Abstract
The Saccharomyces cerevisiae 2-μm plasmid is a multicopy selfish genome that resides in the nucleus. The genetic organization of the plasmid is optimized for stable, high-copy propagation in host-cell populations. The plasmid's partitioning system poaches host factors, including the centromere-specific histone H3-variant Cse4 and the cohesin complex, enabling replicated plasmid copies to segregate equally in a chromosome-coupled fashion. We have characterized the in vivo chromatin topology of the plasmid partitioning locus STB in its Cse4-associated and Cse4-nonassociated states. We find that the occupancy of Cse4 at STB induces positive DNA supercoiling, with a linking difference (ΔLk) contribution estimated between +1 and +2 units. One plausible explanation for this contrary topology is the presence of a specialized Cse4-containing nucleosome with a right-handed DNA writhe at a functional STB, contrasted by a standard histone H3-containing nucleosome with a left-handed DNA writhe at a nonfunctional STB. The similarities between STB and centromere in their nucleosome signature and DNA topology would be consistent with the potential origin of the unusual point centromere of budding yeast chromosomes from the partitioning locus of an ancestral plasmid.
Keywords: CEN evolution, nucleosome topology, reversome
The 2-μm plasmid of Saccharomyces cerevisiae resides in the nucleus at 40 to 60 copies per cell, and propagates itself with nearly the same stability as chromosomes (1, 2). The plasmid is a benign selfish DNA element that seems to provide no advantage to its host, but poses, at its normal copy number, no serious disadvantage either. In haploid cells the plasmid is organized into a cluster of three to five foci, and segregates as a cluster (3). This effective reduction in copy number necessitates an active partitioning system, comprised of two plasmid-coded proteins, Rep1 and Rep2, and a cis-acting partitioning locus STB, to ensure equal or nearly equal plasmid segregation. A decline in plasmid copy number because of rare missegregation events is corrected via DNA amplification triggered by the Flp site-specific recombination system harbored by the plasmid (4, 5). Positive and negative regulatory circuits implemented through the plasmid-coded Raf1 protein and the Rep proteins ensure quick amplification response without the danger of runaway increase in copy number (6–8).
The Rep-STB system channels several host factors involved in chromosome segregation toward the execution of plasmid segregation. These factors include the mitotic spindle, the spindle-associated motor Kip1, the RSC2 chromatin-remodeling complex, the centromere-specific histone H3-variant Cse4 (CenH3), and the yeast cohesin complex (9–15). The de novo assembly of the plasmid-partitioning complex at STB during the G1-S window of each cell cycle (9, 12, 14, 16) is reminiscent of the assembly of the kinetochore complex at centromeres. However, kinetochore components have not been detected at STB by ChIP (13).
The assembly of the plasmid-partitioning complex culminates in the recruitment of cohesin at STB, which pairs sister-plasmid molecules topologically by forming a protein ring around them (11, 13). Several lines of circumstantial evidence are consistent with a chromosome-hitchhiking mechanism for plasmid segregation (3, 10, 13). A plausible scenario invokes replicated sister-plasmid clusters, bridged by cohesin, being tethered to sister chromatids (which are also paired by cohesin). Dissolution of the cohesin bridge in anaphase would then trigger the segregation of sister clusters in unison with sister chromatids.
The association of Cse4, regarded as the signature nucleosome component at the centromere (CEN) (17–19), with STB is intriguing. Furthermore, the presence of Kip1 and the RSC2 complex, as well as the assembly of the cohesin complex, at STB raises the possibility of a potential evolutionary link between the two (11, 12, 20) (see Discussion). We now demonstrate that the STB chromatin, in its functional state in vivo, contributes a nonstandard positive DNA supercoil. This topological equivalence to CEN (21) authenticates the occupancy of STB by a Cse4-containing nucleosome. The plasmid-partitioning complex organized at STB and the kinetochore assembled at CEN may help preserve the Cse4-containing histone core particles at these loci, and potentially foster a reversed chirality with which DNA wraps around such particles.
Our findings lend credence to a possible origin of the unusual point centromere of budding yeast chromosomes by domestication of the STB locus of an ancestral 2-μm related plasmid (20). These findings also speak to the current debate on whether CenH3-containing nucleosomes engender the normal left-handed, or the opposite right-handed DNA writhe (21–23). The in vivo topology of STB revealed in the present investigation and that of CEN deduced from an earlier study (21) are consistent with the right-handed writhe.
Results
STB Chromatin Contributes Positive DNA Supercoiling in Its Functional State.
S. cerevisiae plasmids containing a CEN sequence shift to a higher level of negative supercoiling in vivo when CEN function is inactivated, and by inference, in the absence of a functional Cse4-containing nucleosome (21). The linking difference (ΔLk) between the active and inactive states is close to +2 for one copy of CEN, and roughly doubles with an additional copy of CEN. This difference is consistent with one unit of positive DNA writhe introduced by a single Cse4-containing nucleosome, changing to an equivalent negative writhe when it is replaced by a standard histone H3-containing nucleosome (ΔLk = +1 – (−1) = +2).
Although current evidence is consistent with Cse4 being a nucleosome component at STB (12, 24), a nonnucleosomal role for Cse4 in STB function cannot be ruled out. Cse4 may indeed localize, at low abundance, to highly expressed genes or repeated DNA elements, apparently without engendering kinetochore assembly (25, 26). Because of its rarity, positively supercoiled DNA at STB, if observed, would provide strong support for its occupancy by a CEN-like Cse4 nucleosome or nucleosomes. We therefore undertook a series of topological analyses of the DNA at the STB chromatin in its functional and nonfunctional states.
During characterization of the interaction between Cse4 and the 2-μm plasmid or high-copy STB reporter plasmids, we had previously noticed a significantly lower than molar stoichiometry of Cse4 with respect to STB (24). However, when the copy number of an STB reporter plasmid was reduced to one or close to one (10), Cse4-STB association could be made nearly quantitative. The present assays were performed using nearly or strictly “single”-copy versions of the reporter plasmids (Fig. 1 and SI Appendix, Figs. S2 and S4).
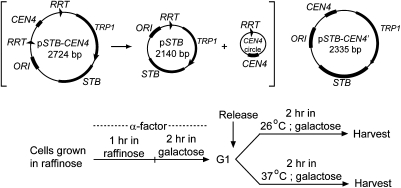
Fig. 1.
Reporter plasmids and protocols for cell-cycle arrest and release. (Upper) The reporter plasmids pSTB-CEN4 and pSTB-CEN4′ used in topology assays are drawn schematically. Recombination by the R recombinase within pSTB-CEN4 would resolve it into the pSTB plasmid plus the CEN4 circle. (Lower) The experimental regimen for obtaining cells for analyses of plasmid topology is schematically indicated. Raffinose was replaced by galactose in G1-arrested cells to induce expression of the R recombinase or the Rep proteins or both, as required by individual assays. Cells were normally released from G1 at 26 °C; they were released at 37 °C to inactivate Ndc10 in the ndc10-1 strain. The predominant fraction of cells was in metaphase, large budded with a single nucleus near the bud neck, at the time of harvest.
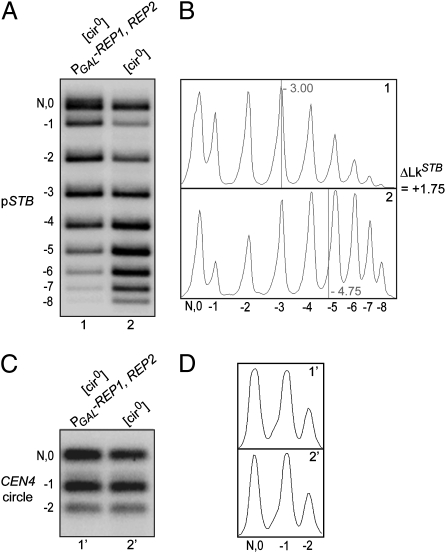
The ∼2.7-kbp pSTB-CEN4 plasmid (Fig. 1), with CEN flanked by two direct copies of the R recombinase target sites, was used in the experiments depicted in Fig. 2. The plasmid was introduced into two essentially isogenic [cir0] strains, both harboring two integrated copies of the R recombinase gene under GAL promoter control, but only one containing a GAL promoter-driven REP1-REP2 expression cassette. Galactose induction would result in the excision of the CEN4-containing small circle (584 bp) (Fig. 1 and SI Appendix, Fig. S1) in both strains, whereas it would also activate STB function in the strain containing the REP1-REP2 cassette. One could thus address the topology of the STB-containing large circle (pSTB plasmid; 2,140 bp) in the STB-active and STB-inactive states. The actual copy number of the “pseudosingle” copy pSTB was estimated to be 2.01 ± 0.61 (SI Appendix, Fig. S2). Electrophoresis in a chloroquine-gel disclosed a ΔLk of +1.75 conferred on pSTB by an active STB (Fig. 2 A and B). The average ΔLk from independent experiments was +1.68. In contrast, the status of STB made no apparent difference to the topology of the CEN circle (Fig. 2 C and D).
Fig. 2.
Topological distributions of a single-copy STB reporter plasmid when STB is maintained functional or nonfunctional. The pSTB-CEN4 reporter plasmid (Fig. 1) was resolved into pSTB and CEN4 circle in G1, and their topologies were analyzed in the ensuing metaphase. After electrophoresis in a 1.5% agarose gel (0.3 μg/mL chloroquine), the pSTB (A) and CEN4 circle (C) bands were revealed by Southern analysis. The topological distributions in A and C are plotted in B and D, respectively. The “0” topoisomer, because of potential overlap with the nicked circle (N), was omitted in deriving their centers (vertical lines).
The observed ΔLk is accommodated by the loss of a Cse4 nucleosome upon inactivation of STB, followed by the organization of a standard histone H3 nucleosome as its replacement (ΔLk = (+1) – (−1) = +2). This interpretation would bring STB in conformity with the explanation posited for the change in writhe from positive to negative between active and inactive forms of CEN (21). However, alternative mechanisms for Cse4-assisted sequestration of net positive supercoiling at STB cannot be excluded (see Discussion).
Positive Writhe of STB Chromatin Is Directly Correlated to its Occupancy by Cse4.
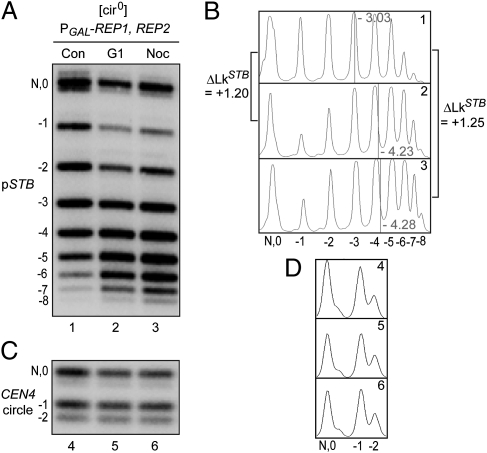
Our previous work showed that STB is free of Cse4 during G1, or when the mitotic spindle is depolymerized (12). In contrast, CEN is occupied by Cse4 in G1-arrested cells or those lacking the spindle (12, 27). We exploited these differences between CEN and STB in their association with Cse4 to further characterize the contribution of Cse4 to the topological status of STB. Control metaphase cells revealed a ΔLk of +1.20 and +1.25 for the pSTB plasmid relative to G1 cells and nocodazole-treated cells, respectively (Fig. 3 A and B). No such difference was noted for the CEN4 circle (Fig. 3 C and D).
Fig. 3.
Topologies of the pSTB plasmid in single copy state in G1 arrested or nocodazole-treated cells. (A–D) The single-copy pSTB and CEN4 circle were generated in G1 by recombination (see Fig. 2). Cells were released from G1 in galactose with the addition of nocodazole (Noc) or without nocodazole as the control (Con). Topology analyses were performed in G1 arrested cells (lane 2) or G2/M cells (Con, lane 1; Noc, lane 3).
The strong correlation between the positive writhe induced by STB DNA and conditions that foster its occupancy by Cse4 suggests that a Cse4-containing nucleosome is responsible for this unique topology.
Magnitudes of the Positive DNA Writhe at CEN and STB.
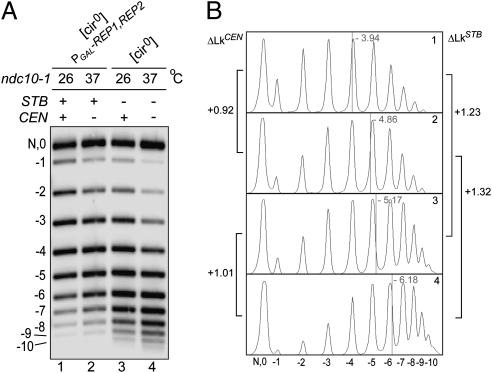
To estimate the ΔLk contribution of STB in reference to CEN, we subjected a reporter plasmid harboring both STB and CEN (pSTB-CEN4′) (Fig. 1) to topological analysis without dissociating CEN from it (Fig. 4). The ndc10-1 (Ts) mutation was used to switch CEN from its active (26 °C) to inactive (37 °C) state.
Fig. 4.
Individual contributions of STB and CEN to the topology of a reporter plasmid harboring both loci. (A and B) The indicated strains harboring the pSTB-CEN4’ reporter plasmid (Fig. 1) were arrested in G1 at 26 °C, conditioned with galactose, and released in galactose at 26 °C or 37 °C. Plasmid topologies were assayed at 2 h after release.
The topological contribution of CEN was derived from topoisomer distributions of adjacent lanes (denoting the shift from 26 °C to 37 °C); that of STB from topoisomer distributions of alternate lanes (denoting the presence or absence of Rep proteins) (Fig. 4). As shown in Fig. 4, the ΔLk resulting from CEN alone was +0.92 (lanes 1 and 2) or +1.01 (lanes 3 and 4); that resulting from STB was +1.23 (lanes 1 and 3) or +1.32 (lanes 2 and 4). The sum of the STB and CEN ΔLk contributions was +2.24 (lanes 1 and 4).
Our estimates of ΔLkCEN from the ndc10-1 strain are smaller than those previously reported (21). Any change in Lk may be partitioned in different ways between writhe (crossings of the helix axis; Wr) and twist (crossings between the two DNA strands; Tw). The plasmid topology displayed in vitro reflects the net balance of Wr and Tw in vivo as a result of protein binding (e.g., nucleosome assembly), the operation of protein machines (e.g., transcription), and the action of DNA relaxing enzymes (topoisomerases). Subtle effects of the ndc10-1 mutation per se and the temperature shift on plasmid topology, as well as potential incomplete inactivation of Ndc10 under the nonpermissive conditions, might account for less-than-expected ΔLk values. Possible interactions between CEN and STB, causing some degree of negative cooperativity in Cse4 recruitment, cannot be ruled out either. A small error is introduced in ΔLk estimates by omitting the “0” (relaxed) DNA band, which could potentially overlap with the nicked plasmid, in marking the centers of topoisomer distributions.
Despite the caveats noted above, the overall similarity between CEN and STB in the sign (+) and magnitude (between 1 and 2) of their ΔLk contributions is apparent. Based on the data from Fig. 3 and table 2 of Furuyama and Henikoff (table 2 in ref. 21), the CEN-induced positive-DNA writhe ranges from 1.33 to 1.85, with a mean of 1.59. In our experiments (Figs. 2–4), STB-promoted positive supercoils vary from 1.20 to 1.75, the mean value being 1.35 (SI Appendix, Table S1).
ΔLk Between Active and Inactive Forms of CEN and STB Estimated After Separation of the Two Loci from the Parent Plasmid Harboring Them.
The interpretations of the results in Fig. 4 were based on the assumption that the observed Lk changes in the reporter plasmid under conditions that maintained CEN or STB or both in their active and inactive states were directly attributable to the individual topologies of these loci. To verify the validity of this assumption, we first unlinked CEN and STB from each other into the pSTB plasmid and the CEN4 circle by recombination within pSTB-CEN4 (as described earlier) (Fig. 2), and then examined the topologies of the two circles separately (SI Appendix, Fig. S3).
The topology of pSTB was shifted toward higher negative supercoiling in the [cir0] background (SI Appendix, Fig. S3, lanes 3 and 4), lacking Rep1 and Rep2, regardless of the temperature, 26 °C or 37 °C. Conversely, the CEN circle became more negatively supercoiled at the nonpermissive temperature (37 °C) (SI Appendix, Fig. S3, lanes 6 and 8), irrespective of the presence or absence of the Rep proteins.
The lower estimates of ΔLk for STB in these assays was likely a result of the shadow bands above and below the −1 and −2 topoisomer positions, the identities of which have not been determined. The slower migrating bands could potentially be the result of a small fraction of the parent plasmid that escaped recombination, and deletion of the CEN circle. The possible source of the faster migrating bands is unknown. The intensities of these spurious bands relative to those of the authentic ones were significant in lanes 3 and 4, broadening the −1 and −2 topoisomer peaks and artificially skewing the distribution to be more positive.
Thus, the independent CEN and STB topologies assayed in the pSTB plasmid and the CEN4 circle are consistent, at least qualitatively, with the composite CEN and STB topologies assayed in their linked state in the pSTB-CEN4 plasmid.
Topology of STB Harbored by an Exact Single-Copy Plasmid.
The copy number of a CEN-containing plasmid only approaches, and is not precisely, 1. As a result, even within a single cell, there might be a distribution in the occupancy by Cse4 of an STB reporter plasmid derived from a CEN-containing precursor plasmid. However, the estimated copy number of the pSTB reporter plasmid assayed in Fig. 1 was ∼2, not significantly different from unity (SI Appendix, Fig. S2). The association of Cse4 with STB is limited to a region proximal to the 2-μm circle replication origin (STB-proximal), comprising five tandem repeats of a 60-bp consensus element (24). Furthermore, a subset of three of these repeats is necessary and sufficient for Cse4 recruitment. Because of the limited size of the DNA region (a maximum of ∼300 bp) to which Cse4 localizes, the potential distribution of Cse4 nucleosomes per plasmid molecule can only be 0, 1, or 2 (see Discussion). As discussed earlier, Cse4 association with the nearly single-copy reporter plasmids used for the topology tests is nearly quantitative. Thus, the “0” class must be negligible. To unequivocally eliminate potential uncertainties introduced by copy-number variations, we have determined the topology of STB maintained precisely at one copy in every cell.
We generated unit-copy circular molecules of pSTB by R recombinase-mediated excision of its linear form integrated into chromosome XV at the HIS3 locale (SI Appendix, Fig. S4). Strategies for pSTB excision in G1 and subsequent topological analysis in metaphase were analogous to those described for the assays using the CEN-STB reporter plasmids. The ΔLk for STB between the Rep1-Rep2 complemented and noncomplemented conditions was estimated as +1.37, nearly the same as that obtained for pseudosingle copy reporter plasmids (+1.35 ± 0.23) (SI Appendix, Table S1).
The concordance of the topological status of STB between its plasmid and integrated forms argues against complications introduced by a distribution of STB copy number within the experimental cell population.
Discussion
The present study reveals an unanticipated positive supercoiling of DNA at the STB locus of the 2-μm plasmid, analogous to the topology recently reported for the yeast centromere (21). It furthermore addresses the potential role of a plasmid-partitioning locus in the origin of the noncanonical point centromere.
The simplest explanation for our results, without invoking significant change in DNA twist, is the trapping of a positive writhe because of one Cse4-containing nucleosome present at STB. As noted earlier, Cse4 is confined to a region of STB composed of five 60-bp tandem repeats, three of which suffice for Cse4 occupancy (24). Based on CEN DNA length (∼125 bp), the likely nucleosome density at STB cannot be more than two. The conserved sign and comparable magnitudes of the Lk contributions by CEN and STB are most parsimoniously accounted for by the same Cse4 stoichiometry: namely, one nucleosome at each of the two loci. However, the possibility that the positive-DNA writhe at STB is constrained collaboratively by an adjacent pair of Cse4-nucleosomes cannot be ruled out entirely.
Centromere Identity: DNA Topology or Nucleosome Structure?
The right-handed DNA writhe of the Cse4-containing nucleosome observed in vitro and implied in vivo (21) suggests a unique topological mechanism for centromere identity. However, it is not certain that the reversed DNA topology applies universally to CenH3-containing nucleosomes. The crystal structure of the subnucleosomal human (CENPA-H4)2 tetramer suggests that physical features, rather than DNA topology, provide the signature of a centromeric nucleosome (23). These features include differences between CENPA-CENPA and H3-H3 interfaces, as well as those between CENPA-H4 and H3-H4 interfaces. In addition, there is a charge reversal within a protruding loop between CENPA and H3. Strikingly, in vitro assembled CENPA-containing nucleosomes induce the conventional negative writhe in DNA. Depending on assembly conditions, DNA can be wrapped in a left- or right-handed fashion around Cse4-containing nucleosomes in vitro (21, 22).
The stoichiometry of the core particle in the centromeric nucleosome is also under debate. Experimental evidence in favor of a histone tetramer [hemisome; H2A/H2B/H4/(CenH3 = Cse4)] or a histone octamer (H2A/H2B/H4/CenH3/CenH3/H4/H2B/H2A) has been reported (22, 25, 28–30). The specter of an unusual hexameric nucleosome core, bereft of H2A and H2B, but including the Cse4-associated nonhistone protein Scm3 [H4/Cse4/Cse4/H4-(Scm3)2], has also been raised (31).
The aforementioned uncertainties notwithstanding, the topological evidence for a finite reduction in negative superhelicity, in vivo in yeast, by a functional CEN is well documented (21). Results from our present study signify a similar attribute of STB when it is associated with Cse4. While this special topology is consistent with positive DNA supercoiling at a Cse4-nucleosome, a recently proposed alternative explanation suggests the possible absence of native nucleosomes (or negative supercoils) adjacent to a Cse4 containing nucleosome, which also harbors negative DNA writhe (32). However, nucleosome mapping data for centromere flanking regions do not support this notion of nucleosome depletion (17, 33). The topology results for plasmid substrates containing two copies of CEN argue against a negative DNA writhe for the Cse4 nucleosome. The number of H3 nucleosomes required to give the ΔLk observed in the absence of Cse4 would be too many to be accommodated within the sizes of the plasmids employed (21). Overall, our results would be consistent with a right handed DNA wrap induced at STB by a Cse4 containing hemisome or some other nucleosome core organization that traps this contrary DNA writhe (a reversome).
Models for the Positive Writhe of an Active STB Chromatin.
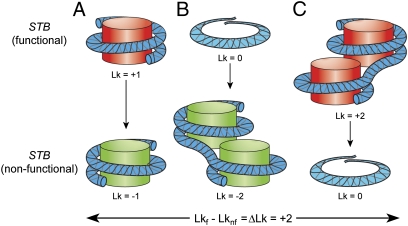
The observed decrease in Lk of 1.35 ± 0.23 (SI Appendix, Table S1) when STB is not occupied by a Cse4 nucleosome is less than the predicted value of 2 if an H3 nucleosome were to take its place quantitatively (Fig. 5A). Perhaps the AT-richness of STB may pose some impediment to nucleosome assembly, resulting in incomplete replacement.
Fig. 5.
Change in DNA topology between functional and nonfunctional STB. A ΔLk of ∼|2| because of the transition of STB between its functional (Lkf) and nonfunctional (Lknf) states may be accommodated in one of at least three ways (A–C). (A) The right-handed DNA wrap around a Cse4-containing nucleosome (red) switches to the standard left-handed wrap around a histone H3-containing nucleosome (green). (B) A nucleosome-free, functional STB switches to a nonfunctional state occupied by two histone H3-containing nucleosomes. (C) Loss of two Cse4-containing nucleosomes, each with a positive-DNA writhe, from a functional STB generates a nucleosome-free, nonfunctional STB. These models assume that the standard twist of B-form DNA is not significantly altered when it is occupied by histone H3- or Cse4-containing nucleosomes.
A formal possibility that a functional STB is nucleosome-free, and a nonfunctional STB accommodates up to two standard nucleosomes (Fig. 5B), is unlikely. STB function depends on its association with Cse4, presumably as a nucleosome component, in a Rep1-Rep2–assisted manner (12, 24, and present study). Furthermore, G1 arrest and spindle disassembly, conditions that do not foster Cse4-STB association (12) but sustain Rep-STB association (3, 9, 14, 16), decrease Lk by the anticipated amount (Fig. 3 A and B).
An alternative scenario for ΔLk = |2| is the presence of two Cse4 nucleosomes at a functional STB and none, Cse4-, or H3-nucleosome, at a nonfunctional STB (Fig. 5C). In principle, two Cse4 hemisomes may jointly constrain one positive writhe, which will be converted to one negative writhe if only one H3 nucleosome substitutes for their absence. In light of the stringent regulation of Cse4 in the nucleus by protein turnover (34–36), its absence from the majority of STB plasmids in their multicopy state (24), and its quantitative occupancy of low-copy plasmids, we favor a functional stoichiometry of one Cse4 nucleosome per STB.
Ancestral STB as the Potential Source of the Budding Yeast Point Centromere.
Eukaryotic centromeres, in general, comprise long DNA stretches, display little or no sequence consensus, and are epigenetically specified (20). Neo-centromeres can arise at chromosomal locales where none existed previously (37, 38). Machineries for RNA interference and heterochromatin formation are important in the establishment of these “regional” centromeres (39). In contrast, the S. cerevisiae point centromere is quite short (∼125 bp), genetically defined, and has a characteristic tripartite DNA organization (40, 41). Point centromeres are unique to members of the Saccharomycetaceae fungal lineage that have lost all or nearly all of the protein components of the RNAi and heterochromatin machineries (42). This lineage also stands apart from the rest of eukaryotes in harboring autonomously replicating plasmids analogous to the 2-μm plasmid. These evolutionary propinquities (20), together with the anomalous positive DNA writhe induced by a functional CEN or STB through a nonstandard nucleosome, argue for the possible origin of CEN from an ancestral STB.
Coupling Between 2-μm Plasmid and Chromosome Segregation Pathways.
The present-day segregation machineries of the budding yeast chromosomes and the 2-μm plasmid likely signify two distinct (and nonconflicting) solutions for stable propagation arrived at by divergence from the same start point. The common components of the two pathways are likely vestiges of their shared evolutionary history. By this reasoning, the scaffold proteins that support the assembly of the kinetochore and plasmid-partitioning complexes must also be bonded by a common ancestry. It is not surprising that this suspected evolutionary kinship is not discernible at the extant DNA or protein sequence levels. The penalty for bearing the plasmid burden (43, 44), however small, would have driven evolution of the chromosome segregation machinery away from the plasmid-partitioning system.
The conservation of Cse4 association and cohesin assembly at CEN and STB perhaps connotes the plasmid's counter-strategy to ensure stable propagation through chromosome-coupled segregation. It is noteworthy that components of the inner kinetochore complex, such as Ndc10 and Ctf13, have no homologs outside of Saccharomycetaceae among fungi and other eukaryotes (20). Similarly, Rep1 and Rep2 homologs are also confined to the family of 2-μm related plasmids. The Rep2 proteins have scant homology among them, perhaps signifying their rapid evolution as an adaptive response to their respective host environments.
Can Nucleosomes Switch Between Left- and Right-Handed DNA Chirality?
Right-handed DNA writhe observed in (H3-H4)2 tetrasomes in vitro (45, 46) suggests a mechanism for the entrapment of a metastable right-handed turn per “reversome” particle in nucleosome arrays subjected to large positive torsional stress (47, 48). In vivo, compensatory positive supercoils generated, for example, as a result of transcription could potentially trigger the flip from nucleosome to reversome. The associated energetic cost may be alleviated with the help of histone chaperones as well as high-order protein interactions: for example, those provided by the kinetochore complex or the plasmid-partitioning complex. Some degree of chiral heterogeneity among Cse4-containing nucleosomes may account for experimentally observed ΔLk of < 2 in the absence of functional Cse4.
The chiral ambiguities of in vitro assembled Cse4 nucleosomes (21, 22) likely arise from conditions that induce alternative histone stoichiometries, modulate their stacking preferences, and promote DNA contacts that are permissive of either left- or right-handed DNA writhe. The deduced in vivo topologies, protected against such vagaries perhaps by the stabilizing influence of the kinetochore and plasmid partitioning complexes, are unlikely to distort the authentic chirality of CEN or STB chromatin.
Materials and Methods
Plasmids.
Reporter plasmids are schematically diagrammed in Fig. 1 and their constructions described in the SI Appendix.
DNA Topology Assays.
The general protocols were based on those described by Furuyama and Henikoff (21) with appropriate modifications. DNA samples were run at 4 °C in 1.5% agarose gels containing 0.3 μg/mL chloroquine (1× TBE buffer; 0.3 μg/mL chloroquine; 2.5 V/cm). The duration of electrophoresis was ∼48 h for resolution of plasmid (∼2,000 bp) topoisomers and ∼24 h for resolution of small circle (∼600 bp) topoisomers. Following Southern hybridization, band intensities were quantitated using a Typhoon Trio phosphorimager and ImageQuant software (GE Healthcare).
Miscellaneous protocols.
Routine experimental protocols, such as bacterial and yeast transformations, total yeast DNA and plasmid DNA preparations, curing of native 2-μm circles from [cir+] yeast strains, and so forth are available upon request.
Supporting Information.
Two tables, one summarizing topological analyses (SI Appendix, Table S1), the other listing experimental strains (SI Appendix, Table S2), four figures (SI Appendix, Figs S1–S4), and relevant details of reporter plasmids are included in the SI Appendix.
Supplementary Material
Acknowledgments
We thank Sue Biggins, Munira Basrai, and Kim Nasmyth for providing us with plasmids and yeast strains, and S. Henikoff, B. Black, S. Sawyer, and members of our group for helpful comments. This work was supported by funds from the National Science Foundation (MCB-1049925); partial support was provided by the Robert F. Welch Foundation award F-1274.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101944108/-/DCSupplemental.
References
- 1.Ghosh SK, Hajra S, Paek A, Jayaram M. Mechanisms for chromosome and plasmid segregation. Annu Rev Biochem. 2006;75:211–241. doi: 10.1146/annurev.biochem.75.101304.124037. [DOI] [PubMed] [Google Scholar]
- 2.Jayaram M, Yang X-M, Mehta S, Voziyanov Y, Velmurugan S. The 2 micron plasmid of Saccharomyces cerevisiae. In: Funnel BE, Phillips G, editors. Plasmid Biology. Washington, DC: ASM Press; 2004. pp. 303–324. [Google Scholar]
- 3.Velmurugan S, Yang X-M, Chan CS, Dobson M, Jayaram M. Partitioning of the 2-microm circle plasmid of Saccharomyces cerevisiae. Functional coordination with chromosome segregation and plasmid-encoded Rep protein distribution. J Cell Biol. 2000;149:553–566. doi: 10.1083/jcb.149.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Futcher AB. Copy number amplification of the 2 micron circle plasmid of Saccharomyces cerevisiae. J Theor Biol. 1986;119:197–204. doi: 10.1016/s0022-5193(86)80074-1. [DOI] [PubMed] [Google Scholar]
- 5.Volkert FC, Broach JR. Site-specific recombination promotes plasmid amplification in yeast. Cell. 1986;46:541–550. doi: 10.1016/0092-8674(86)90879-2. [DOI] [PubMed] [Google Scholar]
- 6.Murray JA, Scarpa M, Rossi N, Cesareni G. Antagonistic controls regulate copy number of the yeast 2 micron plasmid. EMBO J. 1987;6:4205–4212. doi: 10.1002/j.1460-2075.1987.tb02768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reynolds AE, Murray AW, Szostak JW. Roles of the 2 micron gene products in stable maintenance of the 2 micron plasmid of Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:3566–3573. doi: 10.1128/mcb.7.10.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Som T, Armstrong KA, Volkert FC, Broach JR. Autoregulation of 2 micron circle gene expression provides a model for maintenance of stable plasmid copy levels. Cell. 1988;52:27–37. doi: 10.1016/0092-8674(88)90528-4. [DOI] [PubMed] [Google Scholar]
- 9.Cui H, Ghosh SK, Jayaram M. The selfish yeast plasmid uses the nuclear motor Kip1p but not Cin8p for its localization and equal segregation. J Cell Biol. 2009;185:251–264. doi: 10.1083/jcb.200810130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh SK, Hajra S, Jayaram M. Faithful segregation of the multicopy yeast plasmid through cohesin-mediated recognition of sisters. Proc Natl Acad Sci USA. 2007;104:13034–13039. doi: 10.1073/pnas.0702996104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh SK, Huang CC, Hajra S, Jayaram M. Yeast cohesin complex embraces 2 micron plasmid sisters in a tri-linked catenane complex. Nucleic Acids Res. 2010;38:570–584. doi: 10.1093/nar/gkp993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hajra S, Ghosh SK, Jayaram M. The centromere-specific histone variant Cse4p (CENP-A) is essential for functional chromatin architecture at the yeast 2-microm circle partitioning locus and promotes equal plasmid segregation. J Cell Biol. 2006;174:779–790. doi: 10.1083/jcb.200603042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehta S, et al. The 2 micron plasmid purloins the yeast cohesin complex: A mechanism for coupling plasmid partitioning and chromosome segregation? J Cell Biol. 2002;158:625–637. doi: 10.1083/jcb.200204136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehta S, Yang XM, Jayaram M, Velmurugan S. A novel role for the mitotic spindle during DNA segregation in yeast: Promoting 2 microm plasmid-cohesin association. Mol Cell Biol. 2005;25:4283–4298. doi: 10.1128/MCB.25.10.4283-4298.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong MC, Scott-Drew SR, Hayes MJ, Howard PJ, Murray JA. RSC2, encoding a component of the RSC nucleosome remodeling complex, is essential for 2 microm plasmid maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:4218–4229. doi: 10.1128/MCB.22.12.4218-4229.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang XM, Mehta S, Uzri D, Jayaram M, Velmurugan S. Mutations in a partitioning protein and altered chromatin structure at the partitioning locus prevent cohesin recruitment by the Saccharomyces cerevisiae plasmid and cause plasmid missegregation. Mol Cell Biol. 2004;24:5290–5303. doi: 10.1128/MCB.24.12.5290-5303.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci USA. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keith KC, Fitzgerald-Hayes M. CSE4 genetically interacts with the Saccharomyces cerevisiae centromere DNA elements CDE I and CDE II but not CDE III. Implications for the path of the centromere dna around a cse4p variant nucleosome. Genetics. 2000;156:973–981. doi: 10.1093/genetics/156.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 20.Malik HS, Henikoff S. Major evolutionary transitions in centromere complexity. Cell. 2009;138:1067–1082. doi: 10.1016/j.cell.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 21.Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kingston IJ, Yung JS, Singleton MR. Biophysical characterization of the centromere-specific nucleosome from budding yeast. J Biol Chem. 2011;286:4021–4026. doi: 10.1074/jbc.M110.189340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang CC, Hajra S, Ghosh SK, Jayaram M. Cse4 (CenH3) association with the Saccharomyces cerevisiae plasmid partitioning locus in its native and chromosomally integrated states: Implications in centromere evolution. Mol Cell Biol. 2011;31:1030–1040. doi: 10.1128/MCB.01191-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camahort R, et al. Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lefrançois P, et al. Efficient yeast ChIP-Seq using multiplex short-read DNA sequencing. BMC Genomics. 2009;10:37. doi: 10.1186/1471-2164-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearson CG, et al. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr Biol. 2004;14:1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- 28.Dimitriadis EK, Weber C, Gill RK, Diekmann S, Dalal Y. Tetrameric organization of vertebrate centromeric nucleosomes. Proc Natl Acad Sci USA. 2010;107:20317–20322. doi: 10.1073/pnas.1009563107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dalal Y, Furuyama T, Vermaak D, Henikoff S. Structure, dynamics, and evolution of centromeric nucleosomes. Proc Natl Acad Sci USA. 2007;104:15974–15981. doi: 10.1073/pnas.0707648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 32.Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gkikopoulos, T, et al. The SWI/SNF complex acts to constrain distribution of the centromeric histone variant Cse4. EMBO J. 2011;30:1919–1927. doi: 10.1038/emboj.2011.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collins KA, Furuyama S, Biggins S. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr Biol. 2004;14:1968–1972. doi: 10.1016/j.cub.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 35.Hewawasam G, et al. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol Cell. 2010;40:444–454. doi: 10.1016/j.molcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ranjitkar P, et al. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell. 2010;40:455–464. doi: 10.1016/j.molcel.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amor DJ, et al. Human centromere repositioning “in progress”. Proc Natl Acad Sci USA. 2004;101:6542–6547. doi: 10.1073/pnas.0308637101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marshall OJ, Chueh AC, Wong LH, Choo KH. Neocentromeres: New insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet. 2008;82:261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allshire RC. RNA interference, heterochromatin, and centromere function. Cold Spring Harb Symp Quant Biol. 2004;69:389–395. doi: 10.1101/sqb.2004.69.389. [DOI] [PubMed] [Google Scholar]
- 40.Carbon J, Clarke L. Centromere structure and function in budding and fission yeasts. New Biol. 1990;2:10–19. [PubMed] [Google Scholar]
- 41.Cheeseman IM, Drubin DG, Barnes G. Simple centromere, complex kinetochore: Linking spindle microtubules and centromeric DNA in budding yeast. J Cell Biol. 2002;157:199–203. doi: 10.1083/jcb.200201052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aravind L, Watanabe H, Lipman DJ, Koonin EV. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc Natl Acad Sci USA. 2000;97:11319–11324. doi: 10.1073/pnas.200346997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Futcher AB, Cox BS. Maintenance of the 2 microns circle plasmid in populations of Saccharomyces cerevisiae. J Bacteriol. 1983;154:612–622. doi: 10.1128/jb.154.2.612-622.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mead DJ, Gardner DC, Oliver SG. The yeast 2 micron plasmid: Strategies for the survival of a selfish DNA. Mol Gen Genet. 1986;205:417–421. doi: 10.1007/BF00338076. [DOI] [PubMed] [Google Scholar]
- 45.Alilat M, Sivolob A, Révet B, Prunell A. Nucleosome dynamics. Protein and DNA contributions in the chiral transition of the tetrasome, the histone (H3-H4)2 tetramer-DNA particle. J Mol Biol. 1999;291:815–841. doi: 10.1006/jmbi.1999.2988. [DOI] [PubMed] [Google Scholar]
- 46.Hamiche A, et al. Interaction of the histone (H3-H4)2 tetramer of the nucleosome with positively supercoiled DNA minicircles: Potential flipping of the protein from a left- to a right-handed superhelical form. Proc Natl Acad Sci USA. 1996;93:7588–7593. doi: 10.1073/pnas.93.15.7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bancaud A, et al. Nucleosome chiral transition under positive torsional stress in single chromatin fibers. Mol Cell. 2007;27:135–147. doi: 10.1016/j.molcel.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 48.Lavelle C, et al. Right-handed nucleosome: myth or reality? Cell. 2009;139:1216–1217, author reply 1217–1218. doi: 10.1016/j.cell.2009.12.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.