Abstract
HsPKS1 from Huperzia serrata is a type III polyketide synthase (PKS) with remarkable substrate tolerance and catalytic potential. Here we present the synthesis of unnatural unique polyketide–alkaloid hybrid molecules by exploiting the enzyme reaction using precursor-directed and structure-based approaches. HsPKS1 produced novel pyridoisoindole (or benzopyridoisoindole) with the 6.5.6-fused (or 6.6.5.6-fused) ring system by the condensation of 2-carbamoylbenzoyl-CoA (or 3-carbamoyl-2-naphthoyl-CoA), a synthetic nitrogen-containing nonphysiological starter substrate, with two molecules of malonyl-CoA. The structure-based S348G mutant not only extended the product chain length but also altered the cyclization mechanism to produce a biologically active, ring-expanded 6.7.6-fused dibenzoazepine, by the condensation of 2-carbamoylbenzoyl-CoA with three malonyl-CoAs. Thus, the basic nitrogen atom and the structure-based mutagenesis enabled additional C─C and C─N bond formation to generate the novel polyketide-alkaloid scaffold.
Keywords: biosynthesis, enzyme engineering, unnatural natural products
In contrast to the type I (modular type) and type II (subunit type) polyketide synthases (PKSs) of the megaenzyme systems, the structurally and mechanistically simpler type III PKSs catalyze the complete series of decarboxylation, condensation, and cyclization reactions with a single active-site. The enzymes perform C─C bond forming reactions by iterative Claisen-type condensation of CoA thioesters and cyclization of the poly-β-keto intermediates, to produce pharmaceutically and biologically important aromatic polyketides, such as chalcone, stilbene, and curcumin (1–3). The members of the chalcone synthase (CHS) superfamily of type III PKSs share a common three-dimensional overall fold and catalytic machinery with a conserved Cys-His-Asn catalytic triad, and a minor modification of the active-site structure leads to the catalytic diversity of the enzymes (1–3). Interestingly, type III PKSs exhibit remarkable substrate tolerance, accepting a series of nonphysiological substrate analogues to produce chemically and structurally distinct unnatural novel polyketides (4–10). Furthermore, recent structure-based engineering of functionally divergent type III PKSs has significantly expanded the catalytic repertoire of the enzymes and the product diversity (11–17). Due to their remarkable substrate promiscuity and catalytic potential, the structurally and mechanistically simple type III PKS enzymes represent an excellent platform for the further development of unnatural novel biocatalysts with unprecedented catalytic functions (18).
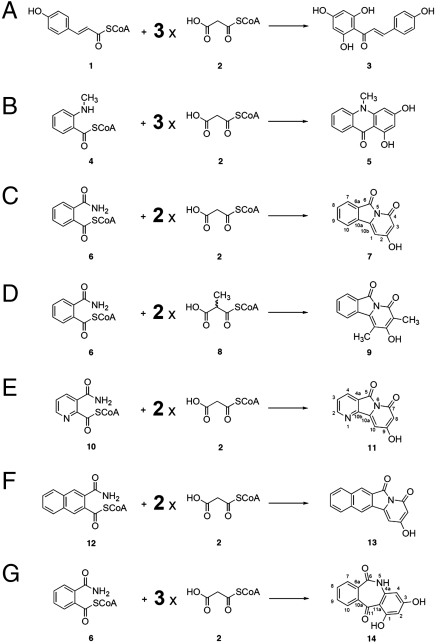
The type III PKSs primarily catalyze C─C bond formation via carbonyl chemistry (1–3). The poly-β-keto intermediates formed by the enzymes are highly reactive and readily react with basic nitrogen atoms to form Schiff bases, which facilitate additional C─C or C─N bond forming chemistry to generate more complex polyketide–alkaloid hybrid molecules. Here we report the synthesis of biologically active, unnatural novel polyketide–alkaloid hybrid molecules by exploiting a type III PKS from a primitive club moss Huperzia serrata (HsPKS1) (19) (Fig. S1), using precursor-directed and structure-based approaches. As previously reported, HsPKS1 is a unique type III PKS that exhibits unusually broad substrate specificity to produce various aromatic polyketides. For example, HsPKS1, which normally catalyzes the sequential condensations of 4-coumaroyl-CoA (1) with three molecules of malonyl-CoA (2) to produce naringenin chalcone (3) (Fig. 1A), also accepts bulky N-methylanthraniloyl-CoA (4) as a starter to produce 1,3-dihydroxy-N-methylacridone (5), after three condensations with malonyl-CoA (19) (Fig. 1B). In contrast, the regular chalcone synthase (CHS) does not accept the bulky substrate, suggesting that HsPKS1 has a larger substrate-binding pocket at the active site.
Fig. 1.
Enzymatic formation of naringenin chalcone and alkaloids by type III PKSs. (A) Naringenin chalcone by CHS or HsPKS1, (B) 1,3-dihydroxy-N-methylacridone by HsPKS1 or ACS, (C) 2-hydroxypyrido[2,1-a]isoindole-4,6-dione by HsPKS1 or its S348G mutant, (D) 2-hydroxy-1,3-dimethylpyrido[2,1-a]isoindole-4,6-dione by HsPKS1 or its S348G mutant, (E) 2-hydroxybenzo[f]pyrido[2,1-a]isoindole-4,6-dione by HsPKS1 or its S348G mutant, and (F) 9-hydroxypyrido[2,3-a]indolizine-5,7-dione by HsPKS1 or its S348G mutant.
Results and Discussion
Precursor-Directed Biosynthesis.
We first designed and synthesized 2-carbamoylbenzoyl-CoA (6) as a starter substrate (Fig. 1C). The CoA thioester has a similar molecular size as N-methylanthraniloyl-CoA and a nitrogen atom that can react with the carbonyl group of the elongating poly-β-keto chain during the enzyme reaction. Here we chose the less reactive carbamoyl group instead of the primary amine to avoid the spontaneous intramolecular cyclization to form the imide with the concomitant removal of CoA-SH. Indeed, our molecular modeling study suggested that the intramolecular cyclization is prevented by hydrogen-bond formation between the carbamoyl proton and the thioester carbonyl oxygen atom.
When incubated with 2-carbamoylbenzoyl-CoA (6) and malonyl-CoA (2) as substrates under the standard assay condition, HsPKS1 efficiently afforded a single product in 6.4% yield (Figs. 1C and 2A and Fig. S2). The product, which was not detected in control experiments performed with the boiled enzyme, gave a UV spectrum (λmax 317 nm) and a parent ion peak [M + H]+ at m/z 214 on LC-ESIMS, indicating triketide formation by sequential condensations of 2-carbamoylbenzoyl-CoA (6) with two molecules of malonyl-CoA (2). The 1H NMR spectrum revealed the presence of four aromatic protons at δ 7.90 (2H, d, J = 7.6 Hz), 7.81 (1H, t, J = 7.6 Hz), and 7.67 (1H, t, J = 7.6 Hz), and two olefinic protons (δ 6.76 and 5.53, each 1H, s). Furthermore, the 13C NMR and heteronuclear correlation spectroscopy (HMQC and HMBC) revealed 12 carbon signals, and the key HMBC correlations from H-10 (δ 7.90) to C-10b (δ135.9), from H-1 (δ6.76) to C-3 (δ91.8), and from H-3 (δ5.53) to C-2 (δ173.0) and C-4 (δ166.8). A structure with the 6.5.6-fused tricyclic ring system, bearing a bridgehead nitrogen atom, was uniquely consistent with both biogenetic reasoning and the spectroscopic data.
Fig. 2.
HPLC elution profiles of enzyme reaction products of the wild-type HsPKS1 and the S348G mutant HsPKS1. The enzyme reaction products from (A) 2-carbamoylbenzoyl-CoA and malonyl-CoA, (B) 2-carbamoylbenzoyl-CoA and (2RS)-malonyl-CoA, (C) 3-carbamoylpicolinoyl-CoA and malonyl-CoA as the substrates, and (D) 3-carbamoyl-2-naphthoyl-CoA and malonyl-CoA.
The structure of the enzyme reaction product was thus unambiguously determined to be 2-hydroxypyrido[2,1-a]isoindole-4,6-dione (7), which is an unnatural, novel alkaloid. Although chemical syntheses of pyridoisoindole derivatives have been reported, no further chemistry has been detailed (20–22). The pyridoisoindole-forming activity was maximal at pH 6.0, with a broad optimal pH within the range of pH 5.5–8.5. Furthermore, analyses of the steady-state kinetics of HsPKS1 revealed a KM = 7.6 μM and a kcat = 4.5 × 10-2 min-1 for 2-carbamoylbenzoyl-CoA, with respect to the pyridoisoindole-forming activity, representing 7- and 453-fold increases in the kcat/KM values of the chalcone- and acridone-forming activities of HsPKS1, respectively (see SI Text). Thus, the enzyme accepted the synthetic substrate much more efficiently than the physiological 4-coumaroyl-CoA (1) and N-methylanthraniloyl-CoA (4), to produce the unnatural novel polyketide-alkaloid hybrid molecule. In contrast, the regular CHS did not accept the bulky substrate. Further, octaketide synthase (OKS) from Aloe arborescens (12), which normally catalyzes iterative condensations of eight molecules of malonyl-CoA to produce the aromatic octaketides SEK4 and SEK4b, only afforded a tetraketide pyrone as a minor product by condensation of 2-carbamoylbenzoyl-CoA with three molecules of malonyl-CoA (Fig. S3).
Interestingly, when (2RS)-methylmalonyl-CoA (8) was used as an extender substrate instead of malonyl-CoA (2), HsPKS1 afforded a dimethylated novel alkaloid, 1,3-dimethyl-2-hydroxypyrido[2,1-a]isoindole-4,6-dione (9), as a single product but with less efficiency, by sequential condensations of 2-carbamoylbenzoyl-CoA (6) with two molecules of (2RS)-methylmalonyl-CoA (8) as an extender, in 2.2% yield (Figs. 1D and 2B and Fig. S4). Furthermore, when pyridine-containing 3-carbamoylpicolinoyl-CoA (10) was employed as a starter substrate, HsPKS1 afforded an unnatural novel alkaloid, 9-hydroxypyrido[2,3-a]indolizine-5,7-dione (11) (Figs. 1E and 2C and Fig. S5), with much less efficiency in 0.5% yield (see SI Text). In contrast, unexpectedly, HsPKS1 efficiently accepted the bulky naphthalene-containing 3-carbamoyl-2-naphthoyl-CoA (12) as a starter to produce 2-hydroxybenzo[f]pyrido[2,1-a]isoindole-4,6-dione (13), a novel alkaloid scaffold with the 6.6.5.6-fused tetracyclic ring system, as a single product in 13% yield (Figs. 1F and 2D and Fig. S6). Analyses of the steady-state kinetics of HsPKS1 revealed a KM = 6.8 μM and a kcat = 3.2 × 10-1 min-1 for 3-carbamoyl-2-naphthoyl-CoA (12) with respect to the benzopyridoisoindole-forming activity, which is an eightfold increase in the kcat/KM values, as compared with the pyridoisoindole-forming activity from 2-carbamoylbenzoyl-CoA (6) (see SI Text). The naphthalene-containing bulky substrate thus fits better into the active-site of the enzyme, to efficiently generate the unnatural novel alkaloid. Notably, the structure of the 6.6.5.6-fused tetracyclic scaffold is quite similar to that of camptothecin, a plant monoterpene indole alkaloid and a DNA topoisomerase I inhibitor, produced by limited angiosperms including Camptotheca acuminata and Nothapodytes foetida (23) (Fig. S6E). The monoterpene indole alkaloid is biosynthesized from tryptamine (the indole-C2N unit) and secologanin (the C10 unit) through the formation of strictosidine (24). Oxidation and rearrangement of the 6.5.6-fused tetrahydro-β-carboline moiety within strictosidine lead to formation of the campthothecin scaffold (24).
Structure-Based Engineering.
It is remarkable that HsPKS1, which normally catalyzes chalcone formation by iterative condensations of 4-coumaroyl-CoA with three molecules of malonyl-CoA, efficiently accepts the bulky carbamoyl-containing synthetic substrates as a starter and performs two condensations with malonyl-CoA, to generate the unnatural novel alkaloids with the 6.5.6- and 6.6.5.6-fused ring systems. To further clarify the structural details of the enzyme-catalyzed alkaloid-forming reactions, we solved the X-ray crystal structure of HsPKS1 at 2.0-Å resolution (Fig. S7 and see also SI Text). The overall structure of HsPKS1 is highly homologous to those of the previously reported plant type III PKSs (1, 2) (rmsds 0.8–0.9 Å), in which the most closely related structural homologue is that of Medicago sativa CHS (25) (Fig. S7). A comparison of the active-site of HsPKS1 with those of the other type III PKSs revealed that the total cavity volume of HsPKS1 is 647 Å3, which is almost as large as that of the chalcone-producing M. sativa CHS (754 Å3) (Fig. 3 A and B). The active-site of HsPKS1 is thus large enough to accommodate the tricyclic and tetracyclic novel polyketide–alkaloids.
Fig. 3.
Comparison of the active-site architecture of wild-type HsPKS1, M. sativa CHS and the S348G mutant HsPKS1. Crystal structures of (A) the wild-type HsPKS1, and (B) M. sativa CHS, and homology model of (C) the S348G mutant HsPKS1. The active-site architectures are represented by surface models, respectively. The substrate entrances are indicated with arrows. Phe275 and Ser348 in wild-type HsPKS1 and the equivalent residues in M. sativa CHS and the S348G mutant HsPKS1 are indicated with blue and purple stick models, respectively. The expanded wall in the S348G mutant HsPKS1 is highlighted with a purple surface. The naringenin molecule bound to the active-site cavity of M.sativa CHS is shown as a green stick model. Three-dimensional models of 2-hydroxybenzo[f]pyrido[2,1-a]isoindole-4,6-dione (7) and 1,3-dihydroxy-5H-dibenzo[b, e]azepine-6,11-dione (14), docked into the active-site cavities of the wild-type and S348G mutant HsPKS1 enzymes, respectively, are shown as green stick models.
We performed docking simulations, using the models of linear and partially cyclized intermediates tethered at the catalytic Cys174 (corresponding to Cys164 in M. sativa CHS), which is the attachment site for the growing poly-β-keto intermediates. The simulations predicted that the hydroxyl group of the neighboring Ser348 (corresponding to Ser338 in M. sativa CHS) forms hydrogen bonds with the C5 carbonyl oxygen in the linear intermediate and with the amide oxygen of the partially cyclized intermediate, respectively (Fig. 4 A and B and Fig. S7), which would be crucial for the enzymatic cyclization of the polyketide intermediates, as described later. Notably, the active-site residue Ser348 is uniquely altered in a number of functionally distinct type III PKSs and is thought to control the polyketide chain elongation reactions and product specificity (1, 2). Therefore, to evaluate the importance of Ser348 in the alkaloid-forming activities of HsPKS1, we constructed a set of site-directed mutants (S348V, S348T, S348C, and S348G) and investigated the effects of the mutagenesis on the enzyme activities.
Fig. 4.
Proposed mechanism for the formation of 2-hydroxypyrido[2,1-a]isoindole-4,6-dione by wild-type HsPKS1 and 1,3-dihydroxy-5H-dibenzo[b, e]azepine-6,11-dione by the S348G mutant HsPKS1. Three-dimensional models of (A) the triketide linear intermediate, (B) heteropentacyclized triketide intermediate, and (C) 2-hydroxypyrido[2,1-a]isoindole-4,6-dione by wild-type HsPKS1, and (E) the tetraketide linear intermediate, (F) heterheptacyclized tetraketide intermediate, and (G) 1,3-dihydroxy-5H-dibenzo[b, e]azepine-6,11-dione by the S348G mutant HsPKS1. Schematic representation of the proposed mechanism for the formation of (D) 2-hydroxypyrido[2,1-a]isoindole-4,6-dione by wild-type HsPKS1 and (H) 1,3-dihydroxy-5H-dibenzo[b, e]azepine-6,11-dione by the S348G mutant HsPKS1. The intermediates and products are highlighted with green stick models.
The mutant proteins were expressed in Escherichia coli at comparable levels to the wild-type enzyme. When incubated with 2-carbamoylbenzoyl-CoA and malonyl-CoA as substrates, the S348G mutant afforded a novel product (1.6% yield) in addition to the 6.5.6-fused pyridoisoindole (3.7% yield) (Figs. 1G and 2A and Fig. S8). The enzyme activity was maximal at pH 6.5–7.0, with a broad pH optimum within the range of pH 5.5–8.5. The novel product gave a UV spectrum (λmax 257, 313, 363, and 376 nm) and a parent ion peak [M + H]+ at m/z 256 on LC-ESIMS, indicating the formation of a tetraketide by sequential condensations of 2-carbamoylbenzoyl-CoA with three molecules of malonyl-CoA. The 1H NMR spectrum revealed the presence of one hydroxyl proton at δ 9.38 (1H, s), one amide proton at δ 8.27 (1H, s), and six aromatic protons at δ 7.91 (1H, d, J = 7.0 Hz), δ 7.51 (2H, m), δ 7.40 (1H, d, J = 7.0 Hz), δ 6.85 (1H, d, J = 1.5 Hz), and δ 5.06 (1H, d, J = 1.5 Hz). Furthermore, the 13C NMR and heteronuclear correlation spectroscopy (HMQC and HMBC) revealed 14 carbon signals, and the key HMBC correlations from H-7 (δ 7.91) to C-6 (δ 165.1) and C-6a (δ 137.0), from H-5 (δ 8.27) to C-6a (δ 137.0), and from H-10 (δ 7.40) to C-11 (δ 188.5). The structure of the novel product was thus unambiguously determined to be 1,3-dihydroxy-5H-dibenzo[b,e]azepine-6,11-dione (14), which is an unnatural, novel alkaloid with the ring-expanded 6.7.6-fused tricyclic ring system (Fig. 1G). The dibenzoazepine scaffold was thus produced by sequential condensations of 2-carbamoylbenzoyl-CoA (6) as a starter substrate with three molecules of malonyl-CoA, through a N/C5 and C6/C1 tandem cyclization reactions (Fig. 4 E and F). The chemical synthesis of dibenzo[b,e]azepine-6,11-dione (morphanthridines-6,11-dione) has been reported (26), and the dibenzo[b,e]azepines (morphanthridines) are reportedly potent activators of the human transient receptor potential ankyrin 1 (TRPA1) channel (27).
A homology model of the S348G mutant predicted that the total cavity volume of the mutant (687 Å3) is slightly larger than that of the wild-type enzyme (647 Å3) (Fig. 3C). Notably, the S348G substitution opens a significant amount of space neighboring the catalytic Cys174, which enables the active site to accommodate the ring-expanded 6.7.6-fused dibenzoazepine. An analysis of the steady-state kinetics of the HsPKS1 S348G mutant revealed a KM = 27.0 μM and a kcat = 3.6 × 10-2 min-1 for 2-carbamoylbenzoyl-CoA with respect to the 6.7.6-dibenzoazepine-forming activity, which is a 4.5-fold decrease in the kcat/KM value, as compared with the 6.5.6-pyridoisoindole-forming activity of the wild-type enzyme (see SI Text). It is remarkable that the single amino acid replacement not only increased the product chain length but also altered the mechanism of the cyclization reaction to generate the ring-expanded dibenzoazepine.
In contrast, the other mutations (S348V, S348T, and S348C) did not alter the product pattern but retained the pyridoisoindole-forming activity. Furthermore, despite the similar molecular size, none of the mutants, including S348G, produced the ring-expanded product from the pyridine-containing 3-carbamoylpicolinoyl-CoA but instead generated the 6.5.6-fused pyridoindolizine with decreased activities (Fig. 2C). On the other hand, although the formation of the ring-expanded product was not detected, the S348G mutant accepted the naphthalene-containing 3-carbamoyl-2-naphthoyl-CoA quite efficiently as a starter substrate, to produce the 6.6.5.6-fused tetracyclic alkaloid (Fig. 2D). The yield was 45%, which is a 3.6-fold increase as compared with that of the wild-type enzyme. This also supports the proposal that the large-to-small S348G substitution opens the space neighboring the catalytic Cys174, the attachment site for the growing polyketide chain, leading to the optimization of the active-site architecture for the production of the bulky tetracyclic product. Widening of the internal active-site pocket is thus one of the key factors for the further engineering of the type III PKS enzymes to generate unnatural, novel molecules.
Proposed Catalytic Mechanism.
On the basis of these findings, we propose that HsPKS1 employs novel catalytic machinery for the production of the newly obtained polyketide-alkaloid hybrid moleculs (Fig. 4A–D). Thus, following the iterative condensations of the 2-carbamoylbenzoyl-CoA starter with two molecules of malonyl-CoA, the active-site residue Ser348 plays a key role in guiding the course of the polyketide chain elongation by the hydrogen bonding with the C5 carbonyl oxygen of the linear intermediate. This facilitates the initial N/C5 cyclization to form the γ-lactam by the nucleophilic attack of the nitrogen on the C5 carbonyl carbon. The hydrogen bonding with Ser348 then switches to the amide carbonyl oxygen, and the resulting carbinolamide is subsequently converted into the enamine by spontaneous dehydration. Here, the interaction of the aromatic moiety with the so-called gatekeeper Phe225 is likely to reinforce the psuedocyclic conformation favored for the orientation of the nitrogen atom toward the C1 carbonyl carbon, which results in the final N/C1 cyclization to produce the 6.5.6-fused ring system (Fig. 4D).
On the other hand, the large-to-small S348G substitution facilitates the polyketide chain elongation reaction up to the tetraketide stage, by enlarging the space neighboring the catalytic Cys174 to produce the ring-expanded 6.7.6-fused dibenzoazepine (Fig. 4 E–H). In this case, it is likely that the active-site residue Asn346, behind Gly348, guides the course of the chain elongation by the hydrogen bonding with the C5 carbonyl oxygen of the tetraketide linear intermediate. This is followed by the nucleophilic attack of the nitrogen on the C5 carbonyl carbon to produce the ring-expanded ϵ-lactam. The interaction with Asn346 then switches to the C7 carbonyl oxygen, and the carbinolamide is subsequently converted into the enamine by spontaneous dehydration. Presumably, the gatekeeper Phe225 would again assist with the final C6/C1 cyclization to generate the 6.7.6-fused tricyclic ring system (Fig. 4H). Indeed, when Phe225 in the HsPKS S348G mutant was substituted with Gly, Ala, Ser, Cys, Leu, and His, respectively, the F225/S348 double mutants completely lost the dibenzoazepine-forming activity and only produced a trace amount of the pyridoisoindole. In contrast, the F225Y/S348G and F225W/S348G double mutants retained the 6.7.6-dibenzoazepine- and 6.5.6-pyridoisoindole-forming activities, suggesting that the aromatic residues play important roles in the formation of the tricyclic ring systems. Thus, the basic nitrogen atom and the structure-based mutagenesis enable the additional C─C and C─N bond forming chemistry, instead of the consecutive C─N bond forming reactions, to generate the ring-expanded 6.7.6-fused dibenzoazepine.
Biological Activities.
Pyridoisoindole derivatives exhibit remarkable physiological properties, such as antibacterial, anti-inflammatory, analgesic, and antitumor cell proliferation activities (28). Furthermore, as previously described, dibenzo[b,e]azepine (morphanthridines) are potent activators of the TRPA1 channel (27). Here 2-hydroxypyrido[2,1-a]isoindole-4,6-dione (7) and 1,3-dihydroxy-5H-dibenzo[b,e]azepine-6,11-dione (14) were tested for their antibacterial activity against methicillin-susceptible Staphylococcus aureus (MSSA), E. coli, and Bacillus cereus, and their cytotoxic activity against murine leukemia P388 cells. Interestingly, although both products lacked significant antibacterial and cytotoxic activities, the dibenzoazepine clearly inhibited biofilm formation by MSSA (MIC = 12.0 μg/mL), which is one of the most important etiological factors responsible for nosocomial infections (29). Biofilms are known to be resistant to conventional antibiotics and host immune responses (30). Although soporific, antipsychotic and anticonvulsant activities have been reported for the dibenzoazepine derivatives (31, 32), this is a previously undescribed demonstration of the antibiofilm activity by a dibenzoazepine alkaloid. These observations suggest that the dibenzoazepine and its derivatives may be antibiotic candidates with inhibitory activity against biofilm formation.
In conclusion, we demonstrated the synthesis of unnatural, novel polyketide-alkaloid scaffolds by exploiting a type III PKS from a primitive club moss, using precursor-directed and structure-based approaches. Thus, HsPKS1 produced the 6.5.6-fused pyridoisoindole (or 6.6.5.6-fused benzopyridoisoindole) scaffold, by the condensation of 2-carbamoylbenzoyl-CoA (or 3-carbamoyl-2-naphthoyl-CoA), a nitrogen-containing nonphysiological starter substrate, with two molecules of malonyl-CoA. Furthermore, the structure-based S348G mutant not only extended the product chain length but also altered the cyclization mechanism to produce a biologically active dibenzoazepine with a ring-expanded 6.7.6-fused ring system, by the condensation of 2-carbamoylbenzoyl-CoA with three malonyl-CoAs. Thus, the basic nitrogen atom and the structure-based mutagenesis facilitated the additional C─C and C─N bond forming chemistry to generate the complex polyketide-alkaloid hybrid molecule. The catalytic versatility and potential of the structurally and mechanistically simple type III PKS enzymes provide an excellent platform for the further development of unnatural novel biocatalysts with unprecedented catalytic functions.
Materials and Methods
Materials and Bioassay Procedures.
Please see SI Materials and Methods.
Enzyme Expression and Purification.
The recombinant HsPKS1, with a GST-tag at the N terminus, was overexpressed in E. coli BL21(DE3)pLysS and purified, as previously reported (33). After removal of the GST-tag, the purified HsPKS1 was used for the enzyme assay and crystallization.
Site-Directed Mutagenesis and Purification of the Mutant Enzymes.
All of the plasmids expressing the S348X and F225X/S348G mutants of HsPKS1 were constructed with a QuikChange Site-Directed Mutagenesis Kit (Stratagene), according to the manufacturer’s protocol. The primers used are listed in SI Materials and Methods. The mutant enzyme was expressed, extracted and purified by the same procedure as for the wild-type HsPKS1, and was used in the enzyme assay.
Standard Enzyme Reaction.
The reaction mixture contained 54 μM of 2-carbamoylbenzoyl-CoA, 3-carbamoylpicolinoyl-CoA, or 3-carbamoyl-2-naphthoyl-CoA, 108 μM of malonyl-CoA or (2RS)-methymalonyl-CoA, and 20 μg of the purified enzyme, in a final volume of 500 μL of 100 mM potassium phosphate (KPB) buffer (pH 6.5), containing 1 mM EDTA. The reactions were incubated at 30 °C for 4 h and stopped by the addition of 5 μL of 20% HCl. The products were then extracted twice with 500 μL of ethyl acetate and were analyzed by an online LC-ESIMS system. The online LC-ESIMS spectral analyses were performed as previously described (19).
For the large-scale enzyme reaction, 20 mg of the purified enzyme was incubated with 10 mg of malonyl-CoA and 5 mg starter CoA in 100 mL of buffer (100 mM KPB buffer, pH 6.0 for 2-carbamoylbenzoyl-CoA reaction, and 100 mM potassium phosphate buffer, pH 6.5 for the 3-carbamoyl-2-naphthoyl-CoA and 3-carbamoylpicolinoyl-CoA reactions), containing 1 mM EDTA either at 30 °C (2-carbamoylbenzoyl-CoA and 3-carbamoylpicolinoyl-CoA reactions) or 45 °C (3-carbamoyl-2-naphthoyl-CoA reaction) for 16 h. The reaction was quenched by the addition of 20% HCl (10 mL) and extracted with ethyl acetate (200 mL × 3). The enzyme reaction products were purified by reverse-phase HPLC (JASCO 880), using the same gradient elution program as that for the standard enzyme reaction. In total, six large-scale reactions were performed to obtain the purified enzyme reaction products. For the LC-HRTOFMS spectral analyses, the same HPLC gradient program as that for the online LC-ESIMS spectral analyses was used. Spectral data for the enzyme reaction products are summarized in SI Materials and Methods.
The yield of 2-hydroxy-1,3-dimethylpyrido[2,1-a]isoindole-4,6-dione, derived from 2-carbamoylbenzoyl-CoA and (2RS)-methylmalonyl-CoA, was calculated by the dried weight of the purified compound obtained from the large-scale reaction. On the other hand, other enzyme reaction product yields were calculated by incubations with [2-14C]-malonyl-CoA (1.8 mCi/mmol) as an extender substrate with the respective corresponding starter substrates, under the enzyme reaction conditions for steady-state kinetic analyses described below.
Enzyme Kinetics.
Steady-state kinetic parameters were determined by using [2-14C]malonyl-CoA (1.8 mCi/mmol) as a substrate. The experiments were performed in triplicate, using four concentrations of 2-carbamoylbenzoyl-CoA, 3-carbamoylpicolinoyl-CoA, 3-carbamoylnaphthoyl-CoA, 4-coumaroyl-CoA, or N-methylanthraniloyl-CoA (86.7, 54.2, 32.5, and 10.8 μM) in the assay mixture, containing 108 μM of malonyl-CoA, 4 μg of purified enzyme, and 1 mM EDTA, in a final volume of 100 μL of 100 mM potassium phosphate buffer, pH 6.0 for 2-carbamoylbenzoyl-CoA, pH 6.5 for 3-carbamoylnaphthoyl-CoA and 3-carbamoylpicolinoyl-CoA, and pH 8.0 for 4-coumaroyl-CoA and N-methylanthraniloyl-CoA, respectively. Incubations were performed for 20 min at 30 °C for 2-carbamoylbenzoyl-CoA, 3-carbamoylpicolinoyl-CoA, 4-coumaroyl-CoA, and N-methylanthraniloyl-CoA, and at 45 °C for 2-carbamoylnaphthaloyl-CoA. The reaction products were extracted and separated by TLC (Merck Art. 1.11798 Silica gel 60 F254; ethyl acetate/hexane/AcOH = 63∶27∶5, v/v/v). Radioactivities were quantified by autoradiography using a bioimaging analyzer BAS-2000II (FUJIFILM). Lineweaver–Burk plots of the data were employed to derive the apparent KM and kcat values (average of triplicates), using Microsoft Excel (Microsoft).
X-ray Crystallography.
Crystallization and X-ray diffraction date collection were performed, as previously reported (33). Data were indexed, integrated, and scaled using the HKL-2000 program (34). The initial phases of the HsPKS1 structure were determined by molecular replacement, using the HsPKS1 structure generated by the SWISS-MODEL package (http://expasy.ch/spdpv/), based on the crystal structure of M. sativa CHS (PDB ID code 1CGK) as a search model. The molecular replacement was performed with CNS (35). Crystallographic refinement and model building were performed with CNS and MIFit (36), respectively. Each refinement cycle was followed by model building, using the σA-weighted 2Fo-Fc and Fo-Fc electron density maps. The water molecules were automatically placed into the difference electron density maps with MIFit and were retained or rejected based on geometric criteria as well as their refined B factors. After several rounds of model building and refinement, the final model was obtained. The final model consists of residues 20–399 of monomer A, two molecules of glycerol, one molecule of SO4, and 168 molecules of water. Details of the data collection, processing, and structure refinement are summarized in Table S3. The quality of the final model was assessed with PROCHECK (37). A total of 91.7% of the residues in the HsPKS1 apo structure are in the most favored regions of the Ramachandran plot, 8.0% in the additional allowed regions, and 0.3% in the generously allowed region. Structure-based similarity was searched by the Dali program (38). The cavity volume and the active-site entrance area were calculated by the CASTP program (http://cast.engr.uic.edu/cast/). All crystallographic figures were prepared with PyMOL (DeLano Scientific, http://www.pymol.org). Crystallization and structure determination of the HsPKS1 complexed with CoA-SH are described in SI Materials and Methods.
Molecular Modeling and Docking Studies.
The three-dimensional models of the CoA thioesters and the Cys-tethered intermediates were generated by the Chem3D Ultra 10 program (CambridgeSoft). The intermediate models were manually swapped with the catalytic Cys174 in the HsPKS1 structure by using MIFit, and the energy minimization calculation by simulated annealing with CNS program was then performed. The parameters of the intermediates for the energy minimization calculation were obtained by the PRODRG server (http://davapc1.bioch.dundee.ac.uk/prodrg/).
Supplementary Material
Acknowledgments.
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to I.A. and H.M.) and grants from the National Project on Protein Structural and Functional Analyses (to S.S. and T.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3AWK, for the apo structure, and 3AWJ, for the CoA-SH complexed structure).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107782108/-/DCSupplemental.
References
- 1.Abe I, Morita H. Structure and function of the chalcone synthase superfamily of plant type III polyketide synthases. Nat Prod Rep. 2010;27:809–838. doi: 10.1039/b909988n. [DOI] [PubMed] [Google Scholar]
- 2.Austin MB, Noel JP. The chalcone synthase superfamily of type III polyketide synthases. Nat Prod Rep. 2003;20:79–110. doi: 10.1039/b100917f. [DOI] [PubMed] [Google Scholar]
- 3.Schröder J. In: Comprehensive Natural Products Chemistry. Sankawa U, editor. Vol 1. Oxford: Elsevier; 1999. pp. 749–771. [Google Scholar]
- 4.Abe I, Morita H, Nomura A, Noguchi H. Substrate specificity of chalcone synthase: Enzymatic formation of unnatural polyketides from synthetic cinnamoyl-CoA analogs. J Am Chem Soc. 2000;122:11242–11243. [Google Scholar]
- 5.Morita H, Takahashi Y, Noguchi H, Abe I. Enzymatic formation of unnatural aromatic polyketides by chalcone synthase. Biochem Biophys Res Commun. 2000;279:190–195. doi: 10.1006/bbrc.2000.3920. [DOI] [PubMed] [Google Scholar]
- 6.Jez JM, Bowman ME, Noel JP. Expanding the biosynthetic repertoire of plant type III polyketide synthases by altering starter molecule specificity. Proc Natl Acad Sci USA. 2002;99:5319–5324. doi: 10.1073/pnas.082590499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abe I, Watanabe T, Noguchi H. Enzymatic formation of long-chain polyketid pyrones by plant type III polyketide synthases. Phytochemistry. 2004;65:2447–2453. doi: 10.1016/j.phytochem.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Abe I, Abe T, Wanibuchi K, Noguchi H. Enzymatic formation of quinolone alkaloids by plant type III polyketide synthase. Org Lett. 2006;8:6063–6065. doi: 10.1021/ol0625233. [DOI] [PubMed] [Google Scholar]
- 9.Katsuyama Y, Funa N, Miyahisa I, Horinouchi S. Synthesis of unnatural flavonoids and stilbenes by exploiting the plant biosynthetic pathway in Escherichia coli. Chem Biol. 2007;14:613–621. doi: 10.1016/j.chembiol.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Wakimoto T, Mori T, Morita H, Abe I. Cytotoxic tetramic acid derivative produced by a plant type-III polyketide synthase. J Am Chem Soc. 2011;133:4746–4749. doi: 10.1021/ja2006737. [DOI] [PubMed] [Google Scholar]
- 11.Abe I, et al. A plant type III polyketide synthase that produces pentaketide chromone. J Am Chem Soc. 2005;127:1362–1363. doi: 10.1021/ja0431206. [DOI] [PubMed] [Google Scholar]
- 12.Abe I, Oguro S, Utsumi Y, Sano Y, Noguchi H. Engineered biosynthesis of plant polyketides: Chain length control in an octaketide-producing plant type III polyketide synthase. J Am Chem Soc. 2005;127:12709–12716. doi: 10.1021/ja053945v. [DOI] [PubMed] [Google Scholar]
- 13.Morita H, et al. Structural insight into chain-length control and product specificity of pentaketide chromone synthase from Aloe arborescens. Chem Biol. 2007;14:359–369. doi: 10.1016/j.chembiol.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Abe I, Watanabe T, Morita H, Kohno T, Noguchi H. Engineered biosynthesis of plant polyketides: manipulation of chalcone synthase. Org Lett. 2006;8:499–502. doi: 10.1021/ol052912h. [DOI] [PubMed] [Google Scholar]
- 15.Abe I, et al. Structure-based engineering of a plant type III polyketide synthase: Formation of an unnatural nonaketide naphthopyron. J Am Chem Soc. 2007;129:5976–5980. doi: 10.1021/ja070375l. [DOI] [PubMed] [Google Scholar]
- 16.Shi S-P, et al. Enzymatic formation of unnatural novel chalcone, stilbene, and benzophenone scaffolds by plant type III polyketide synthase. Org Lett. 2009;11:551–554. doi: 10.1021/ol802606w. [DOI] [PubMed] [Google Scholar]
- 17.Morita H, et al. Structural basis for the one-pot formation of the diarylheptanoid scaffold by curcuminoid synthase from Oryza sativa. Proc Natl Acad Sci USA. 2010;107:19778–19783. doi: 10.1073/pnas.1011499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abe I. Engineered biosynthesis of plant polyketides: Structure-based and precursor-directed approach. Top Curr Chem. 2010;297:45–66. doi: 10.1007/128_2009_22. [DOI] [PubMed] [Google Scholar]
- 19.Wanibuchi K, et al. An acridone-producing novel multifunctional type III polyketide synthase from Huperzia serrata. FEBS J. 2007;274:1073–1082. doi: 10.1111/j.1742-4658.2007.05656.x. [DOI] [PubMed] [Google Scholar]
- 20.Wada M, Nakai H, Sato Y, Kanaoka Y. A removable functional group in a photochemical macrocyclic synthesis: Remote photocyclization with a pair system of phthalimide and 1,3-dithiolanyl groups. Chem Pharm Bull. 1982;30:3414–3417. [Google Scholar]
- 21.Abe Y, Ohsawa A, Igeta H. Synthesis of pyrido[2,1-a]isoindol-6(2H)-one and its analogs. Heterocycles. 1982;19:49–51. [Google Scholar]
- 22.Dura RD, Modolo I, Paquette LA. Effective synthetic routes to 4H- and 10bH-pyrido[2,1-a]isoindol-6-ones. Heterocycles. 2007;74:145–148. [Google Scholar]
- 23.Lorence A, Nessier CL. Camptothecin, over four decades of surprising findings. Phytochemistry. 2004;65:2735–2749. doi: 10.1016/j.phytochem.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Sirikantaramas S, Asano T, Sudo H, Yamazaki M, Saito K. Camptothecin: Therapeutic potential and biotechnology. Curr Pharm Biotechnol. 2007;8:196–202. doi: 10.2174/138920107781387447. [DOI] [PubMed] [Google Scholar]
- 25.Ferrer JL, Jez JM, Bowman ME, Dixon RA, Noel JP. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat Struct Biol. 1999;6:775–784. doi: 10.1038/11553. [DOI] [PubMed] [Google Scholar]
- 26.Werner LH, Ricca S, Mohacsi E, Rossi A, Arya VP. Derivatives of morphanthridine. J Med Chem. 1965;8:74–80. doi: 10.1021/jm00325a016. [DOI] [PubMed] [Google Scholar]
- 27.Gijsen HJ, et al. Analogues of morphanthridine and the tear gas dibenz[b,f][1,4]oxazepine (CR) as extremely potent activators of the human transient receptor potential ankyrin 1 (TRPA1) channel. J Med Chem. 2010;53:7011–7020. doi: 10.1021/jm100477n. [DOI] [PubMed] [Google Scholar]
- 28.Pokholenko AA, Voitenko ZV, Kovtunenko VA. Pyrido- and pyrimidoisoindoles: methods of synthesis and properties. Russ Chem Rev. 2004;73:771–784. [Google Scholar]
- 29.Elward AM, McAndrews JM. Young VL Methicillin-sensitive and methicillin-resistant Staphylococcus aureus: Preventing surgical site infections following plastic surgery. Aesthet Surg J. 2009;29:232–244. doi: 10.1016/j.asj.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 30.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 31.Roth BL, Tanadra S, Burgess LH, Sibley DR, Meltzer HY. D4 Dopamine receptor binding affinity does not distinguish between typical and atypical antipsychotic drugs. Psychopharmacology. 1995;120:365–368. doi: 10.1007/BF02311185. [DOI] [PubMed] [Google Scholar]
- 32.Viti G, et al. New [dibenzo(b, e)azepin-5-yl]-acetamides with anti-convulsant activity. Eur J Med Chem. 1993;28:439–445. [Google Scholar]
- 33.Morita H, et al. Crystallization and preliminary crystallographic analysis of an acridone-producing novel multifunctional type III polyketide synthase from Huperzia serrata. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:576–578. doi: 10.1107/S1744309107026164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 36.McRee DE. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 37.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 38.Holm L, Sander C. Dali: A network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.