Abstract
Nonphotochemical quenching (NPQ) regulates energy conversion in photosystem II and protects plants from photoinhibition. Here we analyze NPQ capacity in a number of rice cultivars. NPQ was strongly induced under medium and high light intensities in rice leaves. Japonica cultivars generally showed higher NPQ capacities than Indica cultivars when we measured a rice core collection. We mapped NPQ regulator and identified a locus (qNPQ1-2) that seems to be responsible for the difference in NPQ capacity between Indica and Japonica. One of the two rice PsbS homologues (OsPsbS1) was found within the qNPQ1-2 region. PsbS protein was not accumulated in the leaf blade of the mutant harboring transferred DNA insertion in OsPsbS1. NPQ capacity increased as OsPsbS1 expression increased in a series of transgenic lines ectopically expressing OsPsbS1 in an Indica cultivar. Indica cultivars lack a 2.7-kb region at the point 0.4 kb upstream of the OsPsbS1 gene, suggesting evolutionary discrimination of this gene.
Keywords: chlorophyll fluorescence, pulse amplitude modulation, quantitative trait loci analysis, rice subclass
Plants have the potential to transform absorbed light energy to chemical energy at relatively high efficiency. However, the actual efficiency is dependent on the inherent capacities of photochemistry and carbon assimilation, and environmental factors including light intensity. The sites at which light energy is absorbed are photosystem I and II (PSI and PSII), which drive photochemistry and production of chemical energy (ATP and NADPH). For PSII, energy transformation processes of chlorophyll excitation energy can be measured by the pulse amplitude modulation technique of chlorophyll fluorescence measurement (1–3).
Chlorophyll deexcitation processes are divided into three groups: photochemistry (photosynthetic electron transport), basal dissipation/ non-light induced quenching (NO), and thermal dissipation, which results in nonphotochemical quenching (NPQ) of chlorophyll fluorescence. Basal dissipation consists of chlorophyll fluorescence, internal conversion, and intersystem crossing. With increasing light intensity, there is a decrease in efficiency of use of excitons in photochemistry and an increase in NPQ. More than half of absorbed energy can be lost through NPQ under high illumination (4, 5). NPQ consists of several components. These can be distinguished by the rate of induction of NPQ in the light and by the rate of its relaxation in the dark (6). The NPQ component that is rapidly induced by illumination and relaxes rapidly in the dark is called qE. qE is the energy-dependent quenching linked to the proton motive force.
Photoinhibition indicates light stress and, under high illumination, excessive energy results in photodamage with inactivation of the PSII machinery. This leads to a decrease in the photochemical rate constant and thermal loss of energy caused by photoinhibition. Photoinhibition results in inactivation of a part of the PSII reaction centers. The degree of photoinhibition and the intrinsic loss of PSII yield can be measured from chlorophyll fluorescence analysis (ratio of variable to maximum fluorescence; Fv/Fm) on dark-adapted leaves. qE safely dissipates excess excitation energy and protects against photoinhibition (7, 8).
We reported that NAD kinase (NADK) is involved in the dissipation of heat by qE through efficient proton delivery in the xanthophyll cycle (9). Furthermore, overexpression of the same gene in rice showed increased photosynthesis by activated photochemistry (10). In this report, we investigate the genetic regulation of qE capacity in rice. Rice is one of the major crops worldwide. More than half the world’s population consumes rice as a staple food (11). Rice is generally cultivated under high illumination, reaching a photosynthetic photon flux density (PPFD) greater than 2,000 μmol m−2 s−1 at noon on sunny days. Even recognizing that rice leaves reduce light intensity per unit area by maintaining relatively vertical leaf angles, qE would always be strongly induced in many parts of the leaf in daytime. There is loss of efficiency of energy use in photochemistry with increasing light, and photosynthesis in rice is saturated well below full sunlight (10, 12). Thus, the ability of qE to safely dissipate excess energy in rice will continue to be important for sustainable growth.
In this study, we identify differences in qE capacity between several rice (Oryza sativa) cultivars, and generally between two rice subclasses, Indica and Japonica. Two loci were identified in QTL analysis between an Indica and a Japonica cultivar. The results suggest one of these loci regulates qE between these subclasses. A PsbS homologue was found within this locus. This PsbS homologue was confirmed to regulate qE in rice, after analyzing a mutant and transformants ectopically expressing this homologue. In some transformants, qE capacity was even increased over the original level. Deletion was found upstream of this gene in Indica cultivars. PsbS accumulation was higher in a Japonica cultivar than in an Indica cultivar, suggesting that PsbS regulates qE difference between rice cultivars.
Results
Measurement of NPQ in Different Rice Cultivars.
We compared chlorophyll fluorescence parameters between various rice cultivars and found significant differences. First we report differences in NPQ between the cultivars Habataki (an Indica cultivar) and Sasanishiki (a Japonica cultivar).
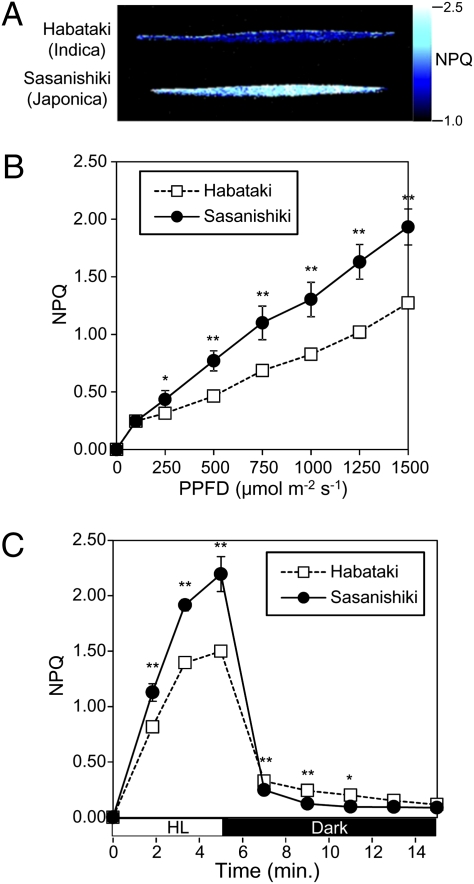
Fig. 1A shows a 2D image of NPQ in the leaf blades of Habataki and Sasanishiki after NPQ induction by high light at a PPFD of 1,500 μmol m−2 s−1 for 5 min. NPQ is higher along the whole leaf blade in Sasanishiki than in Habataki. When we measured NPQ under a range of light intensities, NPQ was consistently higher in Sasanishiki than in Habataki under light intensities higher than a PPFD of 250 μmol m−2 s−1 (Fig. 1B). We also measured the induction of NPQ by high light at 1,500 μmol m−2 s−1 and following relaxation in the dark (Fig. 1C), and found that the magnitude of qE is higher in Sasanishiki than in Habataki.
Fig. 1.
Comparison between Habataki and Sasanishiki. (A) Fully expanded third leaves of the rice cultivars Habataki and Sasanishiki were measured. Image of NPQ values under high illumination (PPFD, 1,500 μmol m−2 s−1) for 5 min is shown. (B) NPQ values under various light intensities. (C) NPQ induction by high light (HL) at a PPFD of 1,500 μmol m−2 s−1 for 5 min and relaxation in the dark for 10 min. Data represent means and SDs. Asterisks indicate significant differences by Student t test (*P < 0.05, **P < 0.01); n = 6.
Asian cultivated rice (O. sativa) comprises two subclasses: Indica and Japonica. The low-NPQ cultivar Habataki belongs to the Indica subclass and the high-NPQ cultivar Sasanishiki belongs to the Japonica subclass. It is reasonable to question whether the difference in NPQ value observed between Habataki and Sasanishiki can also be observed between other Indica and Japonica cultivars. First we compared three Indica and four Japonica cultivars, including Habataki and Sasanishiki, with respect to the parameters Fv/Fm and NPQ, and found that Fv/Fm values tended to be higher in the Japonica cultivars (Fig. S1A). As with Habataki and Sasanishiki, NPQ was significantly higher in the Japonica cultivars under both light intensities of a PPFD of 400 μmol m−2 s−1 and 1,500 μmol m−2 s−1 (Fig. S1B).
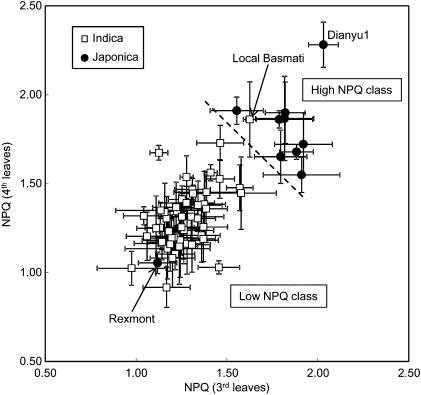
Analysis of the NPQ values at 1,500 μmol m−2 s−1 was extended to other genotypes from the World Rice Core Collection (WRC), including 52 Indica cultivars and 10 Japonica cultivars (Fig. 2). Although the pattern of NPQ size was somewhat different between third and fourth leaves, Indica and Japonica cultivars nevertheless clustered differently when we compared NPQ values in third and fourth leaves (Fig. 2). Based on this measurement, cultivars can be assigned to the high- or low-NPQ classes by using a threshold NPQ value of 1.65, as shown by the dashed line in Fig. 2. It can be concluded that NPQ is generally higher in the Japonica cultivars than in the Indica ones. The Indica subclass includes the aus and indica groups, whereas the Japonica subclass includes the temperate japonica and tropical japonica groups (13). In contrast to the highly significant difference in NPQ between Indica and Japonica, no significant differences in NPQ were observed between the aus and indica groups or between the temperate japonica and tropical japonica groups (Fig. S1C).
Fig. 2.
NPQ values of WRC collection. Fully expanded third leaves from 3-wk-old plants and fully expanded fourth leaves from 4-wk-old plants were measured. NPQ was measured under high illumination (PPFD, 1,500 μmol m−2 s−1) for 5 min. Cultivars were grouped into high- and low-NPQ classes by using an arbitrary NPQ threshold value of 1.65.
Chromosomal Mapping of NPQ Regulator.
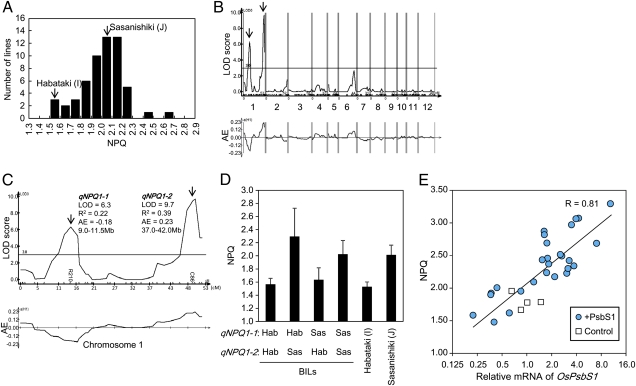
Using backcrossed inbred lines (BILs) between an Indica cultivar (Habataki) and a Japonica cultivar (Sasanishiki), we analyzed genetic loci that regulate the magnitude of NPQ between Indica and Japonica. The frequency distribution of NPQ values measured in 57 BILs is shown in Fig. 3A. Many BILs showed NPQ values similar to Sasanishiki because these BILs were generated by backcrossing to Sasanishiki. Several BILs showed hypersegregation of NPQ values that were even higher than in Sasanishiki.
Fig. 3.
QTL analysis of NPQ regulator. (A) Frequency distribution of NPQ values of BILs. (B) Distribution of LOD scores and AE values along chromosomes. Positive AE values indicate positive effects of the Sasanishiki allele. (C) Magnification of chromosome 1. Maximum LOD scores, explained variances (R2), AE values, and approximate positions are shown for qNPQ1-1 and qNPQ1-2. (D) NPQ values of four BIL groups classified by genotypes of qNPQ1-1 and qNPQ1-2. Genotypes at qNPQ1-1 and qNPQ1-2 are represented by the genotypes of the markers R210 and C86. Habataki genotypes are indicated as “Hab” and Sasanishiki genotypes as “Sas.” Several lines heterozygous for either of the two loci were excluded from the calculation. Data represent means and SDs (n = 3 for each line). (E) Relationship between OsPsbS1 expression and NPQ value. Control (cultivar Kasalath) and transgenic lines harboring the OsPsbS1 gene driven by Ubi1 promoter (+PsbS1) were analyzed. Relative mRNA accumulations of OsPsbS1 and NPQ value under illumination at a PPFD of 1,500 μmol m−2 s−1 for 10 min were measured and plotted. Measurements were made on fully expanded leaves of first transgenic progenies. The solid line represents semilogarithmic regression and correlation coefficient (R).
By quantitative trait loci (QTL) analysis of these data, we identified two loci on chromosome 1 (Fig. 3B). Fig. 3C shows magnification of LOD scores and additive effect (AE) values along chromosome 1. The two loci identified on chromosome 1 were denoted qNPQ1-1 and qNPQ1-2. The locus qNPQ1-1 showed a LOD score peak at approximately 9.0 to 11.5 Mb, and qNPQ1-2 showed a peak at approximately 37.0 to 42.0 Mb. The values of AE were −0.18 for qNPQ1-1 and 0.23 for qNPQ1-2. This means that the Indica allele of qNPQ1-1 increased NPQ by 0.18 (0.36 in the homozygote), whereas the Japonica allele of qNPQ1-2 increased NPQ by 0.23 (0.46 in the homozygote). Thus, qNPQ1-2 seems to increase NPQ in the Japonica cultivars. The Indica allele of qNPQ1-1 seems to increase NPQ only in the presence of the Japonica allele of qNPQ1-2, judging from the NPQ values in the BIL groups, which were classified according to genotypes at qNPQ1-1 and qNPQ1-2 loci (Fig. 3D). The hypersegregation of NPQ over Sasanishiki observed in Fig. 3A is caused by the combination of the Indica allele of qNPQ1-1 and the Japonica allele of qNPQ1-2.
Mapping of Possible NPQ Regulators on Rice Chromosome.
Genetic studies on NPQ regulation have been made in Arabidopsis thaliana (14–19). There is a good possibility that the qNPQ1-1 and qNPQ1-2 genes are rice homologues to the NPQ regulators identified in A. thaliana. Rice homologues to A. thaliana NPQ regulators were listed by a homology search in the Rice Annotation Project Database and mapped on the rice genome (Fig. S2A). No homologue was mapped within the qNPQ1-1 region, so the qNPQ1-1 gene apparently regulates NPQ by some unknown molecular mechanism. On the contrary, the OsPsbS1 gene lies near the center of the qNPQ1-2 region. We found two PsbS homologues in the rice genome (Fig. S2B). These genes (Os01g0869800/LOC_Os01g64960 and Os04g0690800/LOC_Os04g59440) are called OsPsbS1 and OsPsbS2 in the present study. Both these homologues may regulate NPQ if expressed in rice leaves.
Functional Analysis of the OsPsbS1 Gene.
Zulfugarov et al. (20) analyzed a rice mutant harboring transferred DNA insertion in OsPsbS1. mRNA accumulation of OsPsbS1 was not detected in this mutant. This mutant (here denoted psbS1) largely lacks NPQ and is more susceptible to photoinhibition and is shown to set less seeds than controls (21).
We extracted total protein from the leaf blades of control and psbS1 and performed Western blotting analysis by using anti-PsbS antibody. PsbS protein was detected in the control plant but not at all in psbS1 (Fig. S3B). PsbS protein was also detected in CBB staining of control protein, but this band was lost in psbS1. Thus, PsbS accumulates at a relatively high level in rice leaves.
NPQ was measured in psbS1. NPQ was highly induced under PPFDs greater than 500 μmol m−2 s−1 in controls, whereas it was only slightly induced even at a PPFD of 1,500 μmol m−2 s−1 in psbS1 (Fig. S4B). NPQ was lower in psbS1 along the whole leaf blade (Fig. S4A). As described previously (21), qE was also lost in psbS1 under our experimental conditions (Fig. S4C).
We also compared the relationship between OsPsbS1 expression levels and NPQ values by using transgenic lines to further confirm the role of OsPsbS1 in NPQ regulation. For this purpose, OsPsbS1 was ectopically expressed under the control of maize Ubi1 promoter in the Indica cultivar Kasalath, which was suitable for transformation experiments. The transformants showed different levels of OsPsbS1 expression. The relationship between OsPsbS1 mRNA accumulation and NPQ was analyzed (Fig. 3E), and NPQ increased logarithmically with OsPsbS1 expression.
Comparison of OsPsbS1 Between Indica and Japonica.
The DNA sequence corresponding to the coding sequence of OsPsbS1 was nearly the same in Kasalath (Indica), Habataki (Indica), Nipponbare (Japonica), and Sasanishiki (Japonica). Kasalath had one nucleotide substitution, but this mutation did not affect the predicted amino acid sequence. Thus, predicted amino acid sequences are exactly the same in these four cultivars.
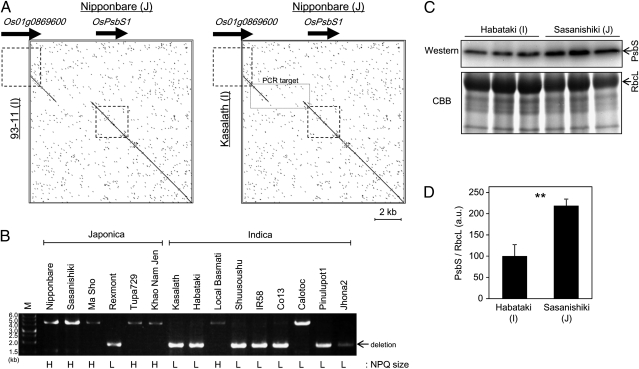
We also compared the genomic DNA sequence around OsPsbS1 between standard Indica cultivars (93–11 and Kasalath) and a standard Japonica cultivar (Nipponbare), whose genomic sequences are available (Fig. 4A). A 2.7-kb region was lost in the Indica cultivars compared with the Japonica one, approximately 400 bp upstream of the translation initiation site of OsPsbS1. Existence of this deletion was also analyzed by PCR in several Japonica and Indica cultivars (Fig. 4B). We found that many Japonica cultivars do not have this 2.7-kb deletion, whereas many Indica cultivars do. Existence of the deletion was correlated with low NPQ values, rather than classification to the Indica subclass. There were a few exceptional cultivars. For example, the Japonica cultivar Rexmont showed low NPQ values and possessed the 2.7-kb deletion. Similarly, an Indica cultivar Local Basmati showed high NPQ values and did not possess the 2.7-kb deletion. Calotoc had a unique pattern. This Indica cultivar showed low NPQ values, although it did not have the 2.7-kb deletion.
Fig. 4.
Comparison of PsbS between Indica and Japonica. (A) Genomic sequences of Indica cultivars (93–11 and Kasalath) and a Japonica cultivar (Nipponbare) were compared by using a dot-plot analysis (http://www.vivo.colostate.edu/molkit/dnadot) with a 9-base frame. Squares with broken lines represent genetic regions of Os01g0869600 and OsPsbS1 (from start codon to stop codon). Rectangle with solid lines in the box (Right) represents the target region of PCR in B. (B) PCR amplification of genomic sequence around the 2.7-kb deletion. M, molecular weight marker. NPQ groups of the cultivars analyzed are indicated below the gel image (L, low-NPQ group; H, high-NPQ group). (C) Total protein was extracted from expanded leaves of Habataki and Sasanishiki. PsbS protein accumulation was compared by Western blotting analysis. Total protein was also stained with CBB. RbcL, large subunit of RuBisCO. (D) Quantification of PsbS band intensity relative to RbcL band intensity. Data represent means and SDs. Values were significantly different by Student t test (P < 0.01); n = 3.
The 2.7-kb deletion near the OsPsbS1 translation initiation site in Indica cultivars may affect OsPsbS1 expression. To test this possibility, we extracted total protein from leaf blades of an Indica cultivar (Habataki) and a Japonica cultivar (Sasanishiki), and compared PsbS protein accumulation by Western blotting. As expected, PsbS accumulation was higher in Sasanishiki than in Habataki (Fig. 4 C and D).
Discussion
In this study, we found differences in NPQ values between many rice cultivars. We first compared NPQ between an Indica cultivar (Habataki) and a Japonica cultivar (Sasanishiki). NPQ was higher in Sasanishiki under moderate and high illuminations. We also compared NPQ and Fv/Fm values in seven cultivars that have undergone much study in Japan. NPQ values were clearly higher in the Japonica cultivars than in the Indica ones. Quantum yields of photochemistry, NPQ, and basal dissipation were also measured by parameters ΦII, ΦNPQ, and ΦNO in these seven cultivars under moderate and high illumination (Fig. S5). ΦII values tend to be lower in Japonica cultivars, which may partly explain the higher NPQ yield of Japonica under moderate illumination. Under high light, ΦII yields were low in both Indica and Japonica cultivars, and NPQ yields were lower in Indica than Japonica cultivars. The results show that the Indica cultivars, compared with the Japonica cultivars, dissipate less energy by NPQ, and more energy by basal processes (NO). This is consistent with a deficiency of PsbS protein in the Indica cultivars, which is required for NPQ (i.e., qE). The Japonica cultivars tended to show slightly higher Fv/Fm values than Indica. The reason for this tendency is not clear at the moment. For example, photoinhibition may be less severe in the Japonica cultivar because of high NPQ under ordinary culture conditions, or otherwise photosynthesis capacity may be slightly higher in Japonica cultivar under low light.
The results of NPQ measurement in numerous rice core collection cultivars confirmed that NPQ capacity is generally higher in Japonica than in Indica. On the contrary, there was no difference between groups within Indica (aus and indica) or within Japonica (temperate japonica and tropical japonica). Cultivated Asian rice (O. sativa) is derived from two different groups within a species of wild rice (Oryza rufipogon), which are known as Indica and Japonica today (22). Indica and Japonica last shared a common ancestor approximately 0.44 Mya (23). The mutation regulating the difference in NPQ between Indica and Japonica will have been inserted relatively soon after the ancestors of Indica and Japonica separated from their common ancestor.
When we performed QTL analysis of NPQ regulators between Indica (cultivar Habataki) and Japonica (cultivar Sasanishiki), two loci were identified. Of these two loci, qNPQ1-2 seems to regulate NPQ between Indica and Japonica, whereas qNPQ1-1 seems to function only in the presence of the positive allele of qNPQ1-2. OsPsbS1 is within the qNPQ1-2 region. We then analyzed the function of the OsPsbS1 gene.
We first characterized the rice psbS1 mutant. We found that PsbS protein was not accumulated in psbS1. This result is consistent with the complete loss of NPQ caused by qE, the energy-dependent quenching in psbS1. PsbS has four transmembrane regions. In addition, two glutamate residues positioned on the lumen side of the PsbS protein are necessary for PsbS function (24). Because these transmembrane regions and glutamate residues are conserved in both of the two rice PsbS homologues, OsPsbS2 is also expected to have the ability to regulate NPQ. NPQ capacity in plants is regulated by environmental conditions (25). OsPsbS2 expression may be induced under special conditions to support growth of mature plants and to improve tolerance to environmental stresses such as heat (26). OsPsbS2 is expressed at least under some conditions because two cDNA are cloned, according to the Knowledge-based Oryza Molecular biological Encyclopedia (accession nos. J023101I15 and D84392).
To obtain further insight into the functional differentiation of PsbS within the plant kingdom, we obtained protein sequences of PsbS from the National Center for Biotechnology Information for various plant species and compared their sequences. Surprisingly, only rice has two obviously distinct PsbS proteins, and a single PsbS sequence was found in other plant species. Two quite similar sequences were found for a few species (maize, soybean, Nicotiana benthamiana, and Selaginella moellendorffii). These sequences may derive from different cultivars. Judging from a phylogenetic tree, the two rice PsbS genes have had different roles since Poaceae separated from the other plant species.
To discover the function of OsPsbS1, we also transgenically introduced the OsPsbS1 gene into an Indica cultivar and obtained independent transformants, which showed different expression levels of OsPsbS1. NPQ capacity increased logarithmically with expression levels of OsPsbS1. This result, together with the observation of psbS1 mutant, confirms that OsPsbS1 expression level is a determinant of NPQ capacity in rice. It is also important that this result shows that NPQ capacity can be increased by transgenic introduction of the PsbS gene not only in dicots but also in monocots, which includes a number of very major crops such as rice, wheat, and maize. Because expression levels of the OsPsbS1 gene are naturally high, many transgenic lines should be generated to allow selection of those showing high levels of OsPsbS1 expression and significantly higher capacity of NPQ. This character was bred through to the next generation, confirming genetic stability of inserted gene expression.
The molecular mechanism and regulatory machinery of NPQ has been a target of genetics in the model dicot A. thaliana. Involvement of the xanthophyll cycle has been deduced from its physiological characteristics (12, 27). Genetics in A. thaliana further identified enzymes regulating the xanthophyll cycle, and NPQ has been largely lost in the mutant of Violaxanthin De-Epoxidase (VDE) gene (15). NPQ is completely lost when a mutation in lutein synthesis is introduced in this mutant, which is indicative of loss of qE (18).
Improvement of “fitness” estimated by the number of seeds set and the aboveground growth estimated by rosette diameter in high-NPQ genotypes is reported in A. thaliana. Such improvements are observed only under growth conditions in which plants are highly illuminated at least for some time within each day (7, 28). This observation has led to the idea that NPQ is a tolerance mechanism against sunflecks. A. thaliana is often grown for experimental purposes under low illumination, e.g., approximately 10% of full sunlight (7). NPQ is only slightly induced under such low illumination, and it is a natural consequence that differences in NPQ capacity would not cause differences in growth under low light conditions. Rice is normally cultivated under high natural illumination, and NPQ is a physiological mechanism that always regulates the conversion of light energy in rice during the day. NPQ was significantly induced under medium and high light intensities in the rice cultivars as demonstrated by our work, thus confirming this idea. Although further studies would be needed, inferior growth and seed set in the psbS1 mutant (Fig. S3A) (21) suggests that NPQ capacity affects rice growth and grain yield significantly.
A 2.7-kb deletion in genomic DNA was found in Indica cultivars, which was upstream of the OsPsbS1 gene. This deletion is highly correlated with NPQ capacity, namely, cultivars without deletion have high NPQ values whereas cultivars with this deletion have low ones. The deletion may decrease the expression of the OsPsbS1 gene in Indica cultivars, which in turn leads to decreased NPQ capacity. In fact, accumulation of the PsbS protein is lower in Habataki compared with Sasanishiki. We speculate that higher OsPsbS1 expression levels result in the higher NPQ capacity of Japonica compared with Indica. Identification of the adaptation mechanisms to high light levels in rice plants overexpressing OsPsbS1 will be the subject of further study.
Materials and Methods
Plant materials, chlorophyll fluorescence, QTL analysis, plasmid preparation, rice callus transformation, mRNA accumulation, genomic DNA and cDNA sequencing, genomic DNA PCR amplification, and Western blot analysis of PsbS are described in detail in SI Materials and Methods. In the text, sequential descriptions of the materials and methods corresponding to each result are reported. For respective methods, references of original sources are added.
Supplementary Material
Acknowledgments
Rice materials and cDNA clones were provided by the Rice Genome Resource Center, Gene Bank of the National Institute of Agrobiological Sciences, and Pohang University of Science and Technology. The Kasalath DNA sequence was provided by Dr. Takashi Matsumoto. We are also grateful to Seiichi Toki, Hiroaki Saika, and Toshiki Ishikawa for advice and technical assistance in rice transformation. This work was supported by Ministry of Agriculture, Forestry and Fishery, Japan, Rice Genome Project Grant IPG0014; the Program for Promotion of Basic and Applied Researchers for Innovations in Bio-oriented Industry; the Funding Program for Next Generation World-Leading Researchers (NEXT Program); and grants from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. G.A. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104809108/-/DCSupplemental.
References
- 1.Lazár D. Chlorophyll a fluorescence induction1. Biochim Biophys Acta. 1999;1412:1–28. doi: 10.1016/s0005-2728(99)00047-x. [DOI] [PubMed] [Google Scholar]
- 2.Heldt HW. Plant Biochemistry. 3rd Ed. London: Elsevier; 2005. The use of energy from sunlight by photosynthesis is the basis of life on earth. [Google Scholar]
- 3.Baker NR. Chlorophyll fluorescence: A probe of photosynthesis in vivo. Annu Rev Plant Biol. 2008;59:89–113. doi: 10.1146/annurev.arplant.59.032607.092759. [DOI] [PubMed] [Google Scholar]
- 4.Kramer DM, Johnson G, Kiirats O, Edwards GE. New fluorescence parameters for the determination of QA redox state and excitation energy fluxes. Photosynth Res. 2004;79:209–218. doi: 10.1023/B:PRES.0000015391.99477.0d. [DOI] [PubMed] [Google Scholar]
- 5.Kasajima I, Takahara K, Kawai-Yamada M, Uchimiya H. Estimation of the relative sizes of rate constants for chlorophyll de-excitation processes through comparison of inverse fluorescence intensities. Plant Cell Physiol. 2009;50:1600–1616. doi: 10.1093/pcp/pcp102. [DOI] [PubMed] [Google Scholar]
- 6.Quick WP, Stitt M. An examination of factors contributing to non-photochemical quenching of chlorophyll fluorescence in barley leaves. Biochim Biophys Acta. 1989;977:287–296. [Google Scholar]
- 7.Külheim C, Ågren J, Jansson S. Rapid regulation of light harvesting and plant fitness in the field. Science. 2002;297:91–93. doi: 10.1126/science.1072359. [DOI] [PubMed] [Google Scholar]
- 8.Li XP, Gilmore AM, Niyogi KK. Molecular and global time-resolved analysis of a psbS gene dosage effect on pH- and xanthophyll cycle-dependent nonphotochemical quenching in photosystem II. J Biol Chem. 2002;277:33590–33597. doi: 10.1074/jbc.M204797200. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi H, et al. Chloroplast NAD kinase is essential for energy transduction through the xanthophyll cycle in photosynthesis. Plant Cell Physiol. 2006;47:1678–1682. doi: 10.1093/pcp/pcl029. [DOI] [PubMed] [Google Scholar]
- 10.Takahara K, et al. Metabolome and photochemical analysis of rice plants overexpressing Arabidopsis NAD kinase gene. Plant Physiol. 2010;152:1863–1873. doi: 10.1104/pp.110.153098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.International Rice Research Institute Rice basics. 2011. Available at: http://irri.org/about-rice/rice-facts/rice-basics.
- 12.Murchie EH, Niyogi KK. Manipulation of photoprotection to improve plant photosynthesis. Plant Physiol. 2011;155:86–92. doi: 10.1104/pp.110.168831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S. Genetic structure and diversity in Oryza sativa L. Genetics. 2005;169:1631–1638. doi: 10.1534/genetics.104.035642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li XP, et al. A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature. 2000;403:391–395. doi: 10.1038/35000131. [DOI] [PubMed] [Google Scholar]
- 15.Niyogi KK, Grossman AR, Björkman O. Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell. 1998;10:1121–1134. doi: 10.1105/tpc.10.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munekage Y, et al. PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell. 2002;110:361–371. doi: 10.1016/s0092-8674(02)00867-x. [DOI] [PubMed] [Google Scholar]
- 17.DalCorso G, et al. A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell. 2008;132:273–285. doi: 10.1016/j.cell.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 18.Niyogi KK, et al. Photoprotection in a zeaxanthin- and lutein-deficient double mutant of Arabidopsis. Photosynth Res. 2001;67:139–145. doi: 10.1023/A:1010661102365. [DOI] [PubMed] [Google Scholar]
- 19.Pogson B, McDonald KA, Truong M, Britton G, DellaPenna D. Arabidopsis carotenoid mutants demonstrate that lutein is not essential for photosynthesis in higher plants. Plant Cell. 1996;8:1627–1639. doi: 10.1105/tpc.8.9.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zulfugarov IS, et al. Dependence of reaction center-type energy-dependent quenching on photosystem II antenna size. Biochim Biophys Acta. 2007;1767:773–780. doi: 10.1016/j.bbabio.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 21.Koo HY, et al. The function of the PsbS protein in relation to non-photochemical energy dependent quenching in rice plants. In: van der Est A, Bruce D, editors. Photosynthesis: Fundamental Aspects to Global Perspectives. Lawrence, KS: Allen; 2004. [Google Scholar]
- 22.Cheng C, et al. Polyphyletic origin of cultivated rice: Based on the interspersion pattern of SINEs. Mol Biol Evol. 2003;20:67–75. doi: 10.1093/molbev/msg004. [DOI] [PubMed] [Google Scholar]
- 23.Ma J, Bennetzen JL. Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci USA. 2004;101:12404–12410. doi: 10.1073/pnas.0403715101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li XP, et al. Regulation of photosynthetic light harvesting involves intrathylakoid lumen pH sensing by the PsbS protein. J Biol Chem. 2004;279:22866–22874. doi: 10.1074/jbc.M402461200. [DOI] [PubMed] [Google Scholar]
- 25.Demming-Adams B, Adams WW. Capacity for energy dissipation in the pigment bed in leaves with different xanthophyll cycle pools. Aust J Plant Physiol. 1994;21:575–588. [Google Scholar]
- 26.Yin Y, et al. Photosystem II photochemistry, photoinhibition, and the xanthophyll cycle in heat-stressed rice leaves. J Plant Physiol. 2010;167:959–966. doi: 10.1016/j.jplph.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 27.Eskling M, Arvidsson PO, Åkerlund HE. The xanthophyll cycle, its regulation and components. Physiol Plant. 1997;100:806–816. [Google Scholar]
- 28.Logan BA, Terry SG, Niyogi KK. Arabidopsis genotypes with differing levels of psbS expression differ in photosystem II quantum yield, xanthophylls cycle pool size, and aboveground growth. Int J Plant Sci. 2008;169:597–604. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.