Abstract
UV light induces DNA lesions, which are removed by nucleotide excision repair (NER). Exonuclease 1 (EXO1) is highly conserved from yeast to human and is implicated in numerous DNA metabolic pathways, including repair, recombination, replication, and telomere maintenance. Here we show that hEXO1 is involved in the cellular response to UV irradiation in human cells. After local UV irradiation, fluorescent-tagged hEXO1 localizes, together with NER factors, at the sites of damage in nonreplicating cells. hEXO1 accumulation requires XPF-dependent processing of UV-induced lesions and is enhanced by inhibition of DNA repair synthesis. In nonreplicating cells, depletion of hEXO1 reduces unscheduled DNA synthesis after UV irradiation, prevents ubiquitylation of histone H2A, and impairs activation of the checkpoint signal transduction cascade in response to UV damage. These findings reveal a key role for hEXO1 in the UV-induced DNA damage response linking NER to checkpoint activation in human cells.
Keywords: local UV damage, UV lesion processing, ssDNA, ubiquitin
UV light generates cyclobutane pyrimidine dimers and (6-4) photoproducts in DNA, which are responsible for the pathogenic effects of sunlight. UV-induced photolesions are repaired by nucleotide excision repair (NER), and mutations in genes coding for NER factors are responsible for inherited disorders, such as xeroderma pigmentosum (XP), Cockayne syndrome, and trichothiodystrophy (1). All patients with NER syndrome are photosensitive, but other clinical features vary. This is most likely a consequence of the mutated NER factors involvement not only in the repair of UV lesions, but also in the control of transcription. A specific NER defect can give rise to XP, whereas a transcription defect may cause Cockayne syndrome or trichothiodystrophy (1).
In response to UV irradiation and other genotoxic treatments, eukaryotic cells activate a surveillance system known as the DNA damage checkpoint, which is involved in cancer protection (2). The mechanism underlying the checkpoint response is a phosphorylation-based signal transduction cascade conserved in all eukaryotes, involving the integrated action of ataxia telangiectasia mutated (ATM) and ataxia telangiectasia- and Rad3- related (ATR) PI3-kinases. In noncycling human cells, ATR acts as the apical kinase in the pathway, leading to checkpoint activation in response to UV irradiation, and ATR activation is responsible for the direct or indirect phosphorylation of several substrates (3). Moreover, it was recently established that H2A is ubiquitylated (uH2A) by the RNF8 E3 ubiquitin ligase in response to UV treatment (4), analogous to what has been found after exposure to ionizing radiation (5). Accumulation of uH2A at sites of local UV-induced DNA damage (LUD) was found to be required for MDC1 and 53BP1 recruitment (4). These findings indicate that although UV lesions and double-strand breaks (DSBs) are processed by different repair pathways, they eventually generate the same epigenetic mark.
We and others have shown that in yeast cells and in resting human fibroblasts, activation of the checkpoint induced by UV irradiation requires a functional NER apparatus (4, 6–9). It is generally assumed that single-stranded (ss) DNA regions represent a common structure required for triggering the DNA damage checkpoint, as the result of damage processing (10, 11). However, NER-mediated excision of UV photoproducts generates short ssDNA gaps (∼30 nt long), which might never be exposed, given that cleavage 3′ to the UV lesion seems to occur only once the refilling of ssDNA gaps by repair synthesis is well under way (12). According to this scenario, it is difficult to predict the presence of long-lived ssDNA regions during NER.
In Saccharomyces cerevisiae cells, Exo1 processes was found to stall NER intermediates, generating longer ssDNA gaps detectable by electron microscopy, the refilling of which by repair synthesis in the presence of BrdU can be monitored by DNA combing (13). Moreover, such processing of NER intermediates is necessary for the recruitment of Mec1, the yeast ortholog of ATR, and for the activation of the DNA damage checkpoint in response to UV irradiation (13).
hEXO1, a member of the Rad2 family of structure-specific nucleases, has 5′-3′ exonuclease and 5′-flap endonuclease activities in vitro (14, 15). Two isoforms of the hEXO1 gene, hEXO1a and hEXO1b, differing in a C-terminal extension of 43 amino acids in hEXO1b, have been described; however, no functional difference between the two products has been found (16). Several studies in yeast and multicellular eukaryotes have implicated this enzyme in various DNA metabolic processes, including meiotic and mitotic recombination, DNA repair, telomere maintenance, mismatch repair, and others (reviewed in ref. 17). The importance of EXO1 is underscored by the finding that Exo1−/− KO mice have impaired DNA damage signaling in response to telomere dysfunction and increased cancer susceptibility (18, 19).
Our findings regarding the role of Exo1 in yeast led us to investigate whether its function in processing NER intermediates was conserved in human cells. Here we show that endogenous hEXO1 interacts with XPA. hEXO1 is recruited at LUDs in a NER-dependent manner, and this accumulation increases when completion of repair synthesis is inhibited. In UV-irradiated nonreplicating human primary fibroblasts, depletion of hEXO1 causes a noticeable reduction in the level of unscheduled DNA synthesis (UDS), impairs H2A ubiquitylation, and affects the checkpoint-signaling cascade. These findings support a general model in which stalling of repair synthesis at particular chromosomal sites allows hEXO1 to process the repair intermediates, generating ssDNA regions that trigger the checkpoint response.
Results
hEXO1a and hEXO1b Accumulate at LUDs and Interact with the NER Preincision Protein XPA.
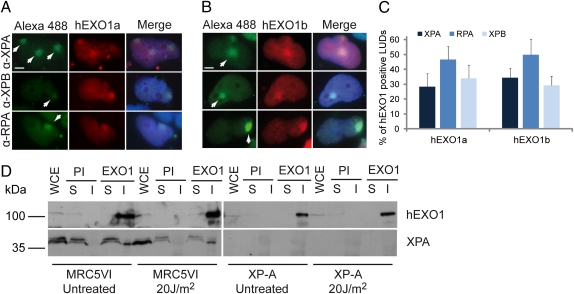
hEXO1 has been implicated in several DNA transactions (16, 17), but its possible role in processing NER intermediates, generating the signal for checkpoint activation after UV irradiation, has not yet been investigated. To analyze the cellular localization of hEXO1 in response to UV treatment, we tagged both hEXO1a and hEXO1b isoforms with mCherry in MRC5VI-transformed human fibroblasts. We found that in S-phase cells (∼15–20% of the cell population), both hEXO1 isoforms accumulate in proliferating cell nuclear antigen-positive foci, as reported previously (20). Application of UV irradiation of confined areas of individual cell nuclei using filters containing pores of a defined size has demonstrated that protein factors involved in the repair of UV lesions localize at LUDs (21). We found that after UV irradiation of cells through 5-μm Isopore filters (Millipore), both hEXO1a and hEXO1b accumulated similarly at LUDs, colocalizing with NER preincision factors (Fig. 1 A and B). Quantification of LUDs in which hEXO1a or hEXO1b colocalized with other NER factors revealed that 30–35% of XPA- and XPB-positive LUDs and 45–50% of RPA-positive LUDs also contained hEXO1 (Fig. 1C). This variation can be related to the reportedly different kinetics of XPA, XPB, and RPA recruitment at UV lesions (21, 22). Cells traversing S-phase and non–S-phase cells can be distinguished after local UV irradiation by staining for ethynyl deoxyuridine (EdU) incorporation. EdU staining is bright and homogenously distributed in the nucleus in S-phase cells, and is less bright and localized at LUDs in non–S-phase cells (Fig. S1). Interestingly, the accumulation of hEXO1at LUDs seems to be restricted to non–S-phase cells; no hEXO1 accumulation at LUDs was detected in 100 S-phase nuclei analyzed (Fig. S1).
Fig. 1.
Accumulation of hEXO1a and hEXO1b at LUDs. (A and B) MRC5VI cells were transfected with hEXO1a-mCherry (A) or hEXO1b-mCherry (B), seeded on coverslips, and locally UV irradiated (40 J/m2) through Isopore filters. After an additional 1-h incubation, cells were fixed and processed for indirect immunofluorescence using Abs against XPA, XPB, and RPA (Alexa Fluor 488), and nuclei were counterstained with DAPI. White arrows indicate LUDs. (Scale bar: 5 μm.) (C) Quantification of immunofluorescence shown in A and B. Histograms represent the mean ± SD of three independent experiments. Approximately 40 LUDs were scored for each experiment. (D) MRC5VI cells were mock- or UV-treated and harvested; lysates were prepared and resolved directly on SDS/PAGE (WCE, whole cell extracts, 10%) and incubated with preimmune serum (PI) as a control or anti-hEXO1 Ab (F15). Western blot analysis was performed with hEXO1 Ab (Ab4) and XPA Ab (12F5). XPA runs as a doublet because of incomplete reduction (32). S, immunoprecipitation flow-through; I, immunoprecipitate.
To gain some insight into the mechanism(s) leading to hEXO1 recruitment at LUDs, we tested whether hEXO1 physically interacts with known NER factors. hEXO1 is expressed at low levels in human nonproliferating tissues (23–26), and the endogenous protein is barely detectable in total crude extracts from cell lines because of its low abundance and limited affinity to the available Abs (23). However, hEXO1 can be recovered and analyzed by Western blot analysis after immunoprecipitation.
As shown in Fig. 1D, XPA, a protein that acts at an early stage of NER (27), coimmunoprecipitates with hEXO1 in the presence or absence of UV treatment. Attempts to detect interaction of hEXO1 with other NER factors by coimmunoprecipitation have so far been unsuccessful (Fig. S2).
hEXO1b Recruitment at LUDs Depends on Assembly of the NER Preincision Complex and Requires the 5′, but Not the 3′, Incision of UV-Damaged DNA.
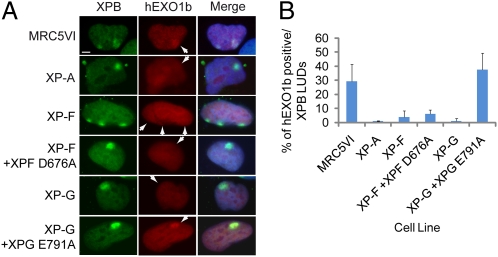
Because hEXO1 accumulates at damage sites together with NER factors, we tested the dependency of hEXO1 recruitment at LUDs on NER functions, by analyzing fibroblasts derived from patients with XP. MRC5VI control cells and XPA, XPF, XPF+XPFD676A (corrected with the catalytically dead endonuclease), XPG, and XPG+XPGE791A (corrected with the catalytically dead endonuclease) were transiently transfected with a construct expressing mCherry-tagged hEXO1b and then locally UV-irradiated. Fig. 2A shows that XPB localizes at LUDs in control and XP-mutated cells. Interestingly, hEXO1b is not recruited at LUDs in the absence of the 5′ incision to the lesion (in the XPA, XPF, XPF+XPFD676A, and XPG cell lines). hEXO1b recruitment at LUDs was restored in the presence of the 5′ incision, even though the 3′ cut could not be executed (XPG+XPGE791A) (12). Quantification of the percentage of XPB-positive LUDs that also contain hEXO1b revealed slightly higher recruitment of hEXO1b in XPG cells expressing the catalytically dead nuclease (XPGE791A) than in MRC5VI control cells, with either dramatically reduced or totally abrogated recruitment of hEXO1b in other XP-mutated cell lines.
Fig. 2.
NER preincision complex and 5′ incision are required for hEXO1b accumulation at LUDs. (A) MRC5VI, XP12RO (XPA), XP2YO (XPF), XP2YO (XPF+XPF D676A), XPCS1RO (XPG), and XPCS1RO (XPG+XPG E791A) cells were transfected with hEXO1b-mCherry, seeded on coverslips, and locally UV-irradiated (40 J/m2) through Isopore filters. After 1 h of incubation, cells were fixed and processed for immunofluorescence against the indicated protein and nuclei were counterstained with DAPI. White arrows indicate the position of LUDs. (Scale bar: 5 μm.) (B) Quantification of immunofluorescence shown in A. Histograms show the mean ± SD of three independent experiments. Approximately 40 LUDs were scored for each experiment.
In conclusion, hEXO1b recruitment at the site of damage requires the NER preincision complex and at least the 5′ incision reaction.
Inhibition of Repair Synthesis Enhances hEXO1 Accumulation at LUDs.
We found that in yeast Exo1 can enlarge NER gaps at a subset of UV-induced lesions when the gap-refilling reaction is somehow slower or impeded. The large ssDNA gaps generated by Exo1 lead to checkpoint activation and are subsequently refilled by repair DNA synthesis (13). If hEXO1 performs a similar function in human cells, then we would expect to see greater recruitment of hEXO1 at LUDs in the presence of 1-β-d-arabinofuranosylcytosine (Ara-C), a DNA synthesis inhibitor. hEXO1 reportedly undergoes degradation after inhibition of replication through hydroxyurea treatment (23), but this did not occur after Ara-C incubation (Fig. S3). Cells locally UV-irradiated and then incubated with Ara-C showed increased accumulation of hEXO1b at XPB-positive LUDs, from 30% to 60% in MRC5VI cells. Such recruitment was lost in XPA cells and was unaffected by Ara-C treatment (Fig. 3). Similar results were obtained for hEXO1a-mCherry (Fig. S4).
Fig. 3.
Accumulation of hEXO1b at LUDs is enhanced when repair synthesis is blocked. MRC5VI and XP12RO (XPA) cells transfected with hEXO1b-mCherry were exposed to local UV irradiation (40 J/m2) and incubated for 1 h at 37 °C in the presence or absence of Ara-C. (A) Histograms indicating the percentage of XPB-positive LUDs that also contained hEXO1b-mCherry, with or without Ara-C treatment. Approximately 40 LUDs were scored in each of three independent experiments; values are mean ± SD. (B) Representative images of cells with hEXO1b-positive LUDs (with or without Ara-C treatment) in the MRC5VI control (Upper) and in the XPA mutated cell lines (Lower). LUDs are indicated by white arrows. (Scale bar: 5 μm.)
Depletion of hEXO1 Reduces UDS and Impairs Checkpoint Signaling.
NER is a multistep process requiring the refilling of ssDNA gaps generated by the processing of the DNA lesions. The UDS DNA resynthesis step requires lesion recognition and processing (27). NER-deficient cells, which are unable to process UV lesions, are UDS-defective. If hEXO1 were involved in extending the NER-induced gaps, similar to what occurs in yeast (13), then we would expect to find lower UDS levels in hEXO1-depleted cells.
A rapid nonradioactive technique for measuring UDS using the nucleoside analog EdU has been reported recently (28). We optimized hEXO1 silencing by using two different siRNA sequences. Human primary fibroblasts were transfected with siRNAs against hEXO1 as a negative control and against luciferase (LUC) and XPA as positive controls. Both XPA and hEXO1 were efficiently down-regulated (Fig. 4B). Mock- or UV-irradiated cells were then incubated with EdU for 3 h, after which EdU incorporation was quantified in nonreplicating cells, which are easily distinguished from S-phase cells (Fig. S5). Fig. 4A shows that EdU incorporation increased 3.6-fold above background after UV irradiation in control LUC-silenced cells, whereas in hEXO1-depleted cells UDS induction was reduced to 1.7-fold, approaching the reduction seen in siXPA cells (1.3-fold). Considering the EdU incorporation measured after UV irradiation in LUC-silenced cells to be 100%, depletion of hEXO1 causes a 52% reduction, and depletion of XPA causes a 65% reduction. These results suggest that hEXO1 activity is indeed involved in the formation of large ssDNA gaps after UV irradiation.
Fig. 4.
Down-regulation of hEXO1 impairs UDS after UV irradiation. (A) Primary 48BR cells were depleted of hEXO1, XPA, or control (LUC); cultured on coverslips; and UVC-irradiated (20 J/m2), followed by incubation with 10 μM EdU for 3 h, fixation, and conjugation of the fluorescent dye to incorporated EdU. The intensity of nuclear fluorescence, which is associated with UDS activity, was measured using a fluorescence microscope and image-processing software. For each sample, at least 100 nuclei were analyzed in three independent experiments; error bars represent SD. (B) Silencing efficiency was monitored by Western blot analysis for XPA (Upper) and by RT-PCR for hEXO1 (Lower).
In response to both DSBs and UV irradiation, RNF8 ubiquitylates histone H2A, and this modification is required for recruiting downstream factors in the damage response (4, 5). Strikingly, when hEXO1 was silenced in quiescent primary fibroblasts and cells were locally UV-irradiated, XPA-positive LUDs were clearly detectable, but H2A ubiquitylation (uH2A) at damage sites was strongly reduced (Fig. 5A). Quantification of various experiments indicates that when hEXO1 was down-regulated (Fig. 5C), uH2A was reduced to ∼40% compared with control (Fig. 5B).
Fig. 5.
hEXO1 modulates a crucial step linking UV-induced lesions to histone H2A ubiquitylation. (A) Quiescent 48BR cells were transfected with siRNA directed against luciferase or hEXO1 (two different sequences, sihEXO1-1 and sihEXO1-2, were used) and locally UV-irradiated. At 1 h after UV irradiation, cells were processed to detect uH2A and XPA by immunofluorescence. Images show one representative nucleus for each treatment. (Scale bar: 5 μm.) (B) Quantification of the percentage (mean ± SD) of XPA-positive LUDs that are also positive for uH2A in siLUC, sihEXO1-1, and sihEXO1-2. (C) Silencing efficiency was monitored by RT-PCR.
Formation of ssDNA regions by DNA damage processing activates a signal transduction cascade (10, 11). Previous work demonstrated that recognition and processing of UV-induced lesions by NER factors is required for checkpoint activation in nondividing yeast and human cells (6, 7). Asynchronously growing XPA primary fibroblasts exhibit checkpoint activation after UV irradiation, as detected by Chk1 Ser317 phosphorylation; however, this effect is limited to cells traversing S phase, which do not require functional NER to trigger the checkpoint response (7). Yeast Exo1 also has been found to be required for an efficient and rapid NER-dependent G1 checkpoint response (13). In quiescent human primary fibroblasts, knockdown of hEXO1 expression by siRNA causes a defect in checkpoint activation after UV irradiation. In fact, ATR-dependent phosphorylation of both Chk1-Ser317 and p53-Ser15 was reduced to a level similar to that found in XPA fibroblasts or in cells in which XPA expression was knocked down by siRNA (Fig. 6A) up to 120 min after UV irradiation (Fig. 6B).
Fig. 6.
hEXO1 is required for checkpoint activation after UV irradiation in quiescent human primary fibroblasts. Quiescent human primary fibroblasts transfected with siRNA against hEXO1 (sihEXO1-1 was used) or XPA were held under low serum conditions for 3 d before mock or UV irradiation. (A) Activation of the DNA damage checkpoint was examined at 1 h after UV irradiation by Western blot analysis using anti-P-Ser-317-Chk1. (B) Cells were UV-irradiated (20 J/m2) and harvested at different time points (1 or 2 h) after treatment. The DNA damage checkpoint was monitored by Western blot analysis using anti–P-Ser-15-p53 Ab. (C) hEXO1 knockdown was analyzed by RT-PCR.
Discussion
A long-standing question in the DNA damage response field is how DNA damage signaling initiated by apical kinases is activated in response to various genotoxic agents (11). It is interesting that although DSBs and UV-induced photoproducts are processed by different DNA repair pathways and trigger signaling responses controlled by distinct apical kinases (ATM/ATR in human and Tel1/Mec1 in yeast cells), they eventually generate the same epigenetic mark involving H2A ubiquitylation (4). ssDNA regions covered by RPA are considered the structures that activate the checkpoint in response to UV irradiation (11). In cycling cells, UV-induced DNA lesions are likely sensed by the DNA replication machinery, whereas in nondividing cells, recognition and processing of UV photoproducts by NER are required to activate the checkpoint (6, 7). However, how the short gaps (∼30 nt long) generated by NER (29) mediate activation of damage signaling is unclear.
In S. cerevisiae cells, Exo1-mediated processing of a subset of NER intermediates is required for rapid checkpoint response and generates extended ssDNA gaps (11). Here we show that in human cells, both isoforms of hEXO1 localize at the sites of UV-induced DNA lesions, and recruitment of the nuclease is dependent on NER functions. Furthermore, endogenous hEXO1 physically interacts with XPA, whereas no interactions of hEXO1 with other NER factors were detected by coimmunoprecipitation analysis; such interactions may be transient and occur with different kinetics, making the analysis quite difficult.
Down-regulation of human hEXO1 impairs UDS, strongly supporting the idea that hEXO1 activity generates large ssDNA gaps, which are later refilled by repair DNA synthesis. We also found that H2A ubiquitylation is strongly reduced when hEXO1 is silenced, indicating that processing of the UV-induced lesions by the nuclease is required to generate this epigenetic modification. H2A ubiquitylation causes chromatin restructuring and creates docking sites for other DNA damage response factors (4). Down-regulation of hEXO1 also strongly reduces phosphorylation of the downstream checkpoint factors Chk1 and p53. Taken together, these data indicate that, as found in yeast, in human cells hEXO1-dependent processing of at least a subset of UV-dependent NER intermediates is an essential step in the formation of the ssDNA structure that triggers the signaling pathway activated by UV-induced lesions in noncycling human cells. This hEXO1 function in the response to UV irradiation must be added to the long list of DNA transactions in which hEXO1 has been implicated (reviewed in ref. 17).
The available data suggest that hEXO1 processing is triggered when the DNA refilling reaction is somehow impaired; indeed, hEXO1 accumulation at LUDs increases when the gap refilling reaction is inhibited by Ara-C treatment. Moreover, the UDS defect in hEXO1-depleted cells is milder than that seen in XPA siRNA-depleted fibroblasts, consistent with the hypothesis that only a subset of UV-induced DNA lesions are processed by hEXO1 in an NER-dependent manner. Finally, we found that for the accumulation of hEXO1 at LUDs, the 5′ incision by XPF is sufficient, even in cases where the 3′ incision by XPG is not executed and a flap structure is generated. Formation of a similar structure is predicted if repair synthesis starts but cannot be completed (12). In agreement with previous in vitro studies (14), we propose that hEXO1 can process in vivo a 5′ flap DNA structure that may arise during the processing of some UV-induced lesions.
In human cells, the DNA refilling reaction during NER is complex and involves different DNA polymerases (30, 31). It is possible to speculate that this unexpected complexity might be linked to the chromatin structure around the UV-induced damage, including the number, location, and reciprocal positioning of lesions on the two DNA strands. Gaining a better understanding of these critical issues is a challenge for future studies.
Materials and Methods
Abs and Chemicals.
The following Abs were used: anti–P-Ser-317-Chk1, anti–P-Ser-15-p53, and RPA70 (Cell Signaling Technology); anti-Chk1 and anti-actin Abs (Santa Cruz Biotechnology); anti-p53 (DO1; GeneSpin); anti-hEXO1 mAb (Ab4; Neomarkers); anti-uH2A (Millipore), rabbit anti-XPA (Sigma-Aldrich); and mouse anti-XPA (12F5; from R. D. Wood, Cancer Center, Smithville, TX), anti-XPB (1B3; from J.-M. Egly, University of Strasbourg), and anti-hEXO1 (F15; from S. Ferrari, University of Zurich). Ara-C (Sigma-Aldrich) was used at a final concentration of 1 mM for 1 h at 37 °C. Secondary Abs were goat anti-mouse or goat anti-rabbit conjugated to HRP (Western blot) or to Alexa Fluor 488 or Alexa Fluor 594 (immunofluorescence).
Cell Culture and Serum Starvation.
MRC5VI, XP12RO, XP2YO, XP2YO+XPF D676A, XPCS1RO, and XPCS1RO+XPG E791A (all SV40-transformed) were cultured in DMEM containing 10% FCS and kept at 37 °C in a humidified atmosphere with 5% CO2 (12). The 48BR primary fibroblasts were cultured in DMEM containing 15% FCS. For serum starvation, cells were incubated on plates for 72 h with medium containing 0.5% FCS.
Plasmid and Constructs.
hEXO1a and hEXO1b were PCR-amplified from hEXO1b-omnitag [kindly provided by S. Ferrari (23)], and then cloned into pmCherry-N1 at BamHI/HindIII sites.
cDNA Transfection.
MRC5VI and XP12RO fibroblasts were transfected using Metafectene Pro (Biontex Labs) following the manufacturer's instructions. XP2YO, XP2YO+XPF D676A, XPCS1RO, and XPCS1RO+XPG E791A were transfected using Ca2PO4 (10 μg DNA each 1.5 × 106 cells) and analyzed at 24–48 h posttransfection.
siRNA Transfection.
Cells were seeded and transfected twice with 50 nM siRNA (MWG siLUC: CGU ACG CGG AAU ACU UCG ATT; sihEXO1-1: GCA CGU AAU UCA AGU GAU GTT; sihEXO1-2: CGU AAA UAG AAG AAU AAU UTT; siXPA: ACC UAG AAG AUG ACA UGU ATT) using HiperFect (Qiagen) in DMEM with or without serum depending on the experiment. Cells were harvested and analyzed at 72 h after the first transfection.
UV Irradiation.
The medium was removed, and cells were washed once with PBS and then irradiated with a UV lamp (254 nm) at a dose of 20 J/m2. The medium was then added back, and cells were returned to culture conditions for different times. LUD was obtained by irradiating cells through a 5-μm Isopore filter (Millipore) with a UV box (254-nm wavelength) at a rate of 0.6 J/m2/s and a final dose of 40 J/m2. The same protocol was used for EdU detection at LUDs.
Immunoprecipitation and Western Blot Analysis.
MRC5VI and XP12RO cells were either mock-transfected or transfected with hEXO1a-mCherry or hEXO1b-mCherry, cultured up to 80% confluence for 24 h, harvested before and after irradiation (20 J/m2 for 1 h) using PBS, and processed as described previously (23). A total of 1.5 mg of protein (as determined by the Bradford method), preimmune serum, and F15 anti-EXO1 Ab were used for the immunoprecipitations, as described previously (23). For total protein extract preparation, cells were lysed in 1% SDS sample buffer [62.5 mM Tris-HCl (pH 6.8), 2% wt/vol SDS, 10% glycerol, 50 mM DTT, 0.01% wt/vol bromophenol blue], sonicated for 10 s, and heated to 95 °C for 5 min. Equal amounts of proteins were analyzed by SDS/PAGE.
Immunofluorescence.
Cells were seeded on a coverslip, transfected, and irradiated as described above. Cells were washed once in PBS, fixed for 20 min with 2% PFA in PBS, and permeabilized for 5 min with ice-cold PBS containing 0.5% Triton X-100. Blocking was performed in 10% BSA in PBS for 30 min, followed by replacement with primary Ab diluted in PBS with 0.1% Tween 20 (PBST 0.1) for 2 h at room temperature. For uH2A staining, permeabilization was performed with PBS-0.2% Triton before fixation, and the Ab was incubated overnight at 4 °C. Coverslips were washed three times in PBST 0.1 for 10 min, after which secondary Ab diluted in PBST 0.1 was added. Nuclei were counterstained with DAPI. Cells were rinsed in PBST 0.1 three times for 10 min and mounted using ProLong Gold (Invitrogen). Images were obtained using a Leica DMRA2 Microscope (with Leica FW4000 software) with a 100× oil immersion objective (1.30 NA). The percentages of hEXO1-positive LUDs were scored, counting RPA, XPA, or XPB accumulation at local damage only in hEXO1-transfected cells. A value of 1 was assigned each time that hEXO1 showed accumulation at local damage and 0 when it did not. Approximately 40 LUDs were scored for each independent experiment.
UDS Measurements.
The procedure used was essentially as described previously (28). EdU was purchased from Invitrogen. The 48BR cells were transfected in 24-well plates and then cultured on coverslips. After UV irradiation, cells were incubated with serum-free DMEM supplemented with 10 μM EdU for 3 h. Cells were then washed with PBS, followed by a chase period of 15 min with thymidine. Fixation, permeabilization, and fluorescent labeling were performed using Click-iT (Invitrogen), according to the manufacturer's instructions. Cell images were captured with a Leica DMRA2 fluorescent microscope (40× objective, 0.75 NA) equipped with a Leica DC300F CCD camera. Captured images were processed and analyzed with Adobe Photoshop software. At least 100 non–S-phase cells were selected at random for each experiment.
Supplementary Material
Acknowledgments
We thank S. Ferrari for the kind gift of the hEXO1 Ab and hEXO1b plasmids, R. D. Wood for the XPA Abs, and J. M. Egly for the XPB Abs. We also thank O. Schaerer for providing XP2YO, XP2YO D676A, XPCS1RO, and XPCS1RO E791A cells and for inspiring discussions, and M. Giannattasio, S. Sabbioneda, M. Stefanini, and all the members of our laboratories for sharing unpublished results and discussions. This work was supported by grants from Associazione Italiana Ricerca sul Cancro and Fondazione Cariplo (to P.P. and M.M.-F.), European Union FP6 Integrated Project DNA Repair (Contract 512113, to P.P.), and Telethon-Italy (Grant GGP030406, to M.M.-F.). S.S. was partially supported by a short-term EMBO Fellowship. Work in A.R.L.'s laboratory was supported by a Medical Research Council Programme Grant.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108547108/-/DCSupplemental.
References
- 1.Andressoo JO, Hoeijmakers JHJ. Transcription-coupled repair and premature ageing. Mutat Res. 2005;577:179–194. doi: 10.1016/j.mrfmmm.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 2.Harper JW, Elledge SJ. The DNA damage response: Ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 3.Lazzaro F, et al. Checkpoint mechanisms at the intersection between DNA damage and repair. DNA Repair (Amst) 2009;8:1055–1067. doi: 10.1016/j.dnarep.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 4.Marteijn JA, et al. Nucleotide excision repair–induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J Cell Biol. 2009;186:835–847. doi: 10.1083/jcb.200902150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mailand N, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 6.Giannattasio M, Lazzaro F, Longhese MP, Plevani P, Muzi-Falconi M. Physical and functional interactions between nucleotide excision repair and DNA damage checkpoint. EMBO J. 2004;23:429–438. doi: 10.1038/sj.emboj.7600051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marini F, et al. DNA nucleotide excision repair–dependent signaling to checkpoint activation. Proc Natl Acad Sci USA. 2006;103:17325–17330. doi: 10.1073/pnas.0605446103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marti TM, Hefner E, Feeney L, Natale V, Cleaver JE. H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc Natl Acad Sci USA. 2006;103:9891–9896. doi: 10.1073/pnas.0603779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Driscoll M, Jeggo PA. Clinical impact of ATR checkpoint signalling failure in humans. Cell Cycle. 2003;2:194–195. [PubMed] [Google Scholar]
- 10.Cortez D. Unwind and slow down: Checkpoint activation by helicase and polymerase uncoupling. Genes Dev. 2005;19:1007–1012. doi: 10.1101/gad.1316905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 12.Staresincic L, et al. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 2009;28:1111–1120. doi: 10.1038/emboj.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giannattasio M, et al. Exo1 competes with repair synthesis, converts NER intermediates to long ssDNA gaps, and promotes checkpoint activation. Mol Cell. 2010;40:50–62. doi: 10.1016/j.molcel.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Lee BI, Wilson DM., 3rd The RAD2 domain of human exonuclease 1 exhibits 5′ to 3′ exonuclease and flap structure–specific endonuclease activities. J Biol Chem. 1999;274:37763–37769. doi: 10.1074/jbc.274.53.37763. [DOI] [PubMed] [Google Scholar]
- 15.Lieber MR. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 16.Liberti SE, Rasmussen LJ. Is hEXO1 a cancer-predisposing gene? Mol Cancer Res. 2004;2:427–432. [PubMed] [Google Scholar]
- 17.Tran PT, Erdeniz N, Symington LS, Liskay RM. EXO1: A multi-tasking eukaryotic nuclease. DNA Repair (Amst) 2004;3:1549–1559. doi: 10.1016/j.dnarep.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 18.Schaetzlein S, et al. Exonuclease-1 deletion impairs DNA damage signaling and prolongs lifespan of telomere-dysfunctional mice. Cell. 2007;130:863–877. doi: 10.1016/j.cell.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei K, et al. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 2003;17:603–614. doi: 10.1101/gad.1060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nielsen FC, Jäger AC, Lützen A, Bundgaard JR, Rasmussen LJ. Characterization of human exonuclease 1 in complex with mismatch repair proteins, subcellular localization and association with PCNA. Oncogene. 2004;23:1457–1468. doi: 10.1038/sj.onc.1207265. [DOI] [PubMed] [Google Scholar]
- 21.Volker M, et al. Sequential assembly of the nucleotide excision repair factors in vivo. Mol Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 22.Luijsterburg MS, et al. Stochastic and reversible assembly of a multiprotein DNA repair complex ensures accurate target site recognition and efficient repair. J Cell Biol. 2010;189:445–463. doi: 10.1083/jcb.200909175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El-Shemerly M, Janscak P, Hess D, Jiricny J, Ferrari S. Degradation of human exonuclease 1b upon DNA synthesis inhibition. Cancer Res. 2005;65:3604–3609. doi: 10.1158/0008-5472.CAN-04-4069. [DOI] [PubMed] [Google Scholar]
- 24.El-Shemerly M, Hess D, Pyakurel AK, Moselhy S, Ferrari S. ATR-dependent pathways control hEXO1 stability in response to stalled forks. Nucleic Acids Res. 2008;36:511–519. doi: 10.1093/nar/gkm1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tishkoff DX, Amin NS, Viars CS, Arden KC, Kolodner RD. Identification of a human gene encoding a homologue of Saccharomyces cerevisiae EXO1, an exonuclease implicated in mismatch repair and recombination. Cancer Res. 1998;58:5027–5031. [PubMed] [Google Scholar]
- 26.Wilson DM, 3rd, et al. Hex1: A new human Rad2 nuclease family member with homology to yeast exonuclease 1. Nucleic Acids Res. 1998;26:3762–3768. doi: 10.1093/nar/26.16.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedberg EC, Walker GC, Siede W. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 2006. [Google Scholar]
- 28.Limsirichaikul S, et al. A rapid non-radioactive technique for measurement of repair synthesis in primary human fibroblasts by incorporation of ethynyl deoxyuridine (EdU) Nucleic Acids Res. 2009;37:e31. doi: 10.1093/nar/gkp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 30.Ogi T, Lehmann AR. The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nat Cell Biol. 2006;8:640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- 31.Ogi T, et al. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell. 2010;37:714–727. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 32.Iakoucheva LM, et al. Aberrant mobility phenomena of the DNA repair protein XPA. Protein Sci. 2001;10:1353–1362. doi: 10.1110/ps.ps.40101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.