Abstract
The histone fold forms the fundamental endoskeleton of the protein core of the nucleosome and is also found in several transcription factors. We have investigated the evolutionary origins of this ubiquitous protein motif, which is found soluble exclusively as an antiparallel (handshake motif) dimer. We introduced a three amino acid insertion into the middle of a homodimeric archaeal histone fold motif. The engineered molecule was found to be a soluble and stable monomer with properties consistent with a four-helix-bundle protein. The experimental evidence presented here support the hypothesis that the handshake association motif characteristic of present-day histone dimers is the evolutionary product of domain swapping between two four-helix bundle domains, each of which derived from the tandem duplication of a primitive helix–strand–helix unit.
Keywords: histone evolution, molecular evolution, gene endoskeleton
The genetic material of the eukaryotic cell (DNA) has to be drastically compacted to fit into the relatively small volume of the nucleus, while at the same time its information must remain available to the various cellular machineries. The compaction is achieved by a cascaded association of DNA with protein factors, beginning with its association with an octamer of the core histones H2A, H2B, H3, and H4.
Histone proteins have been found in all eukaryotes studied, and in almost all Euryarchaeota and some Crenarchaeota (1). Additionally, histone homologues have been reported in some distantly related bacteria (2), suggesting that the histone fold first appeared in a common ancestor of eukaryotes and archaea, and then was introduced to some bacteria, perhaps through lateral gene transfer (3).
The high resolution crystal structure of the histone core of the nucleosome (4) documented that the core histones share similar 3D structures [Protein Data Bank (PDB) ID codes 1HIO and 2HIO] (i.e., they are elongated polypeptides composed of three architectural elements), the histone fold, the extra-fold structured (α-helical) elements, and the flexible termini. Additionally, the histone fold was originally found in transcription factors (TFs) based on sequence searches (5, 6) (before the structure of any histone fold containing TFs was elucidated), which suggests that the histone fold sequence motifs in histones and those in TFs might share a common ancestor. Single, individual histone chains have not been observed in solution. In core histones, they are exclusively found as antiparallel dimers with the characteristic “handshake motif” architecture (5). Rarely, the histone fold sequence occurs as monomer “encased” within the larger protein ensembles within TFs (7).
Each histone-fold motif sequence consists of a short α-helix (Helix I), a β-turn, a long central α-helix of about 30 residues (Helix II), another β-turn, and a second short α-helix (Helix III). Because of the architectural symmetry of this motif, it can also be considered as the sum of two tandemly repeating helix–strand–helix (HSH) units. The two HSH motifs present in one half of the histone dimer (Fig. 1B, Left) are structurally similar to the respective pair of HSH motifs presents in the other dimer half (5). This observation lead to our earlier proposal that a tandem duplication of a primordial HSH element was a step in the evolution of the histone fold sequence (4).
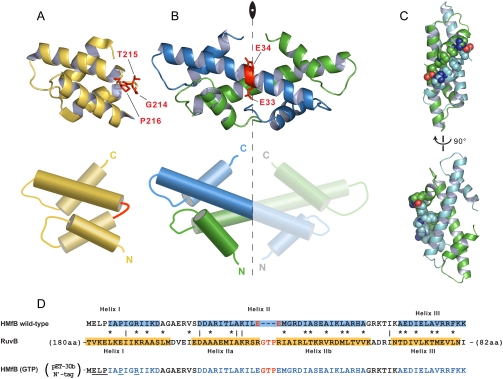
Fig. 1.
Structures and sequences of HMfB and RuvB. (A–B) Structures (Upper) and schemata (Lower) of the four-helix bundle of Thermotoga maritima RuvB (A) and of the Methanothermus fervidus HMfB dimer (B) with the red residues representing the hinge loop and the place of GTP insertion, respectively. The axis of twofold symmetry is also noted in the HMfB dimer. (C) The side-chains of amino acids participating in secondary interface interactions in the HMfB dimer are represented as spheres using CPK coloring for nitrogen and oxygen. The two peptide chains are colored in cyan and green. (D) The sequences of HMfB and the four-helix bundle of RuvB were aligned based on structural comparisons, with identical and similar residues identified by lines and asterisks, respectively. The hinge loop is colored red and helices are highlighted in blue and yellow. The residues participating in secondary interface interactions are underlined in the sequence of the recombinant insert variant.
In the case of archaeal homodimeric histones, which are characterized by a true twofold symmetry, as exemplified by the HMfA and HMfB histones of Methanothermus fervidus (8), the HSH motifs are identical both in sequence and structure. Consequently, the “left” half of the histone homodimer, a four-helix bundle, is identical to that in the right half. This attribute, and the fact that no single histone fold has been found as soluble protein, may imply that the histone fold evolution from an HSH unit to a dimer involved at least two steps. First, a tandem duplication of a primitive HSH unit to yield a soluble four-helix bundle intermediate, and second, a domain swapping event between two such four-helix bundle intermediates that gave rise to the handshake association state of canonical histone dimers.
Three-dimensional domain swapping describes the process by which similar structural elements of two monomers swap their intramolecular associations and establish analogous intermolecular contacts, thus generating dimers or higher oligomers (9). The structure of the subunits in the domain-swapped oligomers remains largely unchanged, except in the segment of the “hinge” that connects the exchanged domains with the rest of the protein. The intramolecular interface in the monomer between the swapped domain and the rest of the protein is identical to the intermolecular interface (10).
Alva et al. (11) used profile Hidden Markov Models (HMMs) of histone fold motifs to search a database of profile HMMs, and identified non-histone-fold sequence homologues of the histone fold, which raised the idea of domain swapping as a key step in the evolution of the histone fold (11). One of the matches was with the helical part (C-domain) of AAA+ ATPase proteins (11, 12). The C-domain could be superimposed onto the four-helix bundle of the histone dimer with root-mean-square deviations ranging from 1.2–1.7 Å. A common feature to all these sequence homologues is the presence of a short “insertion” in the middle of what otherwise would be the long helix in the histone fold. It may be hypothesized that these “insert” residues can serve as a hinge that allows the collapse of the C-terminal half over the N-terminal, and thus shifting the folding equilibrium away from the establishment of the extended helix (Helix II) of the canonical histone fold.
Several analyses and reviews have dealt with the phylogenomics of the nucleosome and the evolution of core histones (13, 14). However, there have been no in vitro studies on the evolution of the histone fold, but the available literature makes a compelling case for such investigation. Therefore, we decided to test experimentally the hypothesis that domain swapping of a primitive four-helix bundle structure could have led to the emergence of the core histone fold dimers.
Results and Discussion
Insertion in the Long Helix Disrupts Dimerization.
Upon inspection of four-helix bundle sequence homologous to the histone fold, a Gly–Thr–Pro (GTP) insertion was identified in the hinge of two different C-domains, namely, Escherichia coli RuvB and E. coli FtsH. The physicochemical properties of the the side-chains of these three residues are consistent with their potential role as hinge residues. Glycine and proline are known helix breakers, whereas proline would also presumably force the chain to take a turn. Threonine has a small side-chain that wouldn’t interfere with the bending of the hinge. Based on this reasoning, we inserted a GTP triplet in the middle of the long helix of the canonical histone fold of HMfB (between Glu33 and Glu34; Fig. 1B) and asked whether this insertion alone would convert the histone homodimers to two four-helix bundle monomers by disrupting the handshake dimerization propensity. The location of the insertion was selected after sequence comparisons between HMfB and the four-helix bundle sequence homologues, and after careful examination of the crystal structures of HMfB (PDB ID code 1A7W) and the four-helix bundle of RuvB (PDB ID code 1IN4) (Fig. 1 A and B).
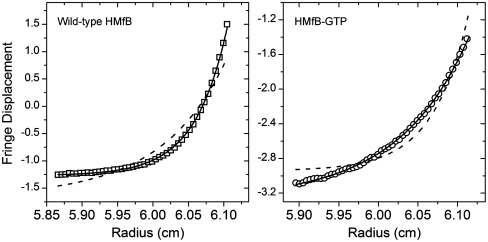
The first and most direct prediction of our experimental design is that the GTP insertion will allow the histone fold to collapse upon itself, and thus the engineered protein should be able to fold into a soluble monomer. Indeed, sedimentation equilibrium in the analytical ultracentrifuge at physiological conditions (0.2 M KCl, 20 mM KPi, pH 7.2) confirmed that this was the case for our HMfB–GTP construct (Fig. 2). Both proteins, the wild-type HMfB and HMfB–GTP, contain exactly identical 44-residue long, His-tag containing amino-terminal tails. Yet, the data in Fig. 2 clearly demonstrate that whereas the mutant is a monomer under these conditions, the wild-type protein is a dimer, as expected. Thus, the amino-terminal tail does not to influence the outcome of oligomerization. This is consistent with the fact that eukaryotic histone sequences with similar tail length (e.g., histone H3) exhibit typical histone fold dimerization.
Fig. 2.
Sedimentation equilibrium in the analytical ultracentrifuge at 35,000 rpm and 20 °C for wild-type HMfB (squares, Left) and HMfB–GTP (circles, Right). Solid lines represent the expected distributions for a dimer (Left) and monomer (Right). Dashed lines show the best fits to the data assuming a monomeric wild-type HMfB (Left) and dimeric HMfB–GTP (Right). The buffer was 0.2 M KCl and 20 mM KPi at pH 7.2.
Our finding that HMfB–GTP exists as a soluble monomer is significant given the knowledge that the canonical histone monomers cannot exist as soluble entities under physiological conditions (15, 16), but have a strong tendency to aggregate in the absence of their preferred dimerization partners. Monotypic preparations of archaeal histones like HMfB resolve this by self-associating, yielding homodimers.
Increases in Salt and Trimethylamine Oxide (TMAO) Concentrations Restore Secondary Structure of HMfB–GTP.
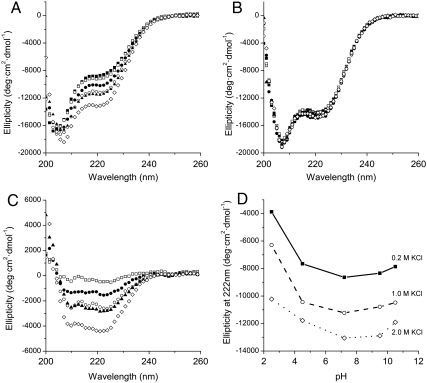
If truly successful in our design, our construct should have a minimal and local effect on the secondary structure of the chain. However, circular dichroism shows that the mutant HMfB in physiological buffer (0.2 M KCl, 20 mM KPi, pH 7.2) has a markedly reduced α-helical content compared to the wild-type protein; at 222 nm the ellipticity of the mutant protein is about -8,500 deg cm2 dmol-1 compared to about -14,500 deg cm2 dmol-1 for the wild-type (Fig. 3 A and B, filled squares).
Fig. 3.
Effect of salt and pH on the secondary structure of wild-type HMfB and HMfB–GTP (A) Far-UV CD spectra of HMfB–GTP as a function of KCl concentration at pH 7.2 (20 mM KPi). The KCl concentrations are, from top to bottom, 0.2 M (▪), 0.5 M (□), 0.75 M (•),1.0 M (○), 1.5 M (▴), and 2.0 M (⋄). (B) Far-UV CD spectra of wild-type HMfB as a function of KCl concentration at pH 7.2. The buffer was 20 mM KPi and the temperature was 20 °C. The KCl concentrations are 0.2 M (▪), 0.5 M (□), 0.75 M (•), 1.0 M (○), 1.5 M (▴), and 2.0 M (⋄). (C) Change in ellipticity of HMfB–GTP compared to 0.2 M KCl upon addition of KCl. The KCl concentrations are, from top to bottom, 0.5 M (□), 0.75 M (•), 1.0 M (○), 1.5 M (▴), and 2.0 M (⋄). (D) Ellipticity of HMfB–GTP at varying pH conditions and 0.2 M (▪), 1.0 M (○), and 2.0 M (⋄) KCl.
It has been well established that increasing the salt concentration of preparations of histones increases their secondary structure and their stability, as well as it promotes their assembly to dimers and tetramers (15, 16). Therefore, we examined the effect of high-salt solutions on the secondary structure of our constructs. Increasing the KCl concentration up to 2 M increases the α-helical content of the mutant HMfB, to about -13,500 deg cm2 dmol-1 (Fig. 3A, open diamonds) whereas the CD signal for the wild type remains largely unchanged (Fig. 3B, open diamonds). This salt-dependant response of the secondary structure of HMfB–GTP is observed throughout the pH range of 2.5–9.5, albeit with varied amplitudes (Fig. 3D).
Given the fact that histones have a preponderance of positively charged residues, this effect could, at first glance, be attributed to charge neutralization. However, for our HMfB–GTP construct, a substantial effect of salt on secondary structure occurs only at elevated salt concentrations, and throughout the pH range of 2.5–10.5 (Fig. 3D). A similar high salt dependence of stabilization of secondary structure has also been observed for the wild-type histone-fold protein HFoB from the mesophile Methanobacterium formicicum (17). Such a high salt response cannot be attributed entirely to simple ionic shielding and requires further investigation.
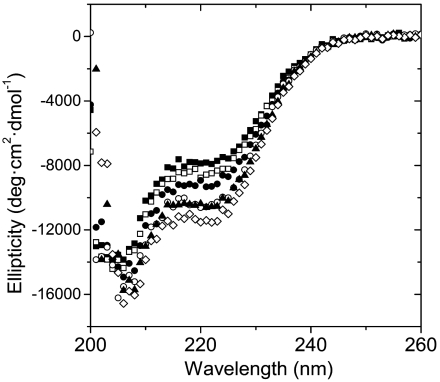
Previous studies of archaeal histones documented that histone HFoB is unstable under physiological conditions and that it is stabilized by TMAO, a naturally occurring osmolyte that has been shown to increase the secondary and tertiary structures of unstable proteins without interfering with their function (18). Adding up to 1 M TMAO in our buffers increased dramatically the α-helical content of our mutant to about -11,500 deg cm2 dmol-1 (Fig. 4). TMAO has no noticeable effect in the secondary structure of the wild-type protein, indicating that the increase of α-helical content in the mutant does not emanate from an ordering of the long His-tag tail, a feature shared by both constructs.
Fig. 4.
Far-UV CD spectra of HMfB–GTP as a function of TMAO concentration at 20 °C (0.2 M KCl, 20 mM KPi, pH 7.2). The TMAO concentrations are, from top to bottom, 0 M (▪), 0.2 M (□), 0.4 M (•), 0.6 M (○), 0.8 M (▴), and 1.0 M (⋄).
An additional consideration that needs to be addressed is the anticipated ellipticity of our construct. Our prediction is that the residues forming α-helices in the wild type will also be in helices in the HMfB–GTP construct. These residues will populate such conformations in a manner governed by statistical probabilities for local helix propensity, and this will be influenced by several factors: the nature of the residue, its sequence neighbors, its spatial neighbors, and its position along as well as the length of a given helix. The last factor will have the greatest impact on the outcome of our studies with HMfB–GTP for two reasons. First, the stability of α-helices is dependant on interactions among neighboring residues along the helix. At the end of helices the number of neighboring group interactions are diminished, and this results in residues spending less time in canonical helical conformation (end-effects) (19, 20). By inserting a tripeptide in the middle of Helix II of HMfB, we have generated two more “helix ends” that can be frayed in the mutant, compared to the wild type. This is significant because there are only about 50 residues altogether that are expected to be in helical conformation. If two to three residues are frayed in each end of the helix, by adding two new ends of helices, we should expect a reduction to the number of residues contributing to the overall helicity reported by the far-UV CD spectrum by as much as 10%. This is in line with our experimental observations. Second, short helices have an “innate” mean CD residual signal lower than longer helices (21). Thus, the decreased ellipticity that we observe in our constructs could also be partially due to the fact that we are breaking a long helix and creating two shorter ones.
HMfB–GTP Does Not Enter into Handshake Motif.
Next, we asked whether an increase in ionic strength promotes the dimerization of our construct, and whether it is the dimerization that is responsible for the salt-dependent increase in secondary structure, as is the case with canonical histones. Analytical ultracentrifugation under increasing KCl concentrations shows that the association state remains unchanged up to 2 M KCl (range tested) for both constructs, that is, the wild type is dimeric whereas the mutant remains monomeric and also soluble through this range (Fig. 5).
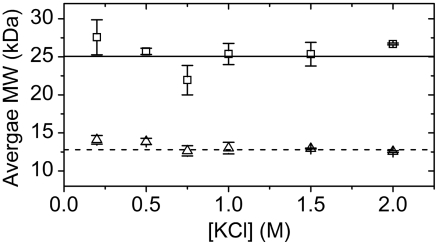
Fig. 5.
Average molecular weight for wild-type HMfB (□) and HMfB–GTP (▵). Measurements were taken at different KCl concentrations in 0.2 M KPi, ph 7.2 buffer and 20 °C using equilibrium sedimentation in the analytical ultracentrifuge at 3 rotor speeds. The error bars represent the standard deviation between the fitted molecular weights at each rotor speed. The solid and dashed horizontal lines represent the calculated MW of wild-type HMfB (25.0 kDa) and HMfB–GTP (12.8 kDa), respectively.
Salt Promotes Thermal Stability of the Mutant.
Several studies have documented the positive correlation between salt concentration and histone stability (22). We investigated next the stability of our constructs by analyzing the temperature dependence of the CD signal at 222 nm. The thermal melting scans show that the stability of the mutant protein is increased by increasing the salt concentration (Fig. 6). The melting profiles are indicative of pronounced “cold denaturation” of HMfB–GTP, a behavior shared with the wild-type HMfB and other archaeal histones (17), and that merits further investigation. At the lowest salt concentration (0.2 M KCl) the Tm of the mutant is about 55 °C and is increased to about 75 °C at 2.0 M KCl.
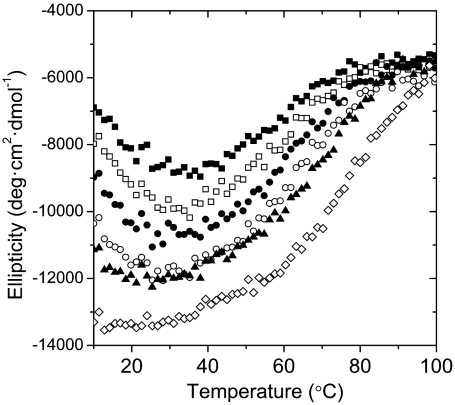
Fig. 6.
Ellipticity of HMfB–GTP at 222 nm as a function of temperature at different KCl concentrations. The KCl concentrations are, from top to bottom, 0.2 M (▪), 0.5 M (□), 0.75 M (•), 1.0 M (○), 1.5 M (▴), and 2.0 M (⋄). Experiments were performed at pH 7.2 in 20 mM KPi.
HMfB–GTP had a considerably lower Tm than the wild type (90 °C at 0.2 M KCl). This finding can be explained when we consider the contribution of the secondary interface interactions. These interactions, which are lost in HMfB–GTP, occur between Helix I at the amino terminal of each histone fold in the HMfB dimer (Fig. 1D), and are driven by two salt bridges and the coalescence of hydrophobic residues (Fig. 1C). Additionally, the major source of stability of the histone fold dimer is the paring via the hydrophobic surfaces between the two Helix II elements (23). As mentioned earlier, we are interrupting these continuous surfaces by inserting three residues, which destabilize the local helical conformation. These two perturbations in the sequence of the histone fold should have a considerable effect on its folding propensity in light of the overall small size of the polypeptides entering into “pairing” and most likely account for the resulting apparent lower thermal stability of HMfB–GTP.
Conclusions
The results we have presented here provided compelling evidence that a minimal and localized disruption in the long helix of the histone fold leads to a folded soluble monomeric protein that has lost the potential to utilize the handshake motif of contemporary histones, for dimerization. This finding supports our starting hypothesis that the evolutionary origin of the histone fold dimers had as first step the tandem duplication of an HSH unit, and this was followed by a domain swapping event. An interesting question is whether the simple monomeric evolutionary intermediate is still in use by present-day organisms and in what function.
Kinetic studies of histone folding showed that eukaryotic histones fold faster than their archaeal ancestors (24). Specifically, the analysis of the folding of histone HMfB dimers suggested the existence of a transient monomeric intermediate. Furthermore, the substitution of a nonnative residue in the middle of the long helix resulted in a more pronounced presence of the folding intermediate (25). A short-lived monomeric intermediate was also observed in folding studies of eukaryotic histones H2A and H2B (26). We propose that the monomeric intermediates detected in those elegant studies should have a structure similar to that of the ancestor of the histone fold as we invoked here, that is, they should transiently exist as four-helix bundles. The physiochemical advantage of the existence of the four-helix bundle intermediate state would be the facilitation of proper histone folding and assembly. By transiently occupying this state, histones temporarily bury the hydrophobic residues that could cause protein aggregation. This state is metastable and once a dimerizing partner appears, the histones assume their energetically favored, domain-swapped dimeric state. This pathway could be particularly advantageous for organisms that lack specialized protein-folding agents, such as chaperones. In this context, one could consider that one half of the histone fold may function as the “chaperone” for the other half. The evolutionary advantage of such a folding pathway may have been diminished with the advent of extended and highly flexible, extra-histone-fold elements, especially the histone tails that decisively improve histone solubility.
The scenario invoked here could be viewed as an outline of the initial steps in the evolutionary process that yielded today’s highly regulated nucleosome core (Fig. 7). In essence, this protein core, the histone octamer, is simply the result of a series of duplication events of covalently linked modules and association patterns, from the HSH to the tripartite organization of eight dimers that form an articulated, proteinaceous “gene endoskeleton” (27). The increased complexity of this assembly endows the system with additional dimensions of regulation by modulation of the interfacial contacts between the dimer and tetramer subunits of the core histone octamer, beyond what is possible through solely histone tail modifications.
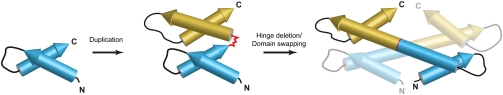
Fig. 7.
A schematic of the proposed evolutionary trajectory of the histone fold structure: A primordial HSH motif forms a stable, soluble four-helix bundle monomer through a tandem duplication (genomic event). Deletion or modification of the hinge sequence (red) connecting the two HSH motifs leads to the formation of the histone fold sequence motif and introduces the thermodynamic potential for its dimerization. The end result of the combined effects of these transitions is a “gain of function” where a HSH motif evolves into the architectural framework of the protein core of the nucleosome.
Materials and Methods
Cloning of HMfB Constructs.
The sequence for wild-type HMfB from Methanothermus fervidus was kindly provided by Lisa Gloss in a pALTER-Ex2 vector (Promega Corp.). The gene was amplified using the M13 set of primers then subcloned in the pET-30b vector (Novagen) in the NcoI and BamHI restriction sites. The vector was then transformed into Escherichia coli XL1-Blue (Stratagene) and, for expression, into E. coli BL21-Star (DE3) (Invitrogen).
The HMfB–GTP variant was produced using a variation of standard PCR protocols for mutagenesis; i.e., three PCRs were performed with four primers. The first set used a forward primer and a reverse primer that included the insertion sequence. The second set included the forward primer with the insertion sequence and a reverse primer located downstream of the HMfB gene. After these two reactions were performed independently, the PCR products were purified and combined. A third PCR was performed to yield the full-length variant using only the end primers for the amplification of the final product. The inserts were then subcloned as described above. Both, the wild type as well as the insert-containing constructs shared the same, 44-residue long amino-terminal tag.
Protein Concentration Determination.
Because the constructs do not contain chromophores, we used the interference optics of the XL-I analytical ultracentrifuge to determine protein concentration. Protein that was dialyzed extensively in buffer overnight was layered on top of the dialysate in a 12 mm synthetic boundary cell in the ultracentrifuge by spinning at 8,000 rpm in a T-60i rotor. Once the menisci in the two cells were aligned, an interference scan was performed and the fringe displacement was measured. We used a factor of 0.3 mg·mL-1·fringe-1 to convert the fringe displacements to protein concentration (28).
Analytical Ultracentrifugation.
Samples were prepared by extensive dialysis overnight at 4 °C. Sedimentation equilibrium experiments were performed in a Beckman XL-I Analytical Ultracentrifuge using either 2- or 6- sector epon-filled charcoal centerpieces and sapphire windows. Equilibrium was confirmed by the subtraction of consecutive scans. Global analysis of the data was performed using an XL-A/XL-I data analysis implementation (from Beckman Instruments, version 4.0) for the MicroCal Origin software (OriginLab). The partial specific volumes of our constructs were calculated from their amino acid composition using SEDNTERP (http://www.rasmb.bbri.org/). SEDNTERP was also used for the calculation of solution densities. We used ProtParam (http://www.expasy.org/tools/protparam.html) to calculate the molecular weights for our constructs.
Circular Dichroism.
Far-UV CD scans were collected in a Jasco-710 instrument equipped with a Peltier PTC-3481 temperature controller. For wavelength scans in the range of 200–260 nm, data were collected using a 1 mm cuvette and a resolution of 0.2 nm, 1.0 nm bandwidth, 50 nm· min-1 scan speed, 4 sec response and accumulated 3 times. Thermal denaturation scans were performed in a range of 10–105 °C and measurements were taken at 222 nm every 0.2 °C, 1.0 nm bandwidth and a 4 sec response. Manipulation of the CD data was performed with J-700 for Windows (version 1.32). The buffers used for the measurements were 10 mM Glycine for pH 2.5, 10 mM Na-Acetate for pH 4.5, 10 mM KPi for pH 7.2, 10 mm N,N-Bis(2-hydroxyethyl)glycine (BICINE) for pH 9.5 and 10 mM N-Cyclohexyl-3-aminopropanesulfonic acid (CAPS) for pH 10.5.
Acknowledgments.
We thank Drs. M. Amzel, D. Barrick, and E. Freire for helpful comments on this manuscript. We thank E. Freire and P. Privalov for allowing access to specialized instrumentation in their laboratories. Part of this work was supported by National Science Foundation Grant MCB0091736 (E.N.M.). We dedicate this paper to the memory of Christian B. Anfinsen, whose 1959 book The Molecular Basis of Evolution provided the foundations on which this work stands.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
References
- 1.Friedrich-Jahn U, Aigner J, Längst G, Reeve JN, Huber H. Nanoarchaeal origin of histone H3? J Bacteriol. 2009;191:1092–1096. doi: 10.1128/JB.01431-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qiu Y, et al. The crystal structure of Aq_328 from the hyperthermophilic bacteria Aquifex aeolicus shows an ancestral histone fold. Proteins. 2006;62:8–16. doi: 10.1002/prot.20590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson KE, et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 4.Arents G, Burlingame RW, Wang BC, Love WE, Moudrianakis EN. The nucleosomal core histone octamer at 3.1 A resolution: A tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci USA. 1991;88:10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arents G, Moudrianakis EN. The histone fold: a ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc Natl Acad Sci USA. 1995;92:11170–11174. doi: 10.1073/pnas.92.24.11170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baxevanis AD, Arents G, Moudrianakis EN, Landsman D. A variety of DNA-binding and multimeric proteins contain the histone fold motif. Nucleic Acids Res. 1995;23:2685–2691. doi: 10.1093/nar/23.14.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birck C, et al. Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell. 1998;94:239–249. doi: 10.1016/s0092-8674(00)81423-3. [DOI] [PubMed] [Google Scholar]
- 8.Decanniere K, et al. Crystal structures of recombinant histones HMfA and HMfB from the hyperthermophilic archaeon Methanothermus fervidus 1. J Mol Biol. 2000;303:35–47. doi: 10.1006/jmbi.2000.4104. [DOI] [PubMed] [Google Scholar]
- 9.Gronenborn AM. Protein acrobatics in pairs—dimerization via domain swapping. Curr Opin Struct Biol. 2009;19:39–49. doi: 10.1016/j.sbi.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlunegger MP, Bennett MJ, Eisenberg D. Oligomer formation by 3D domain swapping: A model for protein assembly and misassembly. Adv Protein Chem. 1997;50:61–122. doi: 10.1016/s0065-3233(08)60319-8. [DOI] [PubMed] [Google Scholar]
- 11.Alva V, Ammelburg M, Söding J, Lupas AN. On the origin of the histone fold. BMC Struct Biol. 2007;7:17. doi: 10.1186/1472-6807-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 13.Malik HS, Henikoff S. Phylogenomics of the nucleosome. Nat Struct Biol. 2003;10:882–891. doi: 10.1038/nsb996. [DOI] [PubMed] [Google Scholar]
- 14.Eirín-López JM, González-Romero R, Dryhurst D, Méndez J, Ausió J. In: Evolutionary Biology. Pontarotti P, editor. Berlin: Springer; 2009. pp. 139–162. [Google Scholar]
- 15.Karantza V, Baxevanis AD, Freire E, Moudrianakis EN. Thermodynamic studies of the core histones: Ionic strength and pH dependence of H2A–H2B dimer stability. Biochemistry. 1995;34:5988–5996. doi: 10.1021/bi00017a028. [DOI] [PubMed] [Google Scholar]
- 16.Karantza V, Freire E, Moudrianakis EN. Thermodynamic studies of the core histones: pH and ionic strength effects on the stability of the (H3-H4)/(H3-H4)2 system. Biochemistry. 1996;35:2037–2046. doi: 10.1021/bi9518858. [DOI] [PubMed] [Google Scholar]
- 17.Li WT, et al. Thermodynamic stability of archaeal histones. Biochemistry. 1998;37:10563–10572. doi: 10.1021/bi973006i. [DOI] [PubMed] [Google Scholar]
- 18.Baskakov I, Bolen DW. Forcing thermodynamically unfolded proteins to fold. J Biol Chem. 1998;273:4831–4834. doi: 10.1074/jbc.273.9.4831. [DOI] [PubMed] [Google Scholar]
- 19.Marqusee S, Baldwin RL. Helix stabilization by Glu-…Lys+ salt bridges in short peptides of de novo design. Proc Natl Acad Sci USA. 1987;84:8898–8902. doi: 10.1073/pnas.84.24.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Presta LG, Rose GD. Helix signals in proteins. Science. 1988;240:1632–1641. doi: 10.1126/science.2837824. [DOI] [PubMed] [Google Scholar]
- 21.Chin D-H, Woody RW, Rohl CA, Baldwin RL. Circular dichroism spectra of short, fixed-nucleus alanine helices. Proc Natl Acad Sci USA. 2002;99:15416–15421. doi: 10.1073/pnas.232591399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Holde KE. Chromatin. 1st Ed. Springer; 1988. [Google Scholar]
- 23.Li W-T, Shriver JW, Reeve JN. Mutational analysis of differences in thermostability between histones from Mesophilic and Hyperthermophilic Archaea. J Bacteriol. 2000;182:812–817. doi: 10.1128/jb.182.3.812-817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Topping TB, Gloss LM. Stability and folding mechanism of mesophilic, thermophilic and hyperthermophilic archael histones: The importance of folding intermediates. J Mol Biol. 2004;342:247–260. doi: 10.1016/j.jmb.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 25.Stump MR, Gloss LM. Unique fluorophores in the dimeric archaeal histones hMfB and hPyA1 reveal the impact of nonnative structure in a monomeric kinetic intermediate. Protein Sci. 2008;17:322–332. doi: 10.1110/ps.073224308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stump MR, Gloss LM. Mutational analysis of the stability of the H2A and H2B histone monomers. J Mol Biol. 2008;384:1369–1383. doi: 10.1016/j.jmb.2008.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arents G, Moudrianakis EN. Topography of the histone octamer surface: repeating structural motifs utilized in the docking of nucleosomal DNA. Proc Natl Acad Sci USA. 1993;90:10489–10493. doi: 10.1073/pnas.90.22.10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Babul J, Stellwagen E. Measurement of protein concentration with interferences optics. Anal Biochem. 1969;28:216–221. doi: 10.1016/0003-2697(69)90172-9. [DOI] [PubMed] [Google Scholar]