Abstract
Viruses have been implicated in the development of neurodegenerative diseases, such as Alzheimer's, Parkinson’s, and multiple sclerosis. Human herpesvirus-6 (HHV-6) is a neurotropic virus that has been associated with a wide variety of neurologic disorders, including encephalitis, mesial temporal lobe epilepsy, and multiple sclerosis. Currently, the route of HHV-6 entry into the CNS is unknown. Using autopsy specimens, we found that the frequency of HHV-6 DNA in the olfactory bulb/tract region was among the highest in the brain regions examined. Given this finding, we investigated whether HHV-6 may infect the CNS via the olfactory pathway. HHV-6 DNA was detected in a total of 52 of 126 (41.3%) nasal mucous samples, showing the nasal cavity is a reservoir for HHV-6. Furthermore, specialized olfactory-ensheathing glial cells located in the nasal cavity were demonstrated to support HHV-6 replication in vitro. Collectively, these results support HHV-6 utilization of the olfactory pathway as a route of entry into the CNS.
Keywords: olfactory system, viral infection
The olfactory system bridges the peripheral environment with the central nervous system and is constantly exposed to a variety of pathogens, such as viruses. A number of neurotropic viruses, including herpes simplex viruses, borna disease virus, rabies virus, influenza A virus, and parainfluenza virus are known to infect the CNS by transmission through the olfactory pathway (1). The olfactory system is connected to and supported by the limbic system, which includes the hippocampus, thalamus, and amygdala (2). Viruses known to use the olfactory system to invade the CNS are primarily found in limbic structures (3, 4). In both HSV encephalitis and rabies encephalitis the limbic system is a major site of damage, and in individuals affected with borna virus, viral DNA has been found in the hippocampus (5–8). Depending on their ability to directly infect olfactory receptor neurons, these viruses are thought to infect cells in the nasal endothelium, thus gaining direct access to the CNS (9, 10).
Olfactory-ensheathing cells (OECs) are a group of specialized glial cells located in the nasal cavity. They possess characteristics of CNS astrocytes and Schwann cells, making them potential targets for viruses known to infect glial cells. The primary function of OECs is to guide newly formed olfactory receptor neurons across the cribiform plate into the CNS to form appropriate connections. OECs are also important in promoting neuro-regeneration. In animal models of spinal transection, transplantation of OECs promotes the restoration of axonal connections as well as remyelination (9). The ability of OECs to remyelinate injured neurons makes them unique and particularly interesting; these cells have recently been the subject of intense research concerning spinal cord injuries, amyotrophic lateral sclerosis, multiple sclerosis (MS), and other neurodegenerative diseases (10). The majority of OEC research has used animal models, and the literature using human OECs is currently limited.
Human herpesvirus-6 (HHV-6) is frequently associated with neurologic diseases, including MS, mesial temporal lobe epilepsy (MTLE), encephalitis, and febrile illness (11). This ubiquitous β-herpesvirus exists in two variants that share 95% sequence homology, HHV-6A and HHV-6B (12). HHV-6B is the etiologic agent for roseola in children and is usually contracted by 2 years of age, the time of exposure and symptoms associated with HHV-6A remains unknown (13). Both variants of HHV-6 have the ability to become latent and remain quiescent in the host following primary exposure (14). One proposed site of HHV-6 latency is in the tonsils and adenoids, where the virus has been shown to be actively shed in saliva (15, 16). In addition, many in vitro studies have demonstrated that HHV-6 has a tropism for glial cells that may also represent an in vivo reservoir of viral latency in the CNS (17–20). Although HHV-6 is increasingly recognized as an important pathogen in a number of neurologic diseases, how the virus gains access to the CNS is not completely understood. Given that HHV-6 has been demonstrated in salivary glands and that a common route of entry into the CNS for many viruses is the olfactory pathway, we investigated whether the olfactory system can support HHV-6 infection. Here we examine (i) the distribution of HHV-6 in the CNS and olfactory tissues at autopsy, (ii) the prevalence of HHV-6 DNA in nasal mucus, and (iii) the ability of human OECs to support HHV-6 infection in vitro.
Results
HHV-6 Is Present in Fresh Brain and CNS Autopsy Material.
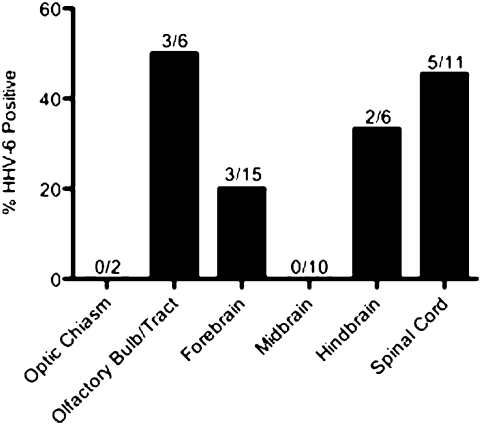
To determine the distribution of HHV-6 in the brain and CNS, we surveyed multiple regions of the brain and spinal cord for HHV-6. A total of 50 brain samples from three patient autopsies were analyzed for the presence of HHV-6 DNA. Because of the low amounts of viral DNA in the tissue, it was necessary to use nested PCR to detect HHV-6. In each patient, 11 to 24 samples were tested and in each individual between 25% and 33% of these samples tested positive for HHV-6 (Table 1). Consistent with previous studies, HHV-6 DNA was amplified from 26% (13 of 50) of total tissue samples tested (21). To evaluate the distribution of HHV-6 in the CNS, brain samples were divided into six categories based on their anatomical locations: optic chiasm; olfactory bulb/tract; forebrain (amygdala, corpus collosum, cortex, frontal lobe, hippocampus, hypothalamus, parietal lobe, pituitary gland, frontal gyrus, temporal lobe, thalamus); midbrain (caudate, internal capsule, palidum, putamen, substantia nigra); hindbrain (cerebellum, dentate, medulla, pons); and spinal cord (cervical spinal cord, lumbar spinal cord/ganglia, thoracic spinal cord/ganglia). Although HHV-6 DNA could be found throughout the brain, percentages of HHV-6 positivity were elevated in the olfactory bulb/tract (50%), spinal cord (45.4%), and hindbrain (33.3%) regions (Fig. 1).
Table 1.
Survey of HHV-6 in the CNS
Fig. 1.
Survey of autopsy brain specimens for HHV-6. Primers specific for the U67 gene region of HHV-6 were used for nested PCR amplification. Tissues from various regions of the brain obtained from three NIH autopsy patients were categorized into the indicated groups based on anatomical locations. The percentages of HHV-6 positivity in each category are expressed on the y axis. The numbers above each region indicate the number of samples positive for HHV-6 of the total number of specimen tested in each category.
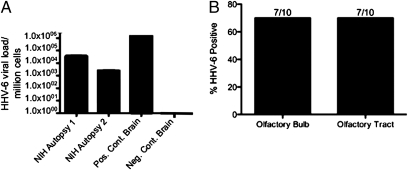
The olfactory bulbs from two of the three National Institutes of Health (NIH) autopsy cases investigated were found to have high enough levels of HHV-6 DNA to be assayed with quantitative real-time TaqMan PCR using variant-specific primers. HHV-6B was detected at high viral loads, 1 × 103 and 4 × 104 virus copies per million cells, in the olfactory bulb tissue samples from the two patients tested (Fig. 2A). By comparison, in HHV-6 associated encephalitis brain biopsies, titers of 1 × 106 virus copies per million cells are found (Fig. 2A). To extend this observation, additional fresh-frozen olfactory bulb and tract materials were obtained from 10 patients, five diagnosed with multiple sclerosis and five with cancer, from the University of California at Los Angeles (UCLA) brain bank and tested for the presence of HHV-6 DNA. The olfactory bulb and tract were tested for HHV-6 using nested PCR because of the low levels of HHV-6 DNA in the tissue. The UCLA olfactory tissues show a high frequency of HHV-6 DNA, 70% (n = 20) (Fig. 2B), and sequencing identified HHV-6B as the variant present in all positive samples. These data confirm what was seen in the olfactory bulb/tract from the initial autopsy cases (Fig. 1). The frequency of HHV-6 positivity did not vary between the MS patients and cancer patients, indicating this may be a common observation not related to disease.
Fig. 2.
Detection of HHV-6 DNA in autopsy olfactory specimens. (A) HHV-6 variant B was amplified by real-time quantitative PCR from olfactory bulb specimens from two NIH autopsy cases (MS and leukemia, respectively) and the insular cortex of a patient with HHV-6–associated encephalitis for comparison. (B) Olfactory bulb (n = 10) and tract (n = 10) specimens ascertained from the UCLA brain bank were tested using nested PCR for the U67 gene of HHV-6. A high prevalence of HHV-6 DNA was detected in the olfactory tissues.
HHV-6 Is Present in the Nasal Cavity.
To evaluate the prevalence of HHV-6 in the nasal passage, a total of 126 nasal mucus specimens were tested for presence of HHV-6 DNA using nested PCR. Nasal mucus specimens are routinely collected from patients with smell loss and their availability allowed us to screen a large number of patients. Given the association of HHV-6 and neurologic conditions, we also collected nasal mucus from MS patients and healthy controls for HHV-6 testing. High frequencies of HHV-6 DNA were detected in nasal mucus samples but were not significantly different among all three cohorts: healthy controls (60.0%), MS (53.8%), and smell loss (35.5%) (Table 2). Sequencing was used to determine the HHV-6 variant in four of the seven positive samples from MS patients and all tested as HHV-6B. Because the saliva is a well-known in vivo reservoir for HHV-6, we also examined the frequency of HHV-6 DNA in saliva of the same patients. As expected, equally high percentages of saliva contained HHV-6 DNA, healthy controls (50.0%), MS (66.7%), and smell loss (40.6%), with no statistically significant differences among the three groups (Table 2). Sequencing was done on 4 of the 13 positive MS patient samples: three tested as HHV-6B and one as HHV-6A. These results demonstrate that the presence of HHV-6 in the nasal cavity and saliva is a common observation and does not appear to be linked to any of the conditions studied. Importantly, these data suggest that the nasal cavity, like the saliva, is an in vivo reservoir for HHV-6.
Table 2.
Demographics of sample cohorts and HHV-6 DNA
| Total subjects | Sex | Mean age ± SD | HHV-6 (+) nasal mucous | HHV-6 (+) saliva | |
| Smell loss | 102 | 53 F/49 M | 56.1 ± 15.0 | 33/93 (35.5%) | 39/96 (40.6%) |
| MS | 21 | 11 F/10 M | 46.2 ± 10.3 | 7/13 (53.8%) | 14/21 (66.7%) |
| Healthy controls | 20 | 13 F/7 M | 31.6 ± 6.8 | 12/20 (60.0%) | 9/18 (50.0%) |
OECs Are Readily Infected with HHV-6.
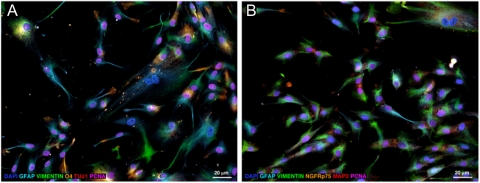
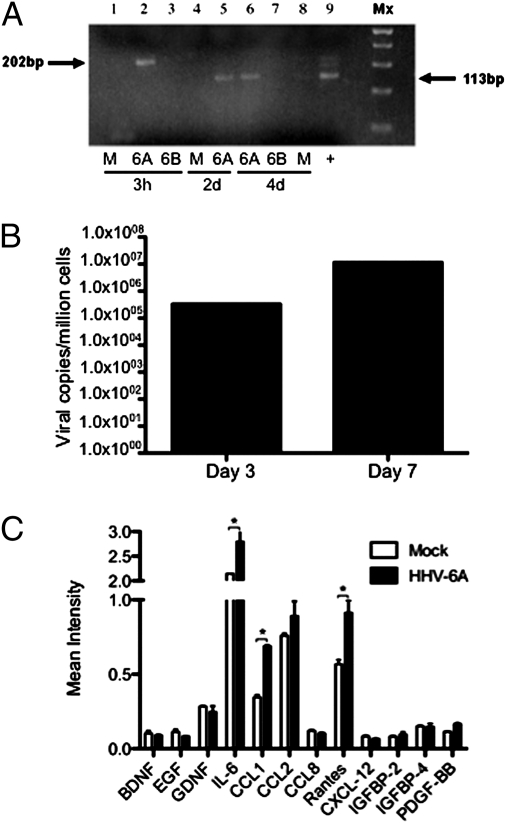
Human OECs are glial cells that reside in the nasal cavity and have properties of astrocytes and Schwann cells. Cultured OECs were histologically characterized by immunofluorescence staining with neuronal, astrocytic, and oligodendrocytic markers (Fig. 3). As shown, no neuronal phenotypes were observed in the cultured human OECs, indicated by the absence of β-tubulin III (TUJ1) or microtubule-associated protein 2 (MAP2). Similarly, positive vimentin staining of intermediate filament expression by immature cells indicated a lack of fibroblasts in the OEC culture. Consistent with previous characterization of OECs, cells in our culture also coexpressed astrocyte, oligodendrocyte, and Schwann cell markers, such as vimentin, glial fibrillary acidic protein (GFAP), O4, and low-affinity nerve growth factor receptor p75 (NGFRp75). Because OECs have properties of astrocytes, which are known to be susceptible to HHV-6 infection (19, 22), we investigated whether OECs could also be productively infected by HHV-6. Cultures of OECs were infected with cell-free viral supernatant containing either HHV-6A (strain U1102) or HHV-6B (strain Z29). Cells were harvested at various time points for evaluation of HHV-6 replication by RT-PCR using primers that can detect de novo RNA synthesis of the U12 gene of HHV-6. De novo RNA synthesis of HHV-6A was found in OECs on days 2 and 4 postinfection (Fig. 4A). OECs harvested days 3 and 7 postinfection with HHV-6A were tested for viral DNA using real-time quantitative Taqman and showed an increased viral load over time (Fig. 4B), suggesting a productive infection of OECs with HHV-6A. In contrast, de novo RNA synthesis was not found in OECs exposed to HHV-6B, suggesting that OECs are not as permissive to infection with this variant. These results are similar to previous studies, which demonstrated that in vitro infection of other glial cells including astrocytes results in a productive infection by HHV-6A, but infection with HHV-6B resulted in either abortive or persistent infections (19, 22). To our knowledge this report is unique in presenting productive infection of human OECs. Because HHV-6A but not HHV-6B could productively infect OEC, we analyzed the cytokine production of OECs infected with HHV-6A early in infection. Infected OECs were found to significantly increase the production of IL-6, and the chemokines CCL-1 and Rantes (Fig. 4C). The cytokine profiles of infected OECs would suggest HHV-6A infection triggers the release of proinflammatory mediators.
Fig. 3.
Characterization of cultured OECs by immunofluorescence staining. (A) Primary human OECs were labeled using anti–vimentin-Alexa Fluor 488 (green), anti–GFAP-AMCA (azure), anti–O4-Alexa Fluor 546 (orange), anti–TUJ1-Alexa Fluor 647 (red), and anti–PCNA-biotin-streptavidin-Alexa Fluor 750 (pink). After imaging the above labels, the remaining AMCA signal was photobleached and the cell nuclei were counterstained with DAPI (blue) to facilitate cell visualization. (B) Primary human OECs from a separate culture probed with anti–vimentin-Alexa Fluor 488 (green), anti-GFAP-Alexa Fluor 350 (azure), anti–NGFPp75-Alexa Fluor 546 (orange), anti–MAP2-Alexa Fluor 647 (red), anti-PCNA-biotin-streptavidin-Alexa Fluor 750 (pink) and DAPI (blue) using the same labeling strategy as in A. Astroglial and oligodendroglial markers such as vimentin, GFAP, NGFRp75, and O4 were widely expressed in OECs. Many of these cells were PCNA+ and thus actively proliferating. Cells of the neuronal lineage were absent in the OEC cultures as indicated by negative TUJ1 and MAP2 staining.
Fig. 4.
HHV-6A infects olfactory ensheathing cells in vitro. (A) OEC cultures were infected with HHV-6A or HHV-6B and cells were harvested for analysis of de novo virus transcript with the U12 primers. The higher 202-bp band corresponds to the nonspliced HHV-6 precursor RNA and the lower 113-bp PCR product represents de novo RNA transcript. Lanes 1, 2, and 3 are 3 h postinfection of mock, OEC+U1102, and OEC+Z29, respectively. Lanes 4 and 5 are 2 d postinfection of mock and OEC+U1102, respectively. Lanes 6, 7, and 8 are 4 d postinfection of OEC+U1102, OEC+Z29, and mock, respectively. Lane 9 is the positive control from HHV-6 infected U251 human astrocyte cell line. (B) HHV-6A infected OECs were harvested 3 and 7 d postinfection for viral load analysis by real time quantitative Taqman. Calculated viral loads were normalized to β-actin. (C) Cytokine production measured on day 3 of HHV-6A infection in OECs. Cell-culture supernatants were analyzed using a cytokine antibody array as described in Methods. The data shown are from a representative experiment and mean intensities were normalized to controls on each membrane (n = 2). Statistical significance was measured using ANOVA (* indicates P < 0.05).
Discussion
Although HHV-6 has been implicated in the development of a wide spectrum of neurologic disorders, including MS, MTLE, and encephalitis, the route of viral entry into the CNS remains unknown (23–29). The olfactory system, a well-known route of entry for many viruses into the CNS, is connected to the limbic structures of the brain, providing a possible path for viruses to infect the CNS. For example, in animal models, Theiler's murine encephalomyelitis virus, influenza A, rabies virus, West Nile virus, and borna disease virus are known to rapidly disseminate throughout the CNS by olfactory transmission (30). In this study, we provided data showing that HHV-6 is able to reside in the olfactory system, making it a possible route of CNS transmission.
HHV-6 infection through the olfactory pathway may result in clinical consequences. For example, HHV-6 DNA has been found in tissues of the limbic system, including the hippocampus and temporal lobe of patients with MTLE (24, 25). In addition, in posttransplant recipients reactivation of HHV-6 in the cerebrospinal fluid led to the development of limbic encephalitis. The presence of HHV-6 in limbic tissues and its ability to cause limbic encephalitis is consistent with other viruses known to enter the CNS through the olfactory system. Interestingly, when the various regions were stratified according to their anatomical locations, the olfactory tissues were one of the brain regions with the highest prevalence of HHV-6 (60%) (Figs. 1 and 2). We also found no correlation between HHV-6 positivity and disease in the olfactory tissue tested, suggesting that the presence of HHV-6 in this tissue is a common observation. Although structures of the limbic system, characterized as forebrain in this study, did not have the highest prevalence of HHV-6, the presence of virus in olfactory bulb and regions beyond the forebrain suggest that the virus may have traveled through the forebrain and established residency elsewhere. HSV-1, another virus known to disseminate into the CNS through the olfactory system, is found in ∼15% of olfactory bulbs at autopsy (5, 31). The comparatively high prevalence of HHV-6 in olfactory bulbs, as well as other regions of the CNS, supports the hypothesis that the olfactory pathway is a route of HHV-6 transmission into the CNS.
In addition to finding virus in the olfactory bulbs, results presented in this article find high frequencies of HHV-6 in the nasal cavity, which allows the virus constant access to the olfactory pathway and thus entry into the CNS. HHV-6 DNA was amplified from 41% of all nasal mucus specimens tested (Table 2). Again we found no correlation between disease and the frequency of HHV-6 positivity in nasal mucus samples. The prevalence of HHV-6 DNA in the nasal cavity is comparable to that found in saliva (46%); saliva is considered an in vivo reservoir of HHV-6 (32). Because of the similarly high incidence of HHV-6 DNA detection in nasal mucus samples and saliva, we suggest that the nasal cavity also be considered an in vivo reservoir of HHV-6. Whether presence of HHV-6 in the nasal cavity is indicative of infection of resident cells in the olfactory pathway in vivo remains to be explored.
Given their similarities to glial cells, such as astrocytes, known to be readily infectable by HHV-6, OECs present an attractive target for HHV-6 infection. The data presented here show that HHV-6A can establish an infection in human OECs in vitro, as evidenced by the presence of de novo viral RNA synthesis in OECs exposed to cell-free HHV-6A (Fig. 4A), indicating active viral replication in these infected OEC cultures. The viral load of the infection increased over time, supporting the presence of an active infection (Fig. 4B). To our knowledge, this article is unique in reporting an in vitro viral infection of human OECs. Although we were not able to infect OECs with HHV-6B in vitro, that does not preclude this from occurring in vivo. Astrocytes, which share many properties of OECs, are less permissive to HHV-6B infection in vitro than in vivo and this may be the case with OECs as well (19, 22). The primary roles of OECs include providing a growth-supportive environment for newly regenerated olfactory neurons and guiding the primary olfactory axons from the nasal cavity across the blood-brain barrier and into the brain (10). Interestingly, recent studies have shown that OECs also have significant neuro-regenerative properties. Transplantation of OECs has been used to promote restoration of axonal connections, as well as remyelination in various animal models of spinal transection (9). Given the increase in proinflammatory cytokine production and slight reduction in neurotrophin production during HHV-6A infection, (Fig. 4C) these functions may be impaired in actively infected OECs. With the important biological properties of OECs, HHV-6 infection in vivo would provide a mechanism for continuous transmission across the blood-brain barrier, resulting in longstanding access to the CNS.
Our study supports the possibility of HHV-6 entry into the CNS via infection of and residence in the olfactory system. Collecting primary OECs from within the CNS and surveying for virus would be ideal to confirm this route of infection. In this study it was shown that, HHV-6 DNA can be amplified from both nasal mucus and olfactory brain tissues, and OECs can be infected productively with HHV-6 in vitro. Data in this study provide evidence that the nasal cavity is a reservoir for HHV-6, and supports the hypothesis that HHV-6 travels from the nasal cavity into the brain, using the olfactory pathway as a route of entry.
Methods
DNA and RNA Extraction.
Informed consent was written and obtained from each subject in accordance with the Declaration of Helsinki. The study was reviewed and approved by the National Institute of Neurological Disorders and Stroke Institutional Review Board. DNeasy Blood and Tissue DNA extraction kit (Qiagen) was used according to the manufacturer's instructions. Fresh brain tissue specimens obtained from three autopsy cases (one MS, one cancer, and one multisystem atrophy) at the NIH, and 10 fresh-frozen olfactory bulb/tract samples from the UCLA brain bank (five MS and five cancer patients) were kept on ice and processed following the protocol for DNA extraction from tissues. Saliva samples from smell-loss patients were collected by aspiration from the parotid gland; from MS patients and healthy controls saliva was collected using a salivette (Sarstedt). Nasal mucus secretions from smell-loss patients, MS patients, and healthy controls were collected over 3 d and pooled for each subject. To decrease the viscosity of nasal secretions, an equal volume of (N-acetyl-l-cysteine) NALC buffer containing 2.94% sodium citrate, 4% sodium hydroxide, and 500 mg of NALC was added before extraction. DNA from nasal secretions with NALC, and saliva, were extracted according to instructions for cell-free materials. RNeasy RNA extraction kit (Qiagen) was used for RNA extraction.
HHV-6–Specific Nested, Real-Time Quantitative TaqMan PCR, and RT-PCR.
Nested PCR detection of HHV-6 DNA was performed with primers against the U67 (major capsid protein-MCP) region of the viral genome, as previously described (29). To avoid PCR contamination, sample extraction, PCR mastermix preparation, and sample loading were performed at three separate locations in the laboratory. HHV-6 viral load was measured using HHV-6A or 6B-specific U90 primers in TaqMan quantitative PCR with 9700HT Fast Real Time PCR System (Applied Biosystems), as previously described (25, 33, 34). Briefly, for cell-free specimens, 5 μL of extracted DNA was amplified with a final concentration of 0.5 μM HHV-6A or 6B-specific primers and probes in a 20-μL reaction using TaqMan Universal Mastermix (Applied Biosystems). Based on the HHV-6A or 6B plasmid standard curve spanning from 107 to 101 copies, the viral load was calculated and expressed as HHV-6 copies per milliliter (33). For brain biopsy samples, 50 ng of DNA was used in each 20-μL TaqMan PCR and viral load per million cells was calculated by normalizing HHV-6 copy numbers to the house-keeping gene β-actin using the following formula: [HHV-6 copy number/(β-actin copy number/2)] × 106.
RT-PCR was performed using the HHV-6 U12 primers, which amplify both HHV-6A and HHV-6B, as previously described (18, 19). RNA samples extracted from cultured human OECs were first treated with DNase I and cDNA was created using the SuperScript III RT kit (Invitrogen). PCR amplifications of cDNA were performed as previously described (19). Primer sequences and PCR cycling conditions are listed in Table 3.
Table 3.
HHV-6 primers/probe sets used in PCR reactions
| Primers/probe sets |
Amplicon size |
|||
| Nucleotide sequence 5′→3′ | DNA | cDNA | PCR cycle conditions | |
| U67 HHV-6 | Ext-F- 5′-GCGTTTTCAGTGTGTAGTTCGGCAG-3′ | 524 | 92 °C:3 min, 92 °C:30 s, 55 °C:30 s, 72 °C:30 s (35 cycles), 72 °C:7 min, 4 °C | |
| Ext-R- 5′-TGGCCGCATTCGTACAGATACGGAGG-3′ | ||||
| Int-F- 5′-GCTAGAACGTATTTGCTGCAGAACG-3′ | 258 | 92 °C:3 min, 92 °C:30 s, 55 °C:30 s, 72 °C:30 s (35 cycles), 72 °C:7 min, 4 °C | ||
| Int-R- 5′-ATCCGAAACAACTGTCTGACTGGCA-3′ | ||||
| U12 HHV-6 | Ext-F-5′-CACTGTCATTGAGCTGTCCAA-3′ | 327 | 238 | 94 °C:5 min, 94 °C:30 s, 57 °C:30 s, 72 °C:30 s (35 cycles), 72 °C:7min, 4 °C |
| Ext-R-5′-ACCACATGAGCACAAAATCG-3′ | ||||
| Int-F-5′-CACTGTCATTGAGCTGTCCAA-3′ | 202 | 113 | 94 °C:5 min, 94 °C:30 s, 57 °C:30 s, 72 °C:30 s (35 cycles), 72 °C:7min, 4 °C | |
| Int-R-5′-TGAGCGTGATTCCGGTAACT-3′ | ||||
| U90 HHV-6A | F-5′-CATGAAGATGATGACAATAAAATG-3′ | 278 | 94 °C:3 min, 94 °C:20 s, 65 °C:50 s (45 cycles), 4 °C | |
| R-5′-TGGAACCATCTTGTTCTGTCC-3′ | ||||
| FAM-CCGCCCAGATCTGTCACTGAGGCG-TAMRA | ||||
| U90 HHV-6B | F-5′-GAGACCGGGTCTGGACAACA-3′ | 145 | 94 °C:3 min, 94 °C:20 s, 65 °C:50 s (45 cycles), 4 °C | |
| R-5′-GAGTTGCTGAGTTGGTAAAGG-3′ | ||||
| FAM-CTCCAAGTGTACCGAAACGCTTCCTGG-TAMRA | ||||
| β-Actin | F-5′-CACACTGTGCCCATCTACGA-3′ | 107 | 94 °C:3 min, 94 °C:20 s, 65 °C:50 s (45 cycles), 4 °C | |
| R-5′-CTCAGTGAGGATCTTCATGAGGTAGT-3′ | ||||
| FAM-ATGCCCTCCCCCATGCCATCCTGCGT-TAMRA | ||||
Maintenance and Infection of Primary Human OECs with HHV-6.
OECs were obtained from A. MacKay-Sim (Griffith University, Brisbane, QLD, Australia) and maintained in poly-l-lysine–coated tissue culture flasks containing 10% FCS and 1% antibiotics, in glutamine supplemented DMEM. OECs were infected with HHV-6A U1102 and HHV-6B Z29 at a ratio of 100 copies of HHV-6–1 cells, as previously described (18). Briefly, cell-free viral supernatant or mock supernatant from noninfected SupT1 cells (human T-cell line) was added to OEC culture that were 60% confluent for 1 d. Supernatants were removed the next day and cells were washed with PBS before replenishing with 5% FCS in DMEM growth medium. Mock and HHV-6 infected OECs were trypsinized and harvested 3 h, 2 d, and 4 d postinfection for analysis of HHV-6 de novo viral RNA synthesis using splice variant specific primers for the HHV-6 U12 gene.
Cytokine Analysis of OEC Culture.
OEC cultures were infected with HHV-6A and supernatant was collected 3 d postinfection. Supernatants from both HHV-6A and mock infections were then analyzed using a cytokine antibody array (RayBiotech, Inc.) as per the manufacturer's instructions. The data were analyzed using densitometry software (Kodak) and the results from different membranes were normalized using positive controls provided. ANOVA was used to determine significant differences.
Characterization of OECs in Culture.
Multiepitope immunolabeling protocols were applied to identify the astroglial, oligodendroglial, and neuronal phenotypes in primary human OEC cultures using different combinations of specific neural lineage-selective markers, as previously described (35). OECs were probed using mixtures of primary antibodies that included mouse IgG1 anti-vimentin (Millipore) to identify immature neural progenitor populations, mouse IgG2b anti-GFAP (Santa Cruz Biotechnology) or chicken IgY anti-GFAP (Aves Labs) to identify the astroglial phenotypes, mouse IgM anti-O4 (Millipore) or mouse IgM targeting the low-affinity NGFRp75 (Santa Cruz Biotechnology) to identify the oligodendrocytic and remyelinating phenotypes, and mouse IgG2b anti-TUJ1 (Sigma) or chicken IgY anti- MAP2 (Millipore) to identify the neuronal phenotypes. All staining protocols also included probing for actively proliferating cells in the aforementioned neural cell phenotypes using mouse IgG2a anti-proliferating cell nuclear antigen (PCNA) antibody (Millipore). Each of the above primary immunoreactions was visualized using appropriate fluorophore-conjugated (AMCA, Alexa Fluor 350, Alexa Fluor 488, Alexa Fluor 546, Alexa Fluor 647, Alexa Fluor 750) secondary antibodies obtained either from Jackson ImmunoResearch or Invitrogen. All fluorescence signals were imaged using an Axiovert 200M inverted fluorescence microscope (Zeiss) equipped with standard Alexa Fluor 350/AMCA, Alexa Fluor 488, Alexa Fluor 546, Alexa Fluor 647, and Alexa Fluor 750 filter sets (Semrock). After imaging, the Alexa Fluor 350/AMCA-labeled epitopes were completely photobleached using prolonged light exposure (1–2 min) through the Alexa Fluor 350/AMCA filter set, and cell nuclei were then counterstained with DAPI and the cells reimaged using the same filter set to facilitate cell visualization.
Acknowledgments
We thank Dr. Mackay-Sim for generously providing the human olfactory ensheathing cells.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
References
- 1.Mori I, Nishiyama Y, Yokochi T, Kimura Y. Olfactory transmission of neurotropic viruses. J Neurovirol. 2005;11(2):129–137. doi: 10.1080/13550280590922793. [DOI] [PubMed] [Google Scholar]
- 2.Bach LM. Regional physiology of the central nervous system. Prog Neurol Psychiatry. 1963;18:46–106. [PubMed] [Google Scholar]
- 3.Chowdhury SI, Lee BJ, Ozkul A, Weiss ML. Bovine herpesvirus 5 glycoprotein E is important for neuroinvasiveness and neurovirulence in the olfactory pathway of the rabbit. J Virol. 2000;74:2094–2106. doi: 10.1128/jvi.74.5.2094-2106.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chowdhury SI, Lee BJ, Onderci M, Weiss ML, Mosier D. Neurovirulence of glycoprotein C(gC)-deleted bovine herpesvirus type-5 (BHV-5) and BHV-5 expressing BHV-1 gC in a rabbit seizure model. J Neurovirol. 2000;6:284–295. doi: 10.3109/13550280009030754. [DOI] [PubMed] [Google Scholar]
- 5.Liedtke W, Opalka B, Zimmermann CW, Lignitz E. Age distribution of latent herpes simplex virus 1 and varicella-zoster virus genome in human nervous tissue. J Neurol Sci. 1993;116(1):6–11. doi: 10.1016/0022-510x(93)90082-a. [DOI] [PubMed] [Google Scholar]
- 6.Conomy JP, Leibovitz A, McCombs W, Stinson J. Airborne rabies encephalitis: Demonstration of rabies virus in the human central nervous system. Neurology. 1977;27(1):67–69. doi: 10.1212/wnl.27.1.67. [DOI] [PubMed] [Google Scholar]
- 7.Winkler WG, Fashinell TR, Leffingwell L, Howard P, Conomy P. Airborne rabies transmission in a laboratory worker. JAMA. 1973;226:1219–1221. [PubMed] [Google Scholar]
- 8.De La Torre JC, et al. Detection of borna disease virus antigen and RNA in human autopsy brain samples from neuropsychiatric patients. Virology. 1996;223:272–282. doi: 10.1006/viro.1996.0479. [DOI] [PubMed] [Google Scholar]
- 9.Mori I, et al. The vomeronasal chemosensory system as a route of neuroinvasion by herpes simplex virus. Virology. 2005;334(1):51–58. doi: 10.1016/j.virol.2005.01.023. [DOI] [PubMed] [Google Scholar]
- 10.Boggian I, et al. Asymptomatic herpes simplex type 1 virus infection of the mouse brain. J Neurovirol. 2000;6:303–313. doi: 10.3109/13550280009030756. [DOI] [PubMed] [Google Scholar]
- 11.Franklin RJ, Barnett SC. Olfactory ensheathing cells and CNS regeneration: The sweet smell of success? Neuron. 2000;28(1):15–18. doi: 10.1016/s0896-6273(00)00080-5. [DOI] [PubMed] [Google Scholar]
- 12.Franssen EH, de Bree FM, Verhaagen J. Olfactory ensheathing glia: Their contribution to primary olfactory nervous system regeneration and their regenerative potential following transplantation into the injured spinal cord. Brain Res Brain Res Rev. 2007;56:236–258. doi: 10.1016/j.brainresrev.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 13.Fotheringham J, Jacobson S. Human herpesvirus 6 and multiple sclerosis: Potential mechanisms for virus-induced disease. Herpes. 2005;12(1):4–9. [PubMed] [Google Scholar]
- 14.Inoue N, Dambaugh TR, Pellett PE. Molecular biology of human herpesviruses 6A and 6B. Infect Agents Dis. 1993;2:343–360. [PubMed] [Google Scholar]
- 15.Asano Y, Yoshikawa T. Human herpesvirus-6 and parvovirus B19 infections in children. Curr Opin Pediatr. 1993;5(1):14–20. doi: 10.1097/00008480-199302000-00003. [DOI] [PubMed] [Google Scholar]
- 16.Caserta MT, Mock DJ, Dewhurst S. Human herpesvirus 6. Clin Infect Dis. 2001;33:829–833. doi: 10.1086/322691. [DOI] [PubMed] [Google Scholar]
- 17.Comar M, Grasso D, dal Molin G, Zocconi E, Campello C. HHV-6 infection of tonsils and adenoids in children with hypertrophy and upper airway recurrent infections. Int J Pediatr Otorhinolaryngol. 2010;74(1):47–49. doi: 10.1016/j.ijporl.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 18.Rhoads MP, Magaret AS, Zerr DM. Family saliva sharing behaviors and age of human herpesvirus-6B infection. J Infect Dis. 2007;54:623–626. doi: 10.1016/j.jinf.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 19.Cassiani-Ingoni R, et al. CD46 on glial cells can function as a receptor for viral glycoprotein-mediated cell-cell fusion. Glia. 2005;52:252–258. doi: 10.1002/glia.20219. [DOI] [PubMed] [Google Scholar]
- 20.Yao K, et al. Differential HHV-6A gene expression in T cells and primary human astrocytes based on multi-virus array analysis. Glia. 2006;53:789–798. doi: 10.1002/glia.20333. [DOI] [PubMed] [Google Scholar]
- 21.Donati D, et al. Variant-specific tropism of human herpesvirus 6 in human astrocytes. J Virol. 2005;79:9439–9448. doi: 10.1128/JVI.79.15.9439-9448.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He J, McCarthy M, Zhou Y, Chandran B, Wood C. Infection of primary human fetal astrocytes by human herpesvirus 6. J Virol. 1996;70:1296–1300. doi: 10.1128/jvi.70.2.1296-1300.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cermelli C, et al. High frequency of human herpesvirus 6 DNA in multiple sclerosis plaques isolated by laser microdissection. J Infect Dis. 2003;187:1377–1387. doi: 10.1086/368166. [DOI] [PubMed] [Google Scholar]
- 24.Ahlqvist J, et al. Differential tropism of human herpesvirus 6 (HHV-6) variants and induction of latency by HHV-6A in oligodendrocytes. J Neurovirol. 2005;11:384–394. doi: 10.1080/13550280591002379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Challoner PB, et al. Plaque-associated expression of human herpesvirus 6 in multiple sclerosis. Proc Natl Acad Sci USA. 1995;92:7440–7444. doi: 10.1073/pnas.92.16.7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donati D, et al. Detection of human herpesvirus-6 in mesial temporal lobe epilepsy surgical brain resections. Neurology. 2003;61:1405–1411. doi: 10.1212/01.wnl.0000094357.10782.f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fotheringham J, et al. Association of human herpesvirus-6B with mesial temporal lobe epilepsy. PLoS Med. 2007;4:e180. doi: 10.1371/journal.pmed.0040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matà S, et al. Human herpesvirus 6-associated limbic encephalitis in adult recipients of unrelated umbilical cord blood transplantation. Bone Marrow Transplant. 2008;42:693–695. doi: 10.1038/bmt.2008.233. [DOI] [PubMed] [Google Scholar]
- 27.Provenzale JM, vanLandingham KE, Lewis DV, Mukundan S, Jr, White LE. Extrahippocampal involvement in human herpesvirus 6 encephalitis depicted at MR imaging. Radiology. 2008;249:955–963. doi: 10.1148/radiol.2492071917. [DOI] [PubMed] [Google Scholar]
- 28.Seeley WW, et al. Post-transplant acute limbic encephalitis: Clinical features and relationship to HHV6. Neurology. 2007;69(2):156–165. doi: 10.1212/01.wnl.0000265591.10200.d7. [DOI] [PubMed] [Google Scholar]
- 29.Yao K, et al. Detection of human herpesvirus-6 in cerebrospinal fluid of patients with encephalitis. Ann Neurol. 2009;65:257–267. doi: 10.1002/ana.21611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wada Y, Fujinami RS. Viral infection and dissemination through the olfactory pathway and the limbic system by Theiler's virus. Am J Pathol. 1993;143:221–229. [PMC free article] [PubMed] [Google Scholar]
- 31.Baringer JR, Pisani P. Herpes simplex virus genomes in human nervous system tissue analyzed by polymerase chain reaction. Ann Neurol. 1994;36:823–829. doi: 10.1002/ana.410360605. [DOI] [PubMed] [Google Scholar]
- 32.Zerr DM, et al. A population-based study of primary human herpesvirus 6 infection. N Engl J Med. 2005;352:768–776. doi: 10.1056/NEJMoa042207. [DOI] [PubMed] [Google Scholar]
- 33.Nitsche A, et al. Human herpesvirus 6A DNA Is detected frequently in plasma but rarely in peripheral blood leukocytes of patients after bone marrow transplantation. J Infect Dis. 2001;183(1):130–133. doi: 10.1086/317651. [DOI] [PubMed] [Google Scholar]
- 34.Fotheringham J, et al. Detection of active human herpesvirus-6 infection in the brain: Correlation with polymerase chain reaction detection in cerebrospinal fluid. J Infect Dis. 2007;195:450–454. doi: 10.1086/510757. [DOI] [PubMed] [Google Scholar]
- 35.Maric D, Maric I, Chang YH, Barker JL. Prospective cell sorting of embryonic rat neural stem cells and neuronal and glial progenitors reveals selective effects of basic fibroblast growth factor and epidermal growth factor on self-renewal and differentiation. J Neurosci. 2003;23:240–251. doi: 10.1523/JNEUROSCI.23-01-00240.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]