Abstract
A chromatin code appears to mark introns and exons with distinct patterns of nucleosome enrichment and histone methylation. We investigated whether a causal relationship exists between splicing and chromatin modification by asking whether splice-site mutations affect the methylation of histone H3K36. Deletions of the 3′ splice site in intron 2 or in both introns 1 and 2 of an integrated β-globin reporter gene caused a shift in relative distribution of H3K36 trimethylation away from 5′ ends and toward 3′ ends. The effects of splice-site mutations correlated with enhanced retention of a U5 snRNP subunit on transcription complexes downstream of the gene. In contrast, a poly(A) site mutation did not affect H3K36 methylation. Similarly, global inhibition of splicing by spliceostatin A caused a rapid repositioning of H3K36me3 away from 5′ ends in favor of 3′ ends. These results suggest that the cotranscriptional splicing apparatus influences establishment of normal patterns of histone modification.
Keywords: pol II, pausing, transcription elongation
Spliceosomes assemble on nascent pre-mRNAs and a significant fraction of splicing occurs cotranscriptionally (1–5) in close proximity to the chromatin template. Several studies have uncovered intriguing correlations between nucleosome localization/modification and the intron-exon structure of genes or the alternative splicing of their transcripts (reviewed in ref. 6). Nucleosomes in internal exons have subtly elevated levels of H3K36me3 relative to adjacent introns, suggesting the existence of a splicing-associated code of histone methylation (7–10).
Cotranscriptional methylation of H3K36 by the SET2 family of methyltransferases is a hallmark of transcribed chromatin that has been implicated in control of transcription elongation, alternative splicing, and mRNA export (11–15). The H3K36me3 modification increases toward the 3′ end of genes and then falls sharply in 3′ flanking regions (16). It is not well understood how this distribution is established but it is noteworthy that the position of the 3′ end of the first intron correlates remarkably well with the beginning of the domain of H3K36 trimethylation (17). It has been suggested that H3K36 methylation is targeted to transcribed regions through association of SETD2 with the Ser2 phosphorylated C-terminal domain (CTD) of pol II in a complex stabilized by Spt6, Iws1, and Aly (13, 18). A critical question raised by these studies is whether a dedicated coupling mechanism functions to connect splicing with chromatin modification and, if so, whether chromatin modification affects splicing or vice versa. In support of the former possibility, numerous alternative splicing changes resulted from knock-down of SETD2 (14). One way in which chromatin modification could affect splicing is through binding of splicing factors to modified histones. A notable example is interaction of the splicing regulator polypyrimidine tract binding protein, PTB1, with the H3K36me3 binding protein, MRG15 (14). Another mechanism by which histone methylation might influence cotranscriptional splicing is by affecting the rate of transcription elongation (19).
Several investigations have implicated H3K36me3 in alternative splice site choice (7, 14, 15), but whether this modification directs changes in splicing remains controversial (17). A local increase in H3K36me3 correlated with exon skipping in neural cell adhesion molecule (15) and fibroblast growth factor receptor 2 pre-mRNAs (14). On the other hand, at CD45 and YPEL5, there is no correlation between alternative splicing and H3K36 trimethylation (17). The latter study suggested H3K36 methylation is linked not to the frequency of splice site use but rather to exon definition (17).
Most previous studies of the mechanism of cross-talk between splicing and chromatin structure have correlated global changes induced by pharmacological inhibitors, overexpression, or knockdown of chromatin factors, with altered splicing of individual genes (14, 20–22). These results are consistent with a “chromatin-affects-splicing” scenario based on the notion that establishment of a particular chromatin state at a particular place will subsequently influence splicing of nascent transcripts in the vicinity. The chromatin-affects-splicing model does not explain how chromatin modifications would be appropriately targeted to affect splicing. An alternative, but not mutually exclusive, idea is that splicing affects chromatin. According to this model, splicing factors acting at the gene can locally influence chromatin modifiers through protein:protein interactions. In this report, we tested the splicing-affects-chromatin model by asking whether splice-site mutations and the splicing inhibitor, spliceostatin A (SSA) (23), affect local chromatin modifications. Our results support this model by showing that splicing is necessary for establishment and/or maintenance of normal patterns of H3K36 trimethylation.
Results
Splice-Site Mutations Reposition H3K36me3 on β-Globin.
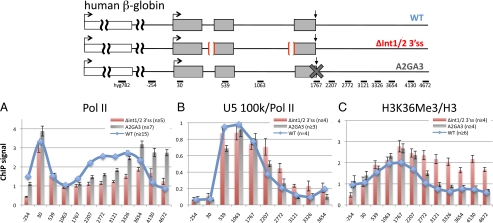
We asked whether mutation of splice sites in a chromosomal reporter gene affects methylation of H3K36. For these experiments, CHO cell lines were constructed with single-copy tet-inducible human β-globin genes integrated at a unique locus by site-specific recombination (Flp-In). To prevent splicing, we deleted the branch-point sequences and 3′ splice sites of both introns (Δint1/2 3′ss) (Fig. 1) and verified that splicing was indeed inhibited (SI Appendix, Fig. S1 A and C). As a control for nonspecific effects of disrupting mRNA processing, including transcript retention at the site of transcription (24, 25), we made an isogenic cell line with a point mutation in the β-globin poly(A) site (A2GA3) that disrupts 3′ end processing (SI Appendix, Fig. S1B). Transcription of the doxycycline-induced WT and mutant β-globin genes was monitored by anti-pol II ChIP at 12 amplicons along the gene (SI Appendix, Table S1). All ChIP signals were normalized to a control amplicon within the TK-hygromycin (hyg) resistance gene integrated upstream of the globin reporter (Fig. 1). The WT and mutant genes were transcribed at approximately equivalent levels, as shown by pol II recruitment to the transcription start site (TSS) (Fig. 1A). In common with many mammalian genes (26, 27, 30), pol II paused on the WT globin gene close to the TSS and downstream of the poly(A) site at the CoTC sequence (amplicons 2207–3326) (28) before termination (amplicons 4130, 4672). In contrast to a similar reporter gene with a mutant SV40 poly(A) site (29), the β-globin poly(A) site mutant did not markedly inhibit transcription initiation as measured by pol II ChIP at the TSS relative to the hyg control (Fig. 1A). The 3′ end pausing and termination were impaired in both the double splice-site mutant and the poly(A) site mutant (Fig. 1A).
Fig. 1.
Splicing inhibition alters distributions of pol II, U5 snRNP, and K36 methylated histone H3. (Upper) Map of the integrated tet-inducible CMV-human β-globin gene and the upstream hygromycin resistant gene (white box) in CHO cells with the Δint1/2 3′ss deletions and poly(A) site mutation (AAGAAA) marked. Arrows mark the TSS and the poly(A) site. PCR amplicons are indicated with their position relative to the TSS. Relative ChIP signals are shown for (A) total pol II normalized to the hygromycin gene integrated about 3-kb upstream of β-globin (hyg 782), (B) U5 100 K normalized to pol II, and (C) H3K36me3 normalized to H3, under doxycycline induced conditions. Means of n PCRs and SEMs are shown.
We determined whether the Δint1/2 3′ss mutant affected cotranscriptional recruitment of a splicing factor by ChIP of the 100 K U5snRNP subunit, huPrp28. ChIP signals obtained with anti-U5 100 K antibody (SI Appendix, Fig. S2) were normalized to pol II to control for variation in pol II density. On the WT β-globin gene, normalized U5 100-K occupancy was low at the TSS, then increased to a maximum in the body of the gene (amplicons +539–1767) and declined in the 3′ flanking region, consistent with previous results for a different U5 snRNP subunit (31) (Fig. 1B). Compared with the WT gene, the Δint1/2 3′ss mutant showed an enrichment of U5 100 K relative to pol II in the 3′ flanking sequence (amplicons 2207–3326). Importantly, this effect was specific to the splice site mutant and was not observed in the poly(A) site mutant (Fig.1B). The fact that deletion of the branch points and 3′ splice sites of introns 1 and 2 did not reduce U5 100-K ChIP signals is consistent with binding of U4/U5/U6 to a 5′ splice site in the absence of U2 and the branch point (32). The 3′ enrichment of U5 100 K when splicing was prevented suggests a defect in release of this factor, possibly because of formation of dead-end spliceosomal complexes.
Having established that the Δint1/2 3′ss mutant is transcribed at a similar level to the WT gene and that it alters cotranscriptional recruitment of a snRNP, we asked whether chromatin modification by H3K36 trimethylation was affected. For these experiments, anti-H3K36me3 ChIP signals were quantified relative to total histone H3 to control for variation in nucleosome occupancy. H3 occupancy was lowest at the promoter, consistent with a nucleosome-depleted region. Remarkably, in the Δint1/2 3′ss double splice-site mutant, H3K36me3 was enriched relative to WT in the 3′ flanking region but not in the body of the gene (Fig. 1C). Elevated H3K36me3 could in principle be caused by an elevation in occupancy of pol II, which can associate with the SETD2 methyltransferase (18). However, the 3′ flanking sequences where H3K36me3 is enriched correspond to the sequences where pol II density is actually lower than in the WT gene (see amplicons 2772–3654) (Fig. 1A). Therefore, the elevation of H3K36 methylation is not accounted for by an increase pol II density. It is also possible that the splice-site mutations altered H3K36 methylation because transcription termination was impaired; however, this possibility is eliminated by the fact that the poly(A) site mutation, which strongly inhibits termination, had little or no effect on H3K36me3 (Fig. 1C). We conclude that inhibition of splicing by double mutation of 3′ splice sites causes redistribution of H3K36me3 toward the 3′ end of the β-globin gene.
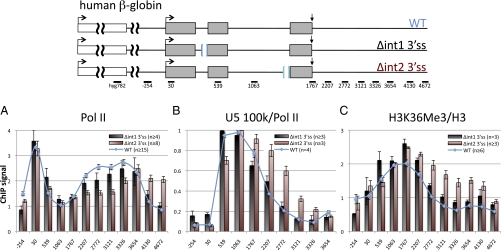
To distinguish whether splicing of one or both introns affects H3K36 trimethylation, we made isogenic CHO cell lines with mutant β-globin genes that delete the branch point and 3′ splice site of either intron 1 (Δint1 3′ss) or intron 2 (Δint2 3′ss). Both mutants inhibited splicing and the Δint2 3′ss mutant also partially inhibited poly(A) site cleavage (SI Appendix, Fig. S1 A and C). ChIP analysis of the Δint1 3′ss mutant revealed little or no detectable effect on the distribution of pol II, U5 100 K, or H3K36me3 relative to WT at the resolution of our analysis (Fig. 2). In contrast, the Δint2 3′ss mutant inhibited 3′ pausing and termination of transcription, as expected (30). Furthermore, the Δint2 3′ss mutant specifically enhanced U5 100-K occupancy in the 3′ flanking region (Fig. 2 A and B), similar to the Δint1/2 3′ss double splice-site mutant (Fig. 1 A and B). Importantly, the Δint2 3′ss mutant caused a specific enrichment of H3K36me3 relative to total H3 in the 3′ flanking region (Fig. 2C), reproducing the effect of the Δint1/2 3′ss double splice-site mutant. We do not fully understand why 3′ splice site mutations of introns 1 and 2 have different effects on histone modification; however, intron 1 is exceptionally short, and inactivation of intron 1 and intron 2 splicing affect other aspects of β-globin gene expression quite differently (25, 30, 33). In summary, we conclude that inhibition of intron 1+2 or intron 2 splicing alone is sufficient to modulate chromatin modification by enhancing the level of H3K36 trimethylation at the 3′ end of an integrated β-globin transcription unit. This effect is probably not a simple consequence of altered transcriptional pausing or termination and it is not correlated with increased pol II density in the region where H3K36me3 is elevated. Enhanced H3K36me3 in the 3′ flanking region of intron 2 3′ss mutants coincided fairly closely with enrichment of the splicing factor U5 100 K. Furthermore U5 100-K retention in the 3′ flank only occurred in the Δint2 and Δint1/2 3′ ss mutants but not in the Δint1 3′ ss or A2GA3 mutants, which had little effect on H3K36 methylation (Figs. 1 and 2).
Fig. 2.
Splicing of intron 2 is required for normal H3K36 methylation. (Upper) Maps of the integrated WT CMV β-globin gene and the intron1 and intron 2 3′ splice site deletions. Relative ChIP signals are shown for (A) pol II relative to the hyg 782 amplicon as in Fig. 1A, (B) U5 100 K normalized to pol II, and (C) H3K36me3 normalized to H3 as in Fig. 1. Means of n PCRs and SEMs are shown.
Widespread Redistribution of H3K36me3 Caused by the Splicing Inhibitor SSA.
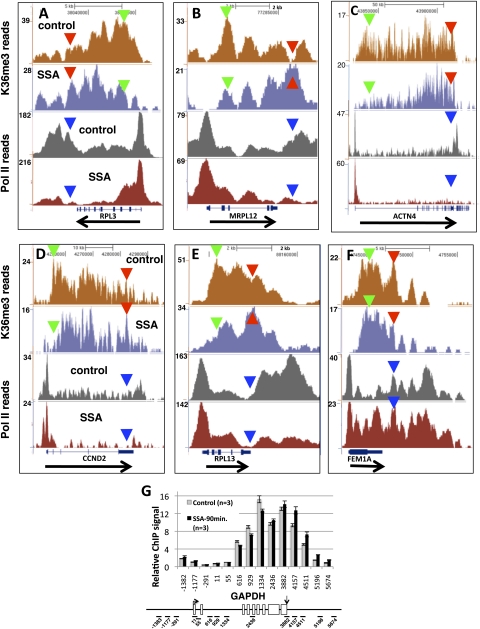
To determine whether splicing influences H3K36me3 patterning on genes other than β-globin, we compared genome-wide distributions of this histone modification before and after inhibition of splicing with SSA. SSA binds the U2 snRNP-associated factor SF3b and blocks spliceosome assembly after U2 binding (23, 33). We mapped H3K36me3 genome-wide by ChIP-seq in control HeLa cells and those treated with SSA for 12 h. SSA strongly inhibited splicing of c-myc transcripts in these cells as determined by RT-PCR (SI Appendix, Fig. S3G). In control cells, peaks of H3K36me3 enrichment often started near the second exon and extending further 3′ as previously observed (16, 17) (SI Appendix, Fig. S3). Remarkably, SSA caused a redistribution of the H3K36me3 mark within genes. This redistribution was characterized by a decrease in 5′ H3K36me3 ChIP signals (green arrowheads) relative to 3′ signals (red arrowheads) on many intron-containing genes (Fig. 3 A–E and SI Appendix, Fig. S4), but not on intronless genes (Figs. 3F and 4B). The SSA-induced 3′ shift in the profile of H3K36me3 did not correlate with a similar shift in pol II distribution (Fig. 3 A–E and SI Appendix, Fig. S4). As we observed for intron 2 splice-site mutants (Figs, 1A and 2A), SSA reduced relative pol II densities in 3′ flanking regions (blue arrowheads, Fig. 3 A–E and SI Appendix, Fig. S4), suggesting a possible link between splicing and 3′ pausing. The effect of inhibiting splicing on the extent of H3K36 trimethylation was detectable on the GAPDH gene within 90 min of SSA treatment (Fig. 3G), showing that localization of this histone modification can be quite rapidly changed in response to acute inhibition of splicing.
Fig. 3.
Inhibition of splicing causes widespread redistribution of histone H3K36me3 toward 3′ ends. Genome browser views of H3K36me3 and total pol II ChIP-seq reads in control HeLa cells and after treatment (20 ng/mL, 12 h) with the splicing inhibitor SSA for five intron-containing genes (A–E) and an intronless gene (F). Green and red or blue arrowheads mark 5′ and 3′ positions, respectively, for comparison of signals in WT and SSA. Note that the apparent 5′ to 3′ shift in H3K36me3 density is not paralleled by a similar shift in pol II density (see also SI Appendix, Fig. S4). (G) Relative H3K36me3 ChIP signals for 12 amplicons spanning the GAPDH gene in control HeLa cells (methanol treated) and after treatment with SSA (20 ng/mL) for 90 min. Relative ChIP signals were normalized to input and to amplicon +55 near the TSS. Means of 3 PCRs and SEMs are shown. Arrows mark the TSS and poly(A) sites in the map. Note the 3′ shift in H3K36me3 positioning similar to that detected in the 12-h treatment.
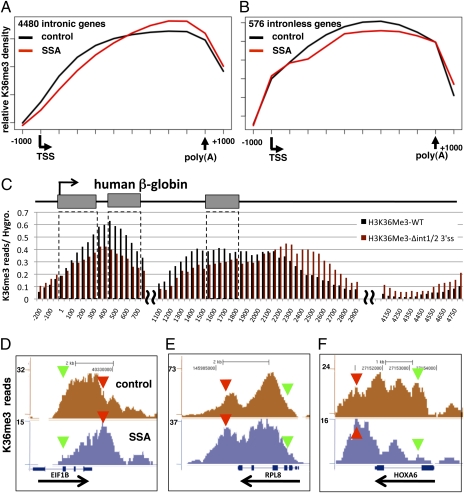
Fig. 4.
SSA mimics the effect of splice site mutants and causes widespread redistribution of H3K36me3. (A and B) Plots of the mean distributions of HeLa H3K36me3 ChIP-seq reads normalized for the total read number for 4,480 intron-containing genes in the first quartile for total read density and 576 intronless genes (excluding histone genes, Dataset S1) in control and SSA samples. The regions from the TSS to the most 3′ poly(A) site (in 10 equal intervals) with 1 kb of sequence upstream and downstream are shown. (C) ChIP-sequencing profiles of H3K36me3 on integrated WT and Δint1/2 3′ss β-globin genes. Read numbers were normalized to the values for a position within the 5′ flanking hygromycin resistance gene and binned into 50-base intervals. For the WT and Δint1/2 3′ss genes, 3,500 and 2,181 reads matching human β-globin were obtained, respectively. Positions of the bins relative to the TSS are given. “∼” Marks repetitive sequences that were excluded. (D–F) Genome browser views of H3K36me3 ChIP-seq reads in control and SSA treated HeLa cells, as in Fig. 3, for three short genes. Note the similarity of the SSA effect to that of the splice site mutants in C.
The effect of inhibiting splicing with SSA is quite general, as shown by the average distribution of H3K36me3 along 4,480 intron-containing genes in the top quartile for ChIP-seq read densities (Fig. 4A). A list of 1,719 genes from within this group with significant SSA-dependent 3′ shifts in H3K36me3 distribution (P < 0.05, Wilcoxon-Ranksum test) is provided in Dataset S1. Only 221 genes in the same group had significant shifts of H3K36me3 in the 5′ direction in response to SSA. Similar results were obtained for transcription units with lower H3K36me3 read densities. In contrast, on a group of 576 intronless genes (Dataset S1), SSA caused no detectable overall bias in the average distribution of H3K36me3 between the 5′ and 3′ ends (Fig. 4B).
To compare the effect of SSA with that of splice-site mutants at high resolution, we performed anti-H3K36me3 ChIP-seq on the CHO cell lines with integrated WT and Δint1/2 3′ss mutant β-globin genes, and normalized the results to the 5′ flanking hygromycin gene, as in Fig. 1. This experiment confirmed the enrichment of H3K36me3 in the 3′ flanking region of the splice-site mutant (Fig. 1C) and revealed a relative loss of H3K36me3 within the gene (Fig. 4C) that was not evident in the lower resolution qPCR analysis. These effects of the β-globin double splice-site mutant closely resemble the effects of SSA on other genes of similar length (Fig. 4 D–F): namely, loss of 5′ H3K36me3 signal (e.g., EIF1B, HOXA6) and gain of 3′ signal, including 3′ flanking sequences (e.g., EIF1B, RPL8, and HOXA6). In summary, these results show that inhibition of splicing by either introduction of splice-site mutations or treatment with SSA has remarkably similar consequences for the repositioning of H3K36me3 along genes.
Discussion
We investigated whether pre-mRNA splicing affects modification of nearby histones by H3K36 methylation. When splicing of β-globin intron 2 or introns 1 and 2 was blocked by deletion of their 3′ splice sites, H3K36me3 was repositioned away from the 5′ end and toward the 3′ end of the gene (Figs. 1, 2, and 4C). This effect was specific to inhibition of intron 2 splicing and was not observed in mutants of intron 1 or the poly(A) site (Figs. 1 and 2). Whether splicing of the last intron is generally a more important determinant of local histone methylation than splicing of the first intron remains to be determined. SSA, an inhibitor of the U2snRNP interacting factor SF3b, also repositioned histone H3K36 trimethylation by shifting its overall distribution away from 5′ positions and toward 3′ ends. This effect was observed on thousands of genes as determined by ChIP-seq (Figs. 3, 4, and Dataset S1), suggesting that the link between splicing and H3K36me3 positioning is quite general. H3K36me3 was repositioned toward the 3′ end of GAPDH within 90 min when splicing was inhibited by SSA (Fig. 3G). Rapid local changes in H3K36me3 could therefore reflect modulations in local splicing activity. Such effects might occur at regulated alternatively spliced exons (7); however, the relationship between H3K36 methylation and alternative splicing appears to be complex (17). Because SSA acts relatively rapidly (Fig. 3G) and shifts H3K36me3 positioning in a very similar manner to splice-site mutations (Fig. 4 C–F), its effect is unlikely to be indirect. Rather, these results argue that cotranscriptional splicing itself helps to direct local deposition and/or removal of the H3K36me3 mark. The 3′ splice site mutations, and probably also SSA, will disrupt exon definition; therefore, our results are consistent with the idea that this early step in splicing is important for establishment of the pattern of H3K36me3 on intron-containing genes (17).
How might defects in exon definition or some other aspect of splicing cause a repositioning of histone methylation marks? One possibility is that components of the splicing apparatus can influence H3 methylation and demethylation. In support of this idea, we observed a coincidence between where H3K36 was hypermethylated and where levels of the U5 snRNP were abnormally elevated downstream of the globin gene when intron 2 splicing was inhibited (Figs. 1B and 2B). Whether this correlation signifies a mechanistic link between splicing-factor recruitment and H3K36 methylation or demethylation remains to be investigated. The 3′ shift in localization of U5 snRNP and H3K36me3 when splicing was prevented could be linked to retention of mis-spliced transcripts at the site of transcription, although the small effects of a defective poly(A) site (Fig. 1 B and C), which also causes transcript retention (24), argue against this possibility. It was previously proposed that a complex of the histone chaperone Spt6, Iws1, and the mRNA export factor, Aly, stabilizes association of SETD2 with pol II elongation complexes phosphorylated on CTD Ser2 residues (13), a hallmark of transcription complexes at 3′ ends. Our results are consistent with the model that prolonged retention of splicing factors on the elongation complex, when splicing fails, stabilizes SETD2 on transcription complexes, resulting in H3K36 hypermethylation. The implication of this model is that under normal conditions, disassembly of splicing complexes at 3′ ends contributes to release of SETD2 and the decline in H3K36me3 that characterizes 3′ flanking regions (16). This prediction could be tested by direct analysis of SETD2; however, we and others (12) have failed to ChIP this protein using commercial or homemade antibodies. The highly reproducible polarity of the shift in H3K36me3 distribution from 5′ to 3′ when splicing is inhibited suggests the possible involvement of transcription elongation, which could influence how this modification is laid down by SETD2. In this context, it is interesting to note that inhibition of splicing by SSA was previously correlated with an increase in overall elongation rate on a reporter gene (35). Consistent with this finding, we observed that SSA reduced pol II pausing downstream of many endogenous genes (Fig. 3 and SI Appendix, Fig. S4). An additional potential link between splicing and elongation rate is the finding that SKIP, a U5 snRNP subunit, interacts with positive transcription elongation factor, PTEFb (36).
Although the mechanisms responsible for modulating the position of histone H3K36 methylation along a gene in response to cotranscriptional splicing activity remain to be elucidated in detail, our results show that one way this cross-talk operates is by a splicing-affects-chromatin mechanism. We suggest that this previously undescribed effect of splicing is one determinant of how the genome-wide distribution of H3K36me3 is established and maintained. Previous investigations that correlated chromatin changes with effects on splicing are consistent with a chromatin-affects-splicing scenario (14, 20–22) and these models are not mutually exclusive. In principle, the splicing-affects-chromatin mechanism of cross-talk helps solve the problem of how a “chromatin code” for splicing is properly targeted through recognition of nascent transcripts by the spliceosome. An attractive hypothesis is that a “chromatin code” for splicing is established by a splicing-affects-chromatin mechanism, which directs appropriate placement of the chromatin marks. Subsequently, the code is maintained and reinforced by one or more chromatin-affects-splicing mechanisms that modulate splicing through chromatin effects on transcription elongation (19, 22), splicing factor recruitment (14, 20), or other processes, such as nuclear positioning (37).
Materials and Methods
Plasmids.
pcDNA5/FRT/TO-β-globin Δint1 3′ss (Δint1 3′ss), pcDNA5/FRT/TO-β-globin Δint1/2 3′ss (Δint1/2 3′ss), and pcDNA5/FRT/TO-β-globin AAGAAA (A2GA3) were derived from pcDNA5/FRT/TO-β-globin WT, which contains the human β-globin gene starting at the ATG with an N-terminal HA tag and 3,034 bases of 3′ flanking sequence beyond the poly(A) cleavage site (38). The intron 1 and intron 2 3′ ss mutants delete 39 bp and 38 bp upstream of the respective 3′ splice sites, including the branch points and replace them by NotI sites.
Cell Lines.
pcDNA5/FRT/TO-β-globin with WT, Δint1 3′ss, Δint2 3′ss, Δint1/2 3′ss, or A2GA3 mutations were integrated into Flp-In-CHO cell line (Invitrogen) using Flp recombinase-mediated site-specific recombination, as previously described (38). The cells were maintained in F12 media supplemented with 10% FBS, 600 μg/mL hygromycin B, 6.5 μg/mL blasticidin, and penicillin/streptomycin. HeLa cells were treated with SSA (20 ng/mL) or methanol vehicle control for 90 min or 12 h.
Antibodies.
Rabbit anti-pan pol II CTD has been described previously (26). Anti-H3K36me3 was raised against the peptide NH2-ATGGVKme3KPHRYC-COOH coupled to KLH. ChIP-seq with the anti-H3K36me3 antibody in HeLa cells gave very similar results to published results using commercial antibody (16) (SI Appendix, Fig. S3). Rabbit anti-U5 100 K (huPRP28) was raised against a His-tagged fragment (amino acids 221–820) expressed from pETM11/U5-100 kDaΔRS (39) and affinity-purified. Cross-reaction with CHO cell U5 100 K was verified by Western blot (SI Appendix, Fig. S2).
Chromatin Immunoprecipitation.
Expression of β-globin was induced with 2 μg/mL of doxycycline overnight. The lysate preparation, ChIP, and real-time PCR with primers (described in SI Appendix, Table S1) using SYBR green reagent (Invitrogen) and Roche LC-480 were carried out as previously described (26). A standard curve was generated each time the real-time PCR was performed and the amplified signal was quantified using absolute quantification/second derivative maximum analysis in the LightCycler 480 Software 1.5.0. Primers for the human β-globin reporter did not yield products with DNA from the parent CHO Flp-In cell line. For each experiment, at least three immunoprecipitations were analyzed. ChIP signals were normalized to input DNA. Pol II ChIP signals on β-globin were normalized to values for the TK-hygromycin resistance gene that is also integrated at the Flp-In locus to correct for any variation in expression levels between cell lines. Where indicated, H3K36me3 ChIP signals were normalized to total H3 and U5 100-K signals were normalized to pol II from the same experiment and analyzed side-by-side. The mean normalized values and the SEM were determined for each amplicon. Where indicated, signals were normalized to the amplicon with the maximum value. The average maximum is not 1.0 in some cases, where it falls at different amplicons in different experiments.
ChIP-Seq.
Immunoprecipitations from 2 mg of cross-linked extract were processed for Illumina library construction and sequenced with the Illumina Genome Analyzer IIx. Single-end 34 base reads (after removing barcodes) were mapped to the hg18 University of California Santa Cruz (UCSC) human genome (March 2006) with Bowtie version 0.12.5 (40). We generated bedgraph profiles using 50-bp bins and 500-bp windows assuming a 200-bp fragment size shifting effect. Results were viewed with the UCSC browser (41). Total matched reads for the control and SSA treated HeLa samples were 11.6 and 8.2 million for anti-H3K36me3 and 6.5 and 5.0 million for anti-pol II. For metagene analysis (Fig. 4 A and B), relative density plots for each transcription unit and 1-kb flanking regions were made using the density program in R (Gaussian kernel smoothing with a default option), and averages were calculated at 10 grid points in the body of each gene and in the flanking regions.
Supplementary Material
Acknowledgments
We thank R. Luhrmann for the U5 100 K expression plasmid; M. Yoshida for spliceostatin A; C. Hittinger for help with Illumina library construction; J. Dover, D. Farrell, and J. Castoe for Illumina sequencing; R. Perales for help with sequencing analysis; and the University of Colorado Denver Cancer Center sequencing facility. This work was supported by National Institutes of Health Grants GM058613 and GM063873 (to D.L.B.); Post-doctoral fellowship PF-07-297-01-GMC from the American Cancer Society and the Clark family (to S.K.); and American Recovery and Reinvestment Act of 2009 Award 3R01GM063873-06S1 (to H.K.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE30895).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109475108/-/DCSupplemental.
References
- 1.Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- 2.Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. RNA. 2009;15:1896–1908. doi: 10.1261/rna.1714509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baurén G, Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. doi: 10.1016/0092-8674(94)90182-1. [DOI] [PubMed] [Google Scholar]
- 4.Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell. 2010;40:571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Han J, Xiong J, Wang D, Fu X-D. Pre-mRNA splicing: Where and when in the nucleus. Trends Cell Biol. 2011;21:336–343. doi: 10.1016/j.tcb.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartz S, Ast G. Chromatin density and splicing destiny: On the cross-talk between chromatin structure and splicing. EMBO J. 2010;29:1629–1636. doi: 10.1038/emboj.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolasinska-Zwierz P, et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 2009;19:1732–1741. doi: 10.1101/gr.092353.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nahkuri S, Taft RJ, Mattick JS. Nucleosomes are preferentially positioned at exons in somatic and sperm cells. Cell Cycle. 2009;8:3420–3424. doi: 10.4161/cc.8.20.9916. [DOI] [PubMed] [Google Scholar]
- 11.Krogan NJ, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoh SM, Lucas JS, Jones KA. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev. 2008;22:3422–3434. doi: 10.1101/gad.1720008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luco RF, et al. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schor IE, Rascovan N, Pelisch F, Alló M, Kornblihtt AR. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc Natl Acad Sci USA. 2009;106:4325–4330. doi: 10.1073/pnas.0810666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Huff JT, Plocik AM, Guthrie C, Yamamoto KR. Reciprocal intronic and exonic histone modification regions in humans. Nat Struct Mol Biol. 2010;17:1495–1499. doi: 10.1038/nsmb.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun XJ, et al. Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J Biol Chem. 2005;280:35261–35271. doi: 10.1074/jbc.M504012200. [DOI] [PubMed] [Google Scholar]
- 19.de la Mata M, et al. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Sims RJ, 3rd, et al. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nogues G, Kadener S, Cramer P, Bentley D, Kornblihtt AR. Transcriptional activators differ in their abilities to control alternative splicing. J Biol Chem. 2002;277:43110–43114. doi: 10.1074/jbc.M208418200. [DOI] [PubMed] [Google Scholar]
- 22.Batsché E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol. 2006;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- 23.Kaida D, et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol. 2007;3:576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- 24.Custódio N, et al. Inefficient processing impairs release of RNA from the site of transcription. EMBO J. 1999;18:2855–2866. doi: 10.1093/emboj/18.10.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Almeida SF, García-Sacristán A, Custódio N, Carmo-Fonseca M. A link between nuclear RNA surveillance, the human exosome and RNA polymerase II transcriptional termination. Nucleic Acids Res. 2010;38:8015–8026. doi: 10.1093/nar/gkq703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gromak N, West S, Proudfoot NJ. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol Cell Biol. 2006;26:3986–3996. doi: 10.1128/MCB.26.10.3986-3996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dye MJ, Proudfoot NJ. Multiple transcript cleavage precedes polymerase release in termination by RNA polymerase II. Cell. 2001;105:669–681. doi: 10.1016/s0092-8674(01)00372-5. [DOI] [PubMed] [Google Scholar]
- 29.Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH. Crosstalk between mRNA 3′ end processing and transcription initiation. Mol Cell. 2010;40:410–422. doi: 10.1016/j.molcel.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Dye MJ, Proudfoot NJ. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol Cell. 1999;3:371–378. doi: 10.1016/s1097-2765(00)80464-5. [DOI] [PubMed] [Google Scholar]
- 31.Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol. 2006;13:815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- 32.Konforti BB, Konarska MM. U4/U5/U6 snRNP recognizes the 5′ splice site in the absence of U2 snRNP. Genes Dev. 1994;8:1962–1973. doi: 10.1101/gad.8.16.1962. [DOI] [PubMed] [Google Scholar]
- 33.Antoniou M, Geraghty F, Hurst J, Grosveld F. Efficient 3′-end formation of human beta-globin mRNA in vivo requires sequences within the last intron but occurs independently of the splicing reaction. Nucleic Acids Res. 1998;26:721–729. doi: 10.1093/nar/26.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roybal GA, Jurica MS. Spliceostatin A inhibits spliceosome assembly subsequent to prespliceosome formation. Nucleic Acids Res. 2010;38:6664–6672. doi: 10.1093/nar/gkq494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brody Y, et al. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol. 2011;9:e1000573. doi: 10.1371/journal.pbio.1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brès V, Gomes N, Pickle L, Jones KA. A human splicing factor, SKIP, associates with P-TEFb and enhances transcription elongation by HIV-1 Tat. Genes Dev. 2005;19:1211–1226. doi: 10.1101/gad.1291705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takizawa T, Meaburn KJ, Misteli T. The meaning of gene positioning. Cell. 2008;135:9–13. doi: 10.1016/j.cell.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fong N, Ohman M, Bentley DL. Fast ribozyme cleavage releases transcripts from RNA polymerase II and aborts co-transcriptional pre-mRNA processing. Nat Struct Mol Biol. 2009;16:916–922. doi: 10.1038/nsmb.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teigelkamp S, Mundt C, Achsel T, Will CL, Lührmann R. The human U5 snRNP-specific 100-kD protein is an RS domain-containing, putative RNA helicase with significant homology to the yeast splicing factor Prp28p. RNA. 1997;3:1313–1326. [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuhn RM, et al. The UCSC Genome Browser Database: Update 2009. Nucleic Acids Res. 2009;37(Database issue):D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.