimer (16). Notably, although these stacking interactions are hydrophobic, they are stabilized by hydrogen bonding and there is no clear hydrophobic core between the HTH domains themselves. There is thus the possibility for rearrangements between the two HTH domains.
Fig. 2.
Dimerization interfaces of EspR. (A) The interactions between the HTH domains are primarily electrostatic in nature with a set of symmetrically stacked arginines and phenylalanines at the center of the interface. (B) Comparison of the CTD of the EspR dimer (gold and green) with the SinR:SinI heterodimer (purple and pink). In both structures, the CTDs form a four-helix bundle and the dimer interface between them is entirely hydrophobic.
In contrast to the HTH domain interactions, the CTD is a four-helix bundle between the two monomers generating a small hydrophobic core composed of I102, L109, V117, I121, and L124 from both monomers along the pseudo-twofold axis. In this region, all of the buried hydrophobic residues are found at the dimer interface. In fact, almost all of the residues between E81 and D130 (43/49 residues) are classified as interacting at the dimer interface by the PISA algorithm (as opposed to 15/80 in the HTH region). This interface is remarkably similar to the interaction between the HTH transcription factor SinR and its inhibitor SinI (8) (Fig. 2B), which Lewis et al. hypothesized might mimic the interactions between two SinR monomers. To test if the CTD region of EspR is required for dimerization, we expressed and purified a truncated form of the protein lacking just the last α-helix (amino acids 108–132). Both gel filtration and light-scattering analyses indicated that this protein was a well-folded, monodisperse monomer, suggesting that the CTD is critical for dimerization (Fig. 1A).
Orientation of HTH Domains.
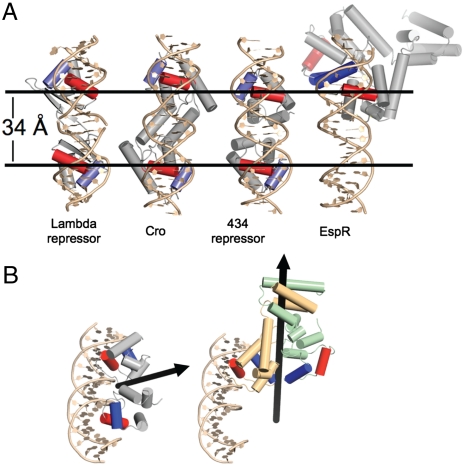
In almost all known cases, HTH proteins form dimers that bind to successive turns of the major groove, with each motif binding one half-site of symmetry-related DNA (13) (Fig. 3A). When we modeled one of the EspR HTH domains onto the DNA-bound structure of the 434 repressor, the HTH of EspR overlaid well onto the 434 HTH, with an rmsd value of 1.8 Å. Indeed, the excellent overlap with the recognition helix suggests that the individual HTH domains adopt a DNA-binding-competent conformation. However, examination of the quaternary structure of the rest of the protein revealed a large deviation from the 434 repressor, with the second HTH of the EspR dimer rotated approximately 120° relative to that of the 434 protein (Fig. 3 A and B). Moreover, manual rotation of the HTH domains could not allow for the recognition helices to be placed onto adjacent major grooves. This spatial orientation of the HTH domains strongly suggests that EspR cannot bind DNA via the standard mode of HTH regulators. In particular, in the absence of dramatic conformational changes, each HTH domain in the EspR dimer cannot simultaneously make contacts between adjacent sequences of DNA.
Fig. 3.
The EspR structure is unique among HTH proteins. (A) Models of the quaternary structure of three HTH domain dimers and the EspR dimer are displayed. The spacing between the DNA-binding domains in the structures of lambda repressor (1LMB), 434 repressor (1PER), and Cro (6CRO) is approximately 34 Å, which corresponds to a single turn of the major groove of DNA and allows for simultaneous binding of two contiguous DNA half-sites. In all structures shown, the dimers are colored gray with the recognition helix in red and the scaffolding helix in blue. Note that alignment of one HTH domain of EspR onto the DNA scaffold results in the other domain to project back into the two-dimensional plane of the diagram. (B) Alignment of EspR with the 434 phage repressor, and subsequent modeling of one EspR HTH monomer onto the DNA, resulted in the second HTH domain to be rotated approximately 120° away from the DNA. The black arrows indicate the twofold axis of symmetry of the 434 repressor and the EspR dimer.
DNA-Binding Experiments.
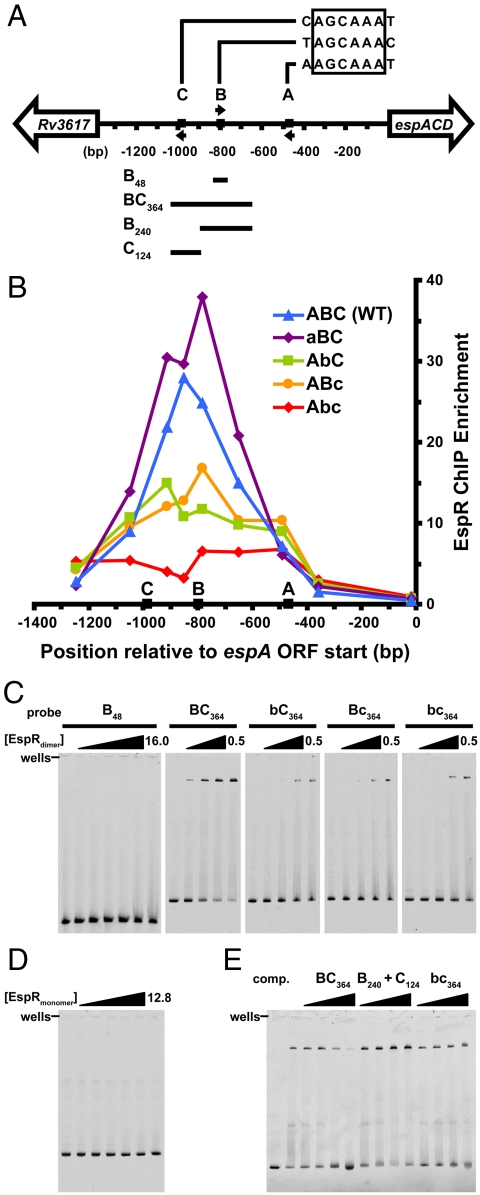
We had shown previously that a maltose-binding protein (MBP)-tagged version of EspR bound to a site approximately 427–520 bp upstream of the translational start site of EspA (site A in Fig. 4A). Similar experiments with untagged, dimeric EspR suggested that this protein bound this region with relatively low affinity in vitro (Fig. S1A). Furthermore, these previous experiments used only part of the sequence immediately upstream of espACD and we reasoned that other elements further upstream may be important for EspR binding. Therefore, we sought to identify EspR operators in the espACD promoter in vivo using a ChIP assay. We integrated the entire intergenic region upstream of espACD (1,357 bp, Fig. 4A) into the Mycobacterium smegmatis genome, expressed 3X-FLAG-tagged M. tuberculosis EspR, cross-linked with formaldehyde, and immunoprecipitated the protein from cell extracts. The relative abundance of DNA fragments within the immunoprecipitate was determined by quantitative PCR using a series of primer sets that spanned the entire intergenic region. Surprisingly, the peak of binding centered on a site approximately 850-bp upstream of the EspA start codon (Fig. 4B). These results, in combination with preliminary genome-wide ChIP-on-Chip studies and the mutational analyses described below, suggested that EspR binds to the DNA sequence AGCAAA. Notably, this sequence is neither palindromic nor present in contiguous repeats, common features of most HTH operators. However, a search of the entire espACD upstream sequence identified three sequence matches, here termed A, B, and C and centered at positions -468, -798, and -983 bp relative to the start site of translation (Fig. 4A). Thus, the peak of EspR binding in vivo centered near the B site, but not the A site.
Fig. 4.
EspR binds far upstream of the espACD promoter and contacts two half-sites separated by 177 bp. (A) Map of the intergenic region upstream of the M. tuberculosis espACD operon, with EspR binding motifs A, B, and C, respectively, located at -468, -798, and -983 bp relative to the start of the ORF. Arrows indicate the orientation of each motif, and the four DNA fragments used for EMSA experiments are indicated below. (B) The wild-type intergenic sequence (ABC, blue triangles), and the same sequence containing scrambled-site mutations in A (aBC, purple diamonds), B (AbC, green squares), C (ABc, orange circles), and B and C together (Abc, red diamonds) were integrated into the M. smegmatis genome. M. tuberculosis EspR-3X-FLAG was expressed in each strain and ChIP-qPCR (quantitative PCR) was performed using anti-FLAG antibodies. Points represent the mean of triplicate qPCR measurements. (C) EMSAs were performed using purified EspR and the indicated 5-FAM-labeled probes (5 nM). For the B48 probe, EspRdimer concentrations were 0, 0.5, 1, 2, 4, 8, 16 µM, and for the BC364 probe and mutants, concentrations were 0, 0.2, 0.3, 0.4, and 0.5 µM. (D) The monomeric form of EspR (amino acids 1–107) was used in EMSAs with the BC364 probe ([EspRmonomer] = 0, 0.4, 0.8, 1.6, 3.2, 6.4, and 12.8 µM). (E) Binding for labeled BC364 probe ([EspRdimer] = 0.3 μM) was competed by adding unlabeled probes in 2-, 8-, 32-, and 128-fold molar excess of the labeled probe.
To test the role of these sequences in EspR binding, we generated scrambled-site mutations in the A, B, and C sites for use in the ChIP assay. As shown in Fig. 4B, mutation of the A site (aBC) had little effect on EspR binding. In contrast, mutation of the B (AbC) or C (ABc) sites inhibited EspR binding, whereas mutation of both sites in combination (Abc) appeared to inhibit binding even more strongly. These data suggest that the B and C sites are required for EspR interaction with the espACD upstream intergenic region in vivo. Furthermore, a 364-bp region centered on the B and C sites (corresponding to probe BC364) was sufficient for complete binding in the ChIP assay (Fig. S2). It is notable that these sites are located extremely far from the promoter, suggesting that any regulatory effects of EspR on RNA polymerase would require constraints on DNA topology.
To begin to decipher the mode of EspR binding to its operators, we performed a series of EMSAs focused on the B and C sites. Although the B site appears to make contact with EspR in vivo, a 48-bp probe containing this sequence failed to bind EspR, even at very high protein concentrations (Fig. 4C, B48 probe, 16 μM). Likewise, probes encompassing only the A or C site also failed to bind the regulator (Fig. S1B). Importantly, a larger probe that encompassed both the B and C sites (Fig. 4C, BC364 probe) bound EspR strongly (estimated Kd = 0.3 μM). Mutation of either the B site (Fig. 4C, bC364 probe) or the C site (Bc364 probe) significantly decreased EspR binding to the labeled probe, though some binding was detectable at higher protein concentrations. Interestingly, a probe containing mutations in both sites (Fig. 4C, bc364 probe) bound EspR similarly to the single mutant probes, indicating that the binding observed at higher protein concentrations does not reflect a site-specific interaction. Binding to these probes may be nonspecific, or it may reflect additional protein–DNA contacts that we do not yet understand. However, these data, combined with the ChIP results, strongly suggest that direct binding of EspR requires both the B and C sites.
The cooperativity between the B and C sites for EspR binding suggests that EspR contacts both sites with a concomitant loop of the intervening sequence. Specifically, because each site is nonpalindromic and cannot bind EspR alone, it indicates that each are equivalent to a half-site with a spacer of 177 bp. In this scenario, cooperative binding would require EspR dimerization as well as having the two operator sites located on a continuous piece of DNA. As shown in Fig. 4D, the monomeric form of EspR (amino acids 1–107) failed to bind the BC364 probe, consistent with dimerization of EspR being required for high-affinity binding. To determine whether EspR binding requires the B and C sites to be on the same molecule of DNA, we performed competition experiments in which unlabeled DNA fragments were added to the binding reaction with labeled BC364 probe. As shown in Fig. 4E, excess unlabeled BC364 DNA, containing both the B and C sites in cis, was able to compete well for EspR binding. However, providing the B and C sites in trans by splitting the competitive DNA fragment at a position between the B and C sites, failed to compete for EspR binding. A similar result was found using the bc364 fragment that contains mutations in both operator sites. Therefore, high-affinity binding of EspR requires both HTH domains to be tethered via the carboxyl-terminal dimerization domain and both DNA recognition sites to be present on the same molecule of DNA.
Discussion
The HTH domain is perhaps the most common DNA-binding motif found in prokaryotes (17). The numerous structures of different repressor–operator complexes show that most HTH proteins adopt approximately similar binding geometries with respect to DNA, with the HTH units lying at similar angles to the major groove (18). In particular, although the base/phosphate contacts differ in each case, the common features of the 434-, lambda-, and cro-repressor complexes discussed here suggest that binding of two closely placed half-sites of DNA sequence represents an important commonality in the way HTH elements are anchored to DNA. EspR thus represents a radical departure from this classic paradigm.
The details of the EspR operators are also unique. The repressors noted above have small operator sites consisting of palindromic half-sites separated by only a few bases. For example, the 434 repressor binds to a 14-bp operator consisting of two 4-mer palindromes separated by 6 bp (19). Our work has shown that the two operator sites contacted by EspR are separated by 177 bp. Because these sites are nonpalindromic and both must be on the same piece of DNA to permit binding, we favor a model in which these sites are equivalent to the half-sites of a single operator normally bound by HTH dimers, with the notable difference being the large “spacer” in between these sites. In this model, one EspR monomer binds to the B site, while the other binds the C site, allowing for cooperative, high-affinity binding of the dimer with concomitant looping of the intervening DNA. This model is consistent with the oblique orientation of the HTH domains in the EspR structure, and the flexibility of the DNA would allow both half-sites to be occupied simultaneously by a single EspR dimer (20). Although this model is most parsimonious with our data, other possibilities certainly exist. For example, it is also possible that each site is bound by an EspR dimer, and that dimer–dimer contacts provide for cooperative binding between the two sites, though we have not detected this in our biophysical and crystallographic studies. We also cannot rule out more complicated scenarios, such as sequential binding of the individual sites and conformational changes upon EspR binding, as well as the contribution of nonspecific contacts of the protein with DNA.
Looping of promoter DNA is a common theme in transcription regulation, with the lactose repressor, LacR, of E. coli being a classic example (21). In this case, the repressor mediates DNA looping via protein–protein contacts between two LacR dimers, creating a bivalent tetramer that contacts two palindromic operators simultaneously (22, 23). Our data support the idea that EspR, like LacR, exerts its effect on transcription by constraining the topology of promoter DNA by binding two sites separated along the DNA. However, the EspR structure and the nature of its operators suggest that the separation of half-sites contacted by the bivalent EspR dimer presents a unique solution to this problem.
Interestingly, another distinctive aspect of genetic regulation by EspR is that it binds an extremely long distance away from the espACD transcriptional start site. Three separate regulators, including EspR, PhoP, and cAMP receptor protein, have been implicated in directly controlling transcription of this operon (3, 24–26). It may be that coordination of multiple regulatory inputs centralized at the large espACD promoter is required to achieve precise control of ESX-1 transcription during infection. Thus, the unique aspects of EspR may allow it to alter transcription by mediating long-distance interactions between other transcriptional regulators and RNA polymerase.
Materials and Methods
Details of bacterial strains, plasmids, oligonucleotides, and ChIP methods are provided in SI Materials and Methods.
Expression, Purification and Crystallization of M. tuberculosis EspR.
Full-length espR was amplified by PCR from M. tuberculosis genomic DNA and inserted via ligation-independent cloning (LIC) into the pLIC-HMK vector to encode the proteins with an amino-terminal 6X-His tag followed by MBP. The protein was induced in 1 mM IPTG at 37 °C in E. coli and cells were lysed by sonication; the cleared lysate was batch bound to Talon resin (Clontech) and eluted with imidazole. After cleavage overnight with tobacco etch virus protease and dialysis into a low ionic strength buffer [20 mM Hepes (pH 6.9), 50 mM NaCl], the protein was further purified via cation exchange chromatography (Bio-Rex 70, Bio-Rad). Peak elution fractions were pooled and concentrated to 30 mg/mL. The best crystals emerged overnight in 35% (vol/vol) 5/4 PO/OH (Hampton Research), 50 mM Hepes (pH 6.9), 200 mM NaCl. This condition was determined to be cryoprotective. Selenomethionine-derived EspR was generated according to the protocol described in ref. 27 and purified equivalently to the native protein, with the exception that 5 mM β-mercaptoethanol was included prior to elution from Talon resin and 10 mM DTT was included in subsequent steps. EMSA experiments were conducted with proteins purified as described above for native EspR, except that the pH3C expression vector was used and 3C protease was used to cleave the 6X-His-MBP tag.
Estimation of Molecular Mass with Light Scattering.
Molecular weight was estimated with size-exclusion chromatography (SEC) through a Superdex 200 10/300 GL analytical column inline with a Viscotek Model 302-050 Tetra Detector Array (Viscotek Ltd., a Malvern Company). SEC experiments were performed at 4 °C in 20 mM Tris (pH 7.4), 100 mM NaCl, and SEC-purified ovalbumin (2 mg/mL, various volumes) was used to calibrate internal instrument constants. All data were acquired and processed using Omnisec software. Protein concentration was determined using a UV spectrophotometer. Right-angle light scattering data, coupled to concentration measured by both refractory index and A280, were used to determine the molecular mass of the eluting protein using the Zimm equation.
Crystal Diffraction and Structure Determination.
Diffraction data for the native and selenomethionine-derived proteins were collected at Advanced Light Source Beamline 8.3.1 (Berkeley, CA) with the assistance of the Elves program suite (28). However, only crystals of the selenomethionine-derived protein were used in the final structure determination because they diffracted to significantly higher resolution than the native EspR crystals. Merging of raw data was carried out with Mosflm (29) and further scaling and processing were done using programs in the CCP4 suite (30). A partial solution was determined in space group P3212 by multiwavelength anomalous dispersion methods using data collected at two wavelengths from a single crystal of selenomethionine substituted EspR. Selenomethionine sites were found by using ShelxD and ShelxE (31) and the initial phases were calculated by using SOLVE (32). The maps were solvent flattened using the program RESOLVE (33) and monomers of an EspR sequence-replaced model (residues 1–80) based on SinR (Protein Data Bank ID code 1BN0) generated by the Phyre server (34) were placed into the initial experimental map and built further with COOT (35). The data were in the point group 32 but one of the dimers in asymmetric unit breaks the twofold symmetry preventing refinement in P3212. We subsequently processed data from a second crystal of selenomethionyl EspR in space group P32. The Rsym for this dataset was somewhat higher than average because of low average I/(σ(I), but still within an acceptable range. All 18 expected monomers were located in the P32 asymmetric unit using Phaser for molecular replacement with a dimer of EspR as a search model (36). The crystal was found to exhibit merohedral twinning and thus least-squares, twinned refinement was carried out using Phenix (36) and the twin operator (-k,-h,-l) and a refined twin fraction of (0.5) (37, 38). Noncrystallographic symmetry was applied according to the default settings in Phenix (36). The final Rfree was 29% (untwinned) and 23% (twinned) (Table S1). Figures were generated using the program PyMOL v. 1.3 (Schrödinger, LLC.)
EspR ChIP.
The full-length espACD upstream intergenic region (1,357 bp, Fig. 4A) was inserted into pMV306.Kan (integrating), and mutations in the EspR binding sequence NAGCAAAN were generated using overlap extension PCR to replace it with the scrambled sequence ATAACCGA. Plasmids were transformed into M. smegmatis expressing M. tuberculosis EspR with a carboxyl-terminal 3X-FLAG tag (Table S2).
ChIP was performed essentially as described previously (39) with several modifications (see SI Materials and Methods). Briefly, 50-mL log-phase M. smegmatis cultures were fixed in 4% (wt/vol) paraformaldehyde. Cell pellets were lysed by sonication, insoluble debris was removed via centrifugation, and extracts were further sonicated to shear DNA fragments to a mean size of 200 bp. Immunoprecipitations were performed using monoclonal anti-FLAG M2 antibody (Sigma) and magnetic Dynabeads (Invitrogen).
Quantitative RT-PCR was performed using primer pairs (Table S3) that spanned the entire espACD upstream intergenic region, using Taq polymerase in the presence of SYBR Green on an Opticon Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.). Immunoprecipitation samples were used as a direct template for PCR, and reactions were quantified using standard curves generated for each primer pair with input material for that sample. Data were normalized to signal from a nonenriched region (16S rRNA) for each sample.
EMSA.
5-FAM-conjugated oligonucleotides (Bioneer, Inc.) were used in PCR reactions to create labeled BC364, B240, and C124 DNA probes. The B48 probe was generated by annealing oligonucleotides oCD166 (5-FAM) and oCD167. Binding reactions were performed with purified protein and 5-nM probe for 30 min at room temperature in EMSA buffer [10 mM Tris (pH 8.0), 50 mM NaCl, 1 mM DTT, 100 μg/mL BSA, 1 mM EDTA, 5% (vol/vol) glycerol, and 30 μg/mL poly(deoxyinosinic-deoxycytidylic) acid (Sigma)]. Reactions were resolved at room temperature on a 4–20% Tris-glycine gel (Invitrogen) and visualized using a Typhoon Fluorescence Imager (GE Healthcare).
Supplementary Material
Acknowledgments.
We thank James Holton at Advanced Light Source for expert technical assistance, and Aaron D. Hernday, Bennett H. Penn, and Robert O. Watson for valuable advice. We thank Alexander Johnson, Paul Sullam, and members of the Cox and Stroud laboratories for helpful discussions, and Tom Alber and Scott Gradia for initial EspR crystallization screens. This work was supported by a National Institutes of Health (NIH) Training Grant T32 AI007641 (to O.S.R.), and by NIH Grants AI081727 and AI051667 (to J.S.C.). J.S.F.-M., R.A.R., and R.M.S. are supported by GM051232.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3R1F).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110242108/-/DCSupplemental.
References
- 1.Dye C, Williams BG. The population dynamics and control of tuberculosis. Science. 2010;328:856–861. doi: 10.1126/science.1185449. [DOI] [PubMed] [Google Scholar]
- 2.Simeone R, Bottai D, Brosch R. ESX/type VII secretion systems and their role in host-pathogen interaction. Curr Opin Microbiol. 2009;12:4–10. doi: 10.1016/j.mib.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Raghavan S, Manzanillo P, Chan K, Dovey C, Cox JS. Secreted transcription factor controls Mycobacterium tuberculosis virulence. Nature. 2008;454:717–721. doi: 10.1038/nature07219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fortune SM, et al. Mutually dependent secretion of proteins required for mycobacterial virulence. Proc Natl Acad Sci USA. 2005;102:10676–10681. doi: 10.1073/pnas.0504922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacGurn JA, Cox JS. A genetic screen for Mycobacterium tuberculosis mutants defective for phagosome maturation arrest identifies components of the ESX-1 secretion system. Infect Immun. 2007;75:2668–2678. doi: 10.1128/IAI.01872-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gangwar SP, Meena SR, Saxena AK. Cloning, purification, crystallization and preliminary X-ray analysis of ESX-1-secreted protein regulator (EspR) from Mycobacterium tuberculosis. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;67:83–86. doi: 10.1107/S174430911004618X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis RJ, Brannigan JA, Offen WA, Smith I, Wilkinson AJ. An evolutionary link between sporulation and prophage induction in the structure of a repressor:anti-repressor complex. J Mol Biol. 1998;283:907–912. doi: 10.1006/jmbi.1998.2163. [DOI] [PubMed] [Google Scholar]
- 9.Beamer LJ, Pabo CO. Refined 1.8 A crystal structure of the lambda repressor-operator complex. J Mol Biol. 1992;227:177–196. doi: 10.1016/0022-2836(92)90690-l. [DOI] [PubMed] [Google Scholar]
- 10.Rodgers DW, Harrison SC. The complex between phage 434 repressor DNA-binding domain and operator site OR3: Structural differences between consensus and non-consensus half-sites. Structure. 1993;1:227–240. doi: 10.1016/0969-2126(93)90012-6. [DOI] [PubMed] [Google Scholar]
- 11.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 12.Saff EB, Kuijlaars ABJ. Distributing many points on a sphere. Math Intell. 1997;19:5–11. [Google Scholar]
- 13.Harrison SC, Aggarwal AK. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- 14.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albright RA, Matthews BW. Crystal structure of lambda-Cro bound to a consensus operator at 3.0 A resolution. J Mol Biol. 1998;280:137–151. doi: 10.1006/jmbi.1998.1848. [DOI] [PubMed] [Google Scholar]
- 16.Flocco MM, Mowbray SL. Planar stacking interactions of arginine and aromatic side-chains in proteins. J Mol Biol. 1994;235:709–717. doi: 10.1006/jmbi.1994.1022. [DOI] [PubMed] [Google Scholar]
- 17.Huffman JL, Brennan RG. Prokaryotic transcription regulators: More than just the helix-turn-helix motif. Curr Opin Struct Biol. 2002;12:98–106. doi: 10.1016/s0959-440x(02)00295-6. [DOI] [PubMed] [Google Scholar]
- 18.Brennan RG, Matthews BW. Structural basis of DNA-protein recognition. Trends Biochem Sci. 1989;14:286–290. doi: 10.1016/0968-0004(89)90066-2. [DOI] [PubMed] [Google Scholar]
- 19.Koudelka GB, Harrison SC, Ptashne M. Effect of non-contacted bases on the affinity of 434 operator for 434 repressor and Cro. Nature. 1987;326:886–888. doi: 10.1038/326886a0. [DOI] [PubMed] [Google Scholar]
- 20.Swigon D, Coleman BD, Olson WK. Modeling the Lac repressor-operator assembly: The influence of DNA looping on Lac repressor conformation. Proc Natl Acad Sci USA. 2006;103:9879–9884. doi: 10.1073/pnas.0603557103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Echols H. Nucleoprotein structures initiating DNA replication, transcription, and site-specific recombination. J Biol Chem. 1990;265:14697–14700. [PubMed] [Google Scholar]
- 22.Kramer H, et al. Lac repressor forms loops with linear DNA carrying two suitably spaced lac operators. EMBO J. 1987;6:1481–1491. doi: 10.1002/j.1460-2075.1987.tb02390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedman AM, Fischmann TO, Steitz TA. Crystal structure of lac repressor core tetramer and its implications for DNA looping. Science. 1995;268:1721–1727. doi: 10.1126/science.7792597. [DOI] [PubMed] [Google Scholar]
- 24.Frigui W, et al. Control of M. tuberculosis ESAT-6 secretion and specific T cell recognition by PhoP. PLoS Pathog. 2008;4:e33. doi: 10.1371/journal.ppat.0040033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JS, et al. Mutation in the transcriptional regulator PhoP contributes to avirulence of Mycobacterium tuberculosis H37Ra strain. Cell Host Microbe. 2008;3:97–103. doi: 10.1016/j.chom.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Rickman L, et al. A member of the cAMP receptor protein family of transcription regulators in Mycobacterium tuberculosis is required for virulence in mice and controls transcription of the rpfA gene coding for a resuscitation promoting factor. Mol Microbiol. 2005;56:1274–1286. doi: 10.1111/j.1365-2958.2005.04609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 28.Holton J, Alber T. Automated protein crystal structure determination using ELVES. Proc Natl Acad Sci USA. 2004;101:1537–1542. doi: 10.1073/pnas.0306241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leslie AGW, Brick P, Wonacott A. Information Quarterly for Protein Crystallography. Vol. 18. Warrington, UK: Daresbury Laboratory; 1986. pp. 33–39. [Google Scholar]
- 30.Anonymous The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 31.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2007;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 32.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaw G, et al. Structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription. Structure. 2008;16:1417–1427. doi: 10.1016/j.str.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeates TO. Detecting and overcoming crystal twinning. Methods Enzymol. 1997;276:344–258. TO () . 276:344-358. [PubMed] [Google Scholar]
- 39.Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 2007;5:e256. doi: 10.1371/journal.pbio.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.