Abstract
The translocase of the mitochondrial outer membrane (TOM) complex is the main import pore for nuclear-encoded proteins into mitochondria, yet little is known about its spatial distribution within the outer membrane. Super-resolution stimulated emission depletion microscopy was used to determine quantitatively the nanoscale distribution of Tom20, a subunit of the TOM complex, in more than 1,000 cells. We demonstrate that Tom20 is located in clusters whose nanoscale distribution is finely adjusted to the cellular growth conditions as well as to the specific position of a cell within a microcolony. The density of the clusters correlates to the mitochondrial membrane potential. The distributions of clusters of Tom20 and of Tom22 follow an inner-cellular gradient from the perinuclear to the peripheral mitochondria. We conclude that the nanoscale distribution of the TOM complex is finely adjusted to the cellular conditions, resulting in distribution gradients both within single cells and between adjacent cells.
Keywords: image analysis, nanoscopy, mitochondrial protein import, protein distribution, super-resolution microscopy
Mitochondria are essential organelles in eukaryotes, occupying a central role in cellular energy metabolism. The activity of mitochondria is adapted to changing cellular conditions: It has been suggested that short-term variations in energy demand may be compensated without modification of the mitochondrial enzyme content, but modulation of the mitochondrial protein content has been observed during long-term adaptations (1).
In human cells, only 13 proteins are encoded by the mitochondrial genome; most mitochondrial proteins are synthesized as precursor proteins in the cytosol and are imported into the organelle. The central entry gate for almost all nuclear-encoded mitochondrial proteins is the translocase of the outer mitochondrial membrane (TOM complex) (for a detailed review see refs. 2–4). After passing through this complex, the precursor proteins follow different routes to their final destinations within the organelle. The TOM complex consists of the receptors Tom20, Tom22, and Tom70, the channel-forming protein Tom40, and several small, associated subunits. Tom20 is the initial recognition site for preproteins with presequences (5, 6) and transfers the preproteins to the central receptor, Tom22 (7, 8). From there, the precursors are inserted into the Tom40 channel.
Although the components of the TOM complex and their molecular functions have been described in great detail, little is known about the distributions of the TOM complexes within the outer membrane, and even less is known about the spatial distributions of the complexes with respect to changing mitochondrial activities. The optical resolution in far-field fluorescence microscopy, arguably the most suitable approach for studying quantitatively the distribution of protein complexes in mitochondria of intact cells, is limited to ∼200 nm by diffraction (9). This resolution is not sufficient to resolve individual TOM complexes in mitochondria (10, 11). To overcome this problem, we used stimulated emission depletion (STED) super-resolution microscopy (12, 13) in combination with automated algorithms for analyzing large image datasets.
Using STED microscopy, we imaged more than 1,000 chemically fixed and immunolabeled cells. We found that the TOM complexes aggregate into clusters and that the number and density of the clusters are regulated tightly and correlate with the activity of the mitochondria and their position in the cell.
Results
STED Microscopy Enables Visualization of Protein Distribution in Mitochondrial Outer Membrane.
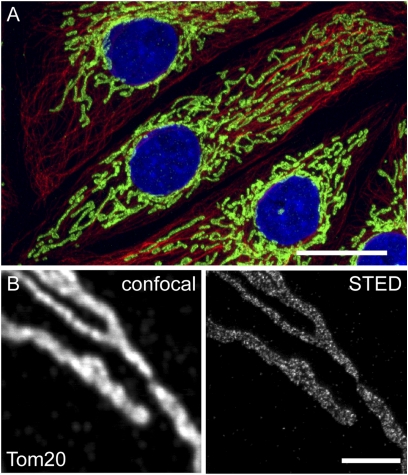
Mitochondria form a highly dynamic network of long, frequently interconnected, branched tubules in the cytoplasm of cultured mammalian cells (Fig. 1A). When chemically fixed intact cells are labeled with an antiserum specific for Tom20, the organelles appear to be stained homogenously when imaged with conventional diffraction-limited fluorescence microscopy (Fig. 1B). With a STED microscope enabling a focal plane resolution of 40–50 nm, we found that the protein is concentrated in clusters (Fig. 1B) (11, 14). Labeling with an antiserum against Tom22, a component of the core complex of the TOM complex, resulted in a very similar, punctuate labeling pattern (Fig. S1). Similar experimental procedures previously have shown that other mitochondrial proteins have different distributions (11, 14, 15).
Fig. 1.
STED microscopy enables the analysis of submitochondrial protein distributions. (A) Mitochondria form a branched network within eukaryotic cells. The mitochondria (green) and the microtubule cytoskeleton (red) of PtK2 cells were labeled with antibodies specific for Tom20 and α-tubulin. The nuclei were labeled with DAPI (blue). (Scale bar: 20 μm.) (B) Mitochondria of PtK2 cells labeled with an antiserum against Tom20. STED microscopy (Right) reveals individual Tom20 clusters, which are blurred and not resolvable when using diffraction-limited confocal microscopy (Left). (Scale bar: 2 μm.)
STED Imaging of Tom20 Distribution in Three Mammalian Cell Lines.
Next, we evaluated which of the TOM complex subunits would be the most appropriate for studying the localization of the complex. Tom40, Tom22, and the small Tom proteins are believed to form the core of the complex. Cryo-electron microscopy of TOM complexes in yeast indicated that three Tom22 subunits are located at the perimeter of the core complex; the Tom20 proteins bind to the periphery of the core complex (16).
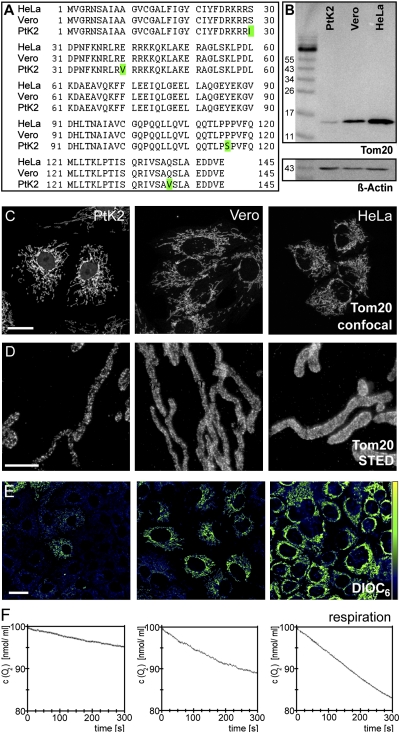
An amino acid sequence alignment of Tom20, Tom22, Tom40, and Tom70 from several vertebrate species suggested that Tom20 is the most conserved of these proteins (Tables S1 and S2). We found that the amino acid sequence of Tom20 [type II isoform (17)] was identical in HeLa and Vero cells and differed from Tom20 in PtK2 cells by only four amino acids (Fig. 2A and Fig. S2).This result is fully in line with a comprehensive Tom20 sequence analysis performed on numerous animal species (17).
Fig. 2.
Analysis of Tom20 in three different cell lines. (A) Amino acid sequence alignment of Tom20 from the three cell lines. Differences are highlighted in green. (B) Western blots of whole-cell extracts of the three cell lines decorated with the antiserum against Tom20. An antiserum against β-actin was used as a loading control. (C) Overview on the shapes of the mitochondrial networks in PtK2, Vero, and HeLa cells labeled with the Tom20 antiserum. Shown are maximal intensity projections of confocal sections. (D) STED images of mitochondrial tubules of the respective cells labeled with the Tom20 antiserum. Shown are representative images. Note that there was substantial heterogeneity within a single cell line. (E) Staining of the cell lines with the mitochondrial membrane potential-sensitive dye DiOC6. High fluorescence intensity indicates a high membrane potential. The same imaging conditions and color tables were used. (Scale bars: 20 μm in C and E; 2 μm in D.) (F) Oxygen consumption of the three lines, as measured with a Clark-type oxygen electrode.
In Western blots, the polyclonal antiserum against human full-length Tom20 recognized only a single band of the predicted size in extracts of these three cell lines (Fig. 2B). Furthermore, the same antiserum provided a very good signal-to-noise ratio when cells of the three lines were immunolabeled (Fig. 2C), further demonstrating its specificity. Both in Western blots of cell extracts (Fig. 2B) and in immunolabeled cells (Fig. S3A), we found the highest expression levels in HeLa cells and the lowest in PtK2 cells.
STED images of PtK2, Vero, and HeLa cells labeled for Tom20 immediately revealed that in all three cell lines Tom20 was concentrated in distinct clusters (Fig. 2D). Importantly, visual inspection of the STED images demonstrated substantial heterogeneity even among cells of the same cell line. This heterogeneity precluded clear conclusions about differences in Tom20 cluster distributions based on the visual inspection of a few images. Hence, for meaningful analysis of data regarding the nanoscale Tom20 distribution, quantitative automated image analysis was required. We decided to develop image-analysis algorithms tailored to this problem on the basis of image data generated using these three cell lines. We anticipated that these nanoscale datasets were a suitable basis for the validation of such algorithms, because the three lines differed in their mitochondrial membrane potential, respiration rates, cytochrome c oxidase activity, and cellular growth rate (HeLa > Vero > PtK2) (Fig. 2E and F and Fig. S3 B and C), suggesting that the distribution of the Tom20 clusters also might differ because of different functional requirements of the mitochondria.
To have a large database enabling statistically significant data, we took 100–200 images from each cell line, each image showing the mitochondrial network in the periphery of a different cell. We choose the periphery of the cells for our analysis because the mitochondria of all three lines exhibited a similar tubular appearance in this region, simplifying an automated analysis. We devised three different image-analysis algorithms to facilitate an unbiased analysis of the obtained large data sets.
Size of the Tom20 Clusters Is Similar in Three Mammalian Cell Lines.
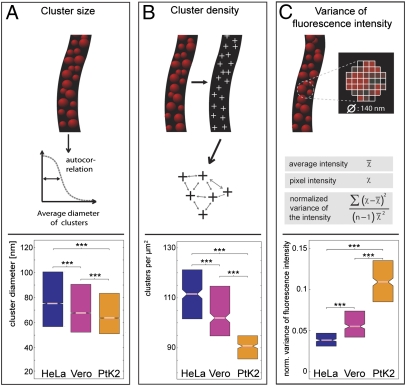
First, we analyzed the diameter of the Tom20 clusters. To this end, quadratic regions with edge lengths of 260 nm were cropped out of the mitochondria automatically (for details on all analysis, see SI Materials and Methods). Then, the size of the Tom20 clusters was determined using a 2D autocorrelation algorithm, an approach that allows the identification of repeating patterns that otherwise are obscured by noise. The analysis in all ∼45,000 quadratic regions indicated slight but statistically significant differences in the median size of the clusters. The antibody-decorated clusters had a median diameter of ∼75 nm in HeLa cells, ∼68 nm in Vero cells, and ∼64 nm in PtK2 cells (Fig. 3A). Because the primary and secondary antibodies enlarge the labeled structure by ∼17.5 nm in each direction (18, 19), we assume that the actual diameter of the Tom20 clusters is ∼30–40 nm. The size distribution of the clusters was broad (Fig. 3A) and overlapped to a large extent in the three cell lines, rendering it unlikely that the small differences in the mean diameter of the clusters, although statistically significant, are of major biological relevance. Nonetheless, the data demonstrate that the Tom20 clusters are substantially larger than a single TOM complex, which has a diameter of about 14.5 nm (16).
Fig. 3.
Quantification of the distributions of the Tom20 clusters in HeLa, Vero, and PtK2 cells. For the analysis, cells were grown to ∼50% confluence, chemically fixed, and labeled with an antiserum against Tom20. More than 120 cells of each line, imaged with STED microscopy, were analyzed. (A) Analysis of the diameter of the protein clusters by an autocorrelation algorithm. Note: The diameters determined for the Tom20 clusters are enlarged by the antibodies used for labeling. (B) Analysis of the density of Tom20 clusters in the mitochondria. (C) Analysis of the normalized variance of the local fluorescence signal intensity on the mitochondria, the most sensitive measure for determining differences in the distributions of the labeled protein. In the boxplots, the central lines represent the median, and the edges of the boxes represent the first and the third quartiles, respectively. Notches indicate P = 0.05 for the medians. ***P = 0.001 for the means (paired t test analysis).
Distribution of Tom20 Clusters Is Cell Line-Specific.
Next, we analyzed the average density of the clusters in the outer membrane. To avoid unwanted edge effects from the part of outer membrane oriented along the optical axis, only the fluorescence signal from the center of the mitochondria (width: 150 nm) was analyzed. We devised an algorithm, analogous to algorithms used in single-particle analyses, that identifies individual clusters and their distributions. Importantly, because the STED microscope provided a diffraction-limited resolution along the optical axis which encompassed the diameter of a single organelle, the obtained images may be regarded as projections of the Tom20 cluster distributions on both sites of the organelle. Therefore, the calculated numbers for cluster density are larger than the numbers for the actual organelle, but the numbers obtained are a robust measure for comparing different cells.
We found statistically significant differences among the three cell lines with respect to the distribution of the clusters. The median densities were ∼112, ∼102, and ∼90 Tom20 clusters per square micrometer in HeLa, Vero, and PtK2 cells, respectively (Fig. 3B).
As a different approach to analyze variations in the distribution of the Tom20 clusters, we next analyzed the normalized local variance of the fluorescence signal within the labeled mitochondria. Such normalized variance values reflect several physical parameters, including the distribution and size of the protein clusters, the cluster-to-mitochondrial background ratio, and others. This range of factors makes translating variance values into a single physical parameter rather difficult but makes them a sensitive measure for determining differences among protein distributions. Generally, dense clustering results in lower variance values, whereas sparse clustering results in higher values. We found normalized variance values of 37 × 10−3, 56 × 10−3, and 109 × 10−3 for HeLa, Vero, and PtK2 cells, respectively (Fig. 3C), fully corroborating the view that, on average, the cluster density is highest in HeLa cells and lowest in PtK2 cells.
Taken together, we found differences in the average size and the average density of the Tom20 clusters between the three cells lines, although there was substantial heterogeneity even between cells of the same line. Although the size distributions of the clusters were broad, and thus the differences in the mean sizes may be of only minor biological relevance, the differences in the nanoscale distribution of the clusters among the three lines were prominent. The Tom20 cluster densities were highest in the cell lines with the highest mitochondrial activity (HeLa cells) and were lowest in the cell lines with the lowest mitochondrial activity (PtK2 cells), as determined by several physiological readouts (Fig. 2 E and F and Fig. S3). The variance values proved to be a very sensitive measure for the Tom20 distribution, and therefore we decided to use them in following as readout.
Cell Density Influences Distribution of Tom20 Clusters.
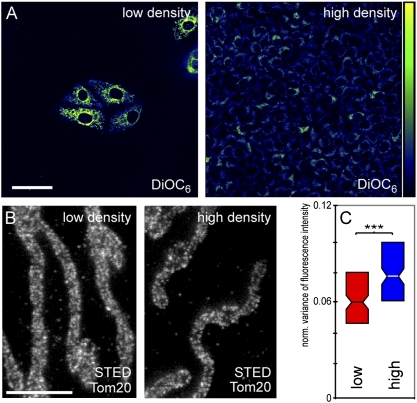
Thus far, we have analyzed the distribution of the Tom20 clusters in cells of different species origin. Although the Tom20 protein in the three cell lines is highly similar or even identical, we cannot fully exclude the possibility that the antibody recognized the proteins to different degrees. Hence to test further the hypothesis of an adaptation of the nanoscale Tom20 cluster distribution to the mitochondrial activity, we used a single cell line, PtK2, but grew the cells to different densities, namely, to >90% or to <10% confluence. Staining with the membrane potential sensitive dye DiOC6 (20) showed that the average DiOC6 signal was three to four times higher in the sparsely seeded cells, indicating an increased membrane potential in the mitochondria of these cells (Fig. 4A and Fig. S4). After decoration with the Tom20 antiserum, more than 100 cells grown under both conditions were imaged by STED microscopy and the whole dataset was automatically analyzed with respect to the variance of the fluorescence signal (Fig. 4 B and C). We found that the normalized variance of the fluorescence signal was lower in the sparsely seeded cells (60 × 10−3) than in the dense cells (77 × 10−3) (Fig. 4C), strongly indicating that the density of the Tom20 clusters is higher in the mitochondria of the more active, sparsely grown cells than in the less active, confluent cells. This adaptation was relatively rapid, because it already was manifested 1 d after the cells were seeded to a low or high density.
Fig. 4.
Growth conditions influence the distribution of the Tom20 clusters. PtK2 cells were seeded to low density (<10% confluence) or high density (>90% confluence) on coverslips. (A) Cells were stained with the membrane potential-sensitive dye DiOC6. (Left) Low confluence. (Right) High confluence. High fluorescence intensity indicates a high membrane potential.(Scale bar: 50 μm.) (B) STED imaging of cells labeled with a Tom20-specific antiserum. Shown are representative images of mitochondria of cells grown to low or high confluence, respectively. (Scale bar: 2 μm.) (C) Boxplots summarizing the quantitative analysis of 200 cells imaged by STED microscopy. Significance levels are as in Fig. 3.
Distribution of Tom20 Clusters Depends on Individual Cell Location Within a Microcolony.
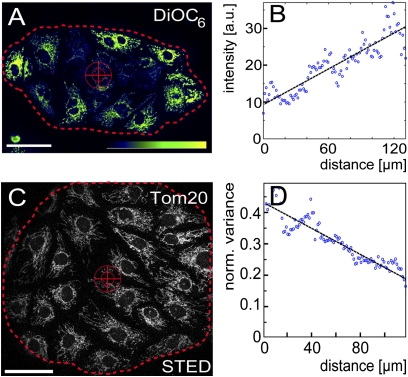
An even more subtle difference in the immediate cellular environments was generated by allowing the cells to grow into small spherical microcolonies on the coverslip. In general, the microcolonies had a diameter of ∼200 μm and contained ∼25 cells. Staining a microcolony with DiOC6 revealed that the mitochondrial membrane potential of the cells at its rim generally was higher than that of the cells in the interior (Fig. 5 A and B). We next analyzed whether this gradient in mitochondrial activity within a microcolony correlates to differences in the distribution of the Tom20 clusters. To this end, we plotted the mean variance values of the fluorescence signals radiating from the center of the microcolony to its rim (Fig. 5 C and D). We found a gradient in the normalized variance of the fluorescence signal in all the colonies analyzed (n = 5). That is, the variance of the Tom20 fluorescence signal was lower in the mitochondria of cells at the rim than in cells in the interior of the microcolony. This finding strongly indicates that, in the region where the mitochondrial activity is the highest (the rim of the colony), the Tom20 cluster density is highest as well. Hence, the distribution and the number of Tom20 clusters vary even between adjacent cells when they face a different environment.
Fig. 5.
Densities of Tom20 clusters and the membrane potentials form radial gradients in microcolonies. PtK2 cells were seeded sparsely on a coverslip and allowed to grow into microcolonies of about 25 cells. (A) Cells were stained with the membrane potential-sensitive dye DiOC6. Shown is a single microcolony. The cross indicates the center, and the dotted line indicates the rim of the microcolony as used for the subsequent analysis. (Scale bar: 50 μm.) (B) Plot of the average DiOC6 fluorescence intensities from the center to the rim of the microcolony shown in A. Blue circles represent 100 bins that pool the ∼4 × 104 individual fluorescence intensity values. The black line indicates the linear fit to the individual fluorescence intensity values. (C) STED image of a microcolony labeled with an antiserum against Tom20. (Scale bar: 50 μm.) (D) Plot of the normalized variance of the fluorescence signal from the center to the rim of the microcolony shown in C. Blue circles represent 100 bins that pool the ∼5 × 106 individual variance values. The black line indicates the linear fit to the individual variance values. The lower variance values at the rim of the colony indicate a higher density of Tom20 clusters.
Distribution of Tom20 Clusters Depends on Subcellular Location of Individual Mitochondria.
Previous work indicated some heterogeneity in the respiratory activities of the mitochondria of an individual cell (21–24). Indeed, the DiOC6 staining of living cells, as shown in Figs. 2E, 4A, and 5A, generally indicated that the membrane potential of perinuclear mitochondria was higher than that of mitochondria at the periphery of the cell. Mitochondria generally are closer to each other around the nucleus than in the periphery of the cell. Hence we were concerned that the apparent gradient in the brightness of the DiOC6 signals was caused by the density of the mitochondria and not by the membrane potential. Therefore, we targeted the red fluorescent protein DsRed to the mitochondrial matrix to label the mitochondria. After using the DsRed fluorescence intensity distribution to normalize the DiOC6 fluorescence intensity distribution, we found a gradient of the DiOC6 signal from the perinuclear mitochondria to the mitochondria at the cell periphery (Fig. S5) in the majority of the cells analyzed (>70%, n = 25), strongly indicating a gradient in membrane potential from the mitochondria close to the nucleus to those at the periphery.
We next asked whether the mitochondria of a single cell exhibit distinct distributions of Tom20 clusters depending on the specific location of mitochondria within the cell. To this end, we plotted the mean variance values of the Tom20 fluorescence signal radiating from the nucleus to the cell periphery (Fig. 6 A–C), essentially as done previously for an entire microcolony. Initially, this single-cell analysis was performed with all cells of the microcolony shown in Fig. 5C. Within the microcolony analyzed, we found an increase in the normalized variance values from the mitochondria around the nucleus to the mitochondria at the periphery in 23 of the 25 cells (Fig. 6A and Fig. S6). To validate this finding further, we analyzed ∼300 additional cells. In 90% of all cells analyzed we found a gradient in the distribution of the normalized variance of the Tom20 fluorescence signal, with the smallest variance values close to the nucleus. This result demonstrates that the density of the Tom20 clusters is higher in the presumably more active mitochondria close to the nucleus than in mitochondria at the periphery of the cell.
Fig. 6.
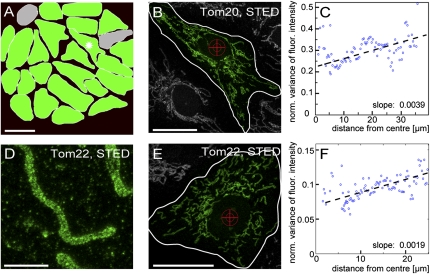
Density of TOM clusters is higher in the perinuclear mitochondria than in mitochondria at the cellular periphery. (A) Graphical representation of all cells of the microcolony shown in Fig. 5C. Green indicates cells in which the normalized variance of the fluorescence signal is lower in the perinuclear mitochondria (corresponding to a higher density of the TOM clusters) than in the cellular periphery. (For detailed data, see Fig. S6). Gray indicates cells in which the linear fits to the variance values have negative slopes. (Scale bar: 50 μm.) (B) Representative cell used for the analysis shown in C. Its location in the microcolony is indicated by an asterisk in A. The cross indicates the center, and the white line indicates the border of the cell as used for the subsequent analysis. (Scale bar: 20 μm.) (C) The normalized local variance values of the fluorescence signals radiating from the center of the cell to its border. Blue circles indicate 100 bins that pool the ∼3 × 105 individual variance values. The black line indicates the linear fit based on the individual variance values. The positive slope of the curve indicates that the density of the TOM clusters is higher in mitochondria around the nucleus. (D) STED image of a mitochondrion of a Vero cell labeled with an antiserum against Tom22. (Scale bar: 2 μm.) (E) Representative Vero cell labeled with a Tom22 antiserum. (Scale bar: 20 μm.) (F) Analysis of the distribution of Tom22 in the cell shown in E. The analysis was performed as in C.
Moreover, the slopes of the linear fits of the mean variance values radiating from the cell nucleus to the periphery of the cells were lower, on average, in the center of the microcolony than at the rim. This finding implies that even the inner-cellular gradients of the Tom20 cluster distribution are finely tuned to the specific cellular environment.
We next investigated whether the Tom22 cluster distribution also shows such a gradient from the periplasmatic region to the periphery of the cell. We labeled Vero cells with an antiserum against Tom22 and analyzed the distribution of the variance of the fluorescence signal as before (Fig. 6 D–F). In the majority of all cells analyzed (∼95%, n = 127), we found a gradient in the variance of the fluorescence signal, exhibiting slopes similar to those observed for Tom20 and further supporting the view that the distribution of Tom20 reflects the distribution of the TOM complex. We conclude that the nanoscale distribution of the TOM complexes is finely tuned to the activity of individual mitochondria within a single cell.
Discussion
Using immunolabeling of chemically fixed intact cells in combination with super-resolution STED microscopy and automated image analysis of >1,000 cells, we report a clustering of a large fraction of the Tom20 proteins in the mitochondrial outer membrane, although our data do not exclude the existence of a sizeable pool of Tom20 proteins that is not associated in a cluster. In the majority of cells, we observed a gradient in the density of these Tom20 clusters from the perinuclear regions to the periphery of the cells. Labeling with an antiserum against Tom22, a component of the TOM core complex, resulted in similar subcellular gradients, strongly supporting the view that the observed Tom20 clusters reflect the distribution of the TOM complexes.
Our findings suggest that the average diameter of the TOM clusters, without the decorating Tom20 antibodies, is between ∼30 nm and ∼40 nm. Single-particle cryo-electron microscopy data previously showed that the budding yeast TOM complex (including Tom20) is triangular, measuring 14.5 nm on edge (16). Our data indicate that if TOM complexes are packaged tightly in a cluster, a single cluster contains 8–14 TOM complexes on average. Because the TOM complexes might be associated with further proteins in these clusters or might not be tightly packaged, we propose that 8–14 TOM complexes per cluster may be regarded as an upper limit.
Many mRNAs that encode mitochondrial proteins are localized to mitochondrion-bound polysomes (25). It has been suggested that in budding yeast the RNA-binding protein Puf3 stabilizes the localization of the transcripts at the mitochondria, so that the mitochondrial-targeting sequences emerging from the ribosomes bind efficiently to Tom20 and are imported cotranslationally via the TOM complex (26). Given the model for mRNA anchoring in proximity to the TOM complex and the prospect of polysomes translating multiple mitochondrial preproteins that are close together spatially, such coupling might explain the clustering of the TOM complexes.
Upon import of preproteins, the TOM complex forms a complex with the presequence translocase of the inner membrane (TIM complex) (27). Preproteins in transit can span both mitochondrial membranes at translocation contact sites where both membranes are in close proximity (28–31). Interestingly, several studies using electron microscopy determined the lateral size of contact sites as 13–30 nm (29, 32, 33). These values are smaller than our estimate of the average size of a Tom20 cluster (∼30–40 nm without decorating antibodies). Furthermore, biochemical studies demonstrated that TOM complexes are four times more frequent than TIM23 complexes in budding yeast mitochondria (34). Hence one could speculate that only a fraction of the TOM complexes in a cluster are engaged in interactions with TIM23 complexes forming the actual contact site and that the remaining TOM complexes are at the rim of the contact site and do not interact with a TIM23 complex. This hypothesis clearly requires further experimental verification.
In this study we have shown that the density of the TOM clusters is higher in mitochondria with a higher membrane potential than in those with a lower membrane potential. This correlation was demonstrated in cells of different species origin having distinct physiological activities (i.e., different respiratory rates, doubling times, and cytochrome c oxidase activities) as well as in cells grown to different densities. In microcolonies of a few cells, a positive gradient in mitochondrial activities from the center to the rim of the microcolony matches a gradient in the density of the TOM clusters. We even found a gradient in the density of TOM clusters within single cells that was matched by a gradient in the membrane potential.
The molecular mechanisms that regulate the distribution of the TOM complexes on the nanoscale are unknown. Likewise, although the components of the translocation machineries and their molecular functions have been described in great detail, relatively little is known about the mechanisms that regulate protein import into mitochondria. For some proteins of the intermembrane space, mitochondria use redox signals for the regulation of translocation (35, 36), and a metabolite-dependent regulation of protein import has been shown for fumarase (37). Recently, it has been demonstrated that the cytosolic kinases casein kinase 2 (CK2) and protein kinase A (PKA) exert stimulatory and inhibitory effects on the biogenesis and the function of the TOM complex and thus contribute to the regulation of protein import into mitochondria (38), demonstrating that regulation of import can take place at the level of the TOM complex. Hence one may speculate that the distribution of the TOM complexes also is involved in the regulation of mitochondrial import and that the dynamic adjustment of the nanoscale distribution of the TOM complexes is an adaptation mechanism to the different import needs of mitochondria. The data shown here may spur the search for the signaling mechanisms that determine the distribution of the TOM complexes on the nanoscale.
Materials and Methods
Cell Culture.
Cell lines were cultivated in DMEM with Glutamax and 4.5% (wt/vol) glucose (Invitrogen), supplemented with 50 u/mL penicillin, 50 μg/mL streptomycin, 1 mM Na-pyruvate, and 10% (vol/vol) FCS (Invitrogen) at 37 °C, 7% CO2. For transfection with the plasmid pDsRed2-Mito (Clontech), cells were seeded onto coverslips 12 h before the experiment. Transfections were carried out using the Nanofectin Kit (PAA) according to the manufacturer’s recommendations.
Sample Preparation.
For immunolabeling, cultured mammalian cells were grown on coverslips overnight, fixed with 8% (wt/vol) formaldehyde in PBS (137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, pH 7) for 5 min at 37 °C, extracted with 0.5% (vol/vol) Triton X-100 in PBS, blocked with 5% (wt/vol) BSA in PBS, and incubated with polyclonal rabbit antibodies against Tom20 (Santa Cruz Biotechnology) or with mouse monoclonal antibodies against Tom22 (Sigma-Aldrich) or mouse monoclonal antibodies against α-tubulin (Sigma-Aldrich). The primary antibodies were detected with secondary antibodies (sheep anti-mouse and goat anti-rabbit; Jackson ImmunoResearch Laboratories) custom-labeled with ATTO532, ATTO647N (AttoTec), or KK114 [compound 6 (39)]. After immunolabeling, the samples were mounted in Mowiol supplemented with 0.1% (wt/vol) 1,4-diazabicyclo[2.2.2]octane (Sigma Aldrich) and 2.5 μg/mL DAPI (Sigma-Aldrich). For further details, see ref. 40.
Fluorescence Microscopy.
For the generation of overview images, for the quantification of the cytochrome c oxidase activity, and for the imaging of DiOC6-labeled cells, an epifluorescence microscope (DM6000; Leica Microsystems) or a confocal microscope (TCS SP5; Leica) was used. For STED and the corresponding confocal microscopy, a custom-built STED microscope (41) was used.
Image Analysis.
For the automated analysis of the STED images, several algorithms were devised: (i) an autocorrelation algorithm that allows the estimation of the size of protein aggregates within the mitochondria; (ii) an algorithm that identifies individual aggregates within mitochondria and calculates their densities and distances; and (iii) an algorithm that evaluates the normalized variance of the local fluorescence signal. All algorithms were programmed using MATLAB (Mathworks). For further details on the quantitative image analyses, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank K. Kolmakov and V. Belov for providing the fluorophore KK114, M. Kollmar for help with the sequence alignments, T. Gilat and S. Löbermann for excellent technical assistance, and J. Jethwa for helpful comments on the manuscript. This work was supported by the Gottfried Wilhelm Leibniz Program of the Deutsche Forschungsgemeinschaft (to S.W.H.) and by the Deutsche Forschungsgemeinschaft through Grant JA1129/3 and through the DFG-Research Center for Molecular Physiology of the Brain (both to S.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. N.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107553108/-/DCSupplemental.
References
- 1.Devin A, Rigoulet M. Mechanisms of mitochondrial response to variations in energy demand in eukaryotic cells. Am J Physiol Cell Physiol. 2007;292:C52–C58. doi: 10.1152/ajpcell.00208.2006. [DOI] [PubMed] [Google Scholar]
- 2.Hoogenraad NJ, Ward LA, Ryan MT. Import and assembly of proteins into mitochondria of mammalian cells. Biochim Biophys Acta. 2002;1592:97–105. doi: 10.1016/s0167-4889(02)00268-9. [DOI] [PubMed] [Google Scholar]
- 3.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 4.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: Machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Söllner T, Griffiths G, Pfaller R, Pfanner N, Neupert W. MOM19, an import receptor for mitochondrial precursor proteins. Cell. 1989;59:1061–1070. doi: 10.1016/0092-8674(89)90762-9. [DOI] [PubMed] [Google Scholar]
- 6.Saitoh T, et al. Tom20 recognizes mitochondrial presequences through dynamic equilibrium among multiple bound states. EMBO J. 2007;26:4777–4787. doi: 10.1038/sj.emboj.7601888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiebler M, et al. The mitochondrial receptor complex: A central role of MOM22 in mediating preprotein transfer from receptors to the general insertion pore. Cell. 1993;74:483–492. doi: 10.1016/0092-8674(93)80050-o. [DOI] [PubMed] [Google Scholar]
- 8.van Wilpe S, et al. Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature. 1999;401:485–489. doi: 10.1038/46802. [DOI] [PubMed] [Google Scholar]
- 9.Born M, Wolf E. Principles of Optics. , 7th Ed. Cambridge: Cambridge Univ Press; 2002. [Google Scholar]
- 10.Jakobs S. High resolution imaging of live mitochondria. Biochim Biophys Acta. 2006;1763:561–575. doi: 10.1016/j.bbamcr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Donnert G, et al. Two-color far-field fluorescence nanoscopy. Biophys J. 2007;92:L67–L69. doi: 10.1529/biophysj.107.104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: Stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 13.Hell SW. Far-Field Optical Nanoscopy. Single Molecule Spectroscopy in Chemistry, Physics and Biology. Berlin: Springer; 2009. pp. 298–365. [Google Scholar]
- 14.Schmidt R, et al. Spherical nanosized focal spot unravels the interior of cells. Nat Methods. 2008;5:539–544. doi: 10.1038/nmeth.1214. [DOI] [PubMed] [Google Scholar]
- 15.Neumann D, Bückers J, Kastrup L, Hell SW, Jakobs S. Two-color STED microscopy reveals different degrees of colocalization between hexokinase-I and the three human VDAC isoforms. PMC Biophys. 2010;3:4. doi: 10.1186/1757-5036-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Model K, Meisinger C, Kühlbrandt W. Cryo-electron microscopy structure of a yeast mitochondrial preprotein translocase. J Mol Biol. 2008;383:1049–1057. doi: 10.1016/j.jmb.2008.07.087. [DOI] [PubMed] [Google Scholar]
- 17.Likić VA, et al. Patterns that define the four domains conserved in known and novel isoforms of the protein import receptor Tom20. J Mol Biol. 2005;347:81–93. doi: 10.1016/j.jmb.2004.12.057. [DOI] [PubMed] [Google Scholar]
- 18.Weber K, Rathke PC, Osborn M. Cytoplasmic microtubular images in glutaraldehyde-fixed tissue culture cells by electron microscopy and by immunofluorescence microscopy. Proc Natl Acad Sci USA. 1978;75:1820–1824. doi: 10.1073/pnas.75.4.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dyba M, Jakobs S, Hell SW. Immunofluorescence stimulated emission depletion microscopy. Nat Biotechnol. 2003;21:1303–1304. doi: 10.1038/nbt897. [DOI] [PubMed] [Google Scholar]
- 20.Galluzzi L, et al. Methods for the assessment of mitochondrial membrane permeabilization in apoptosis. Apoptosis. 2007;12:803–813. doi: 10.1007/s10495-007-0720-1. [DOI] [PubMed] [Google Scholar]
- 21.Collins TJ, Berridge MJ, Lipp P, Bootman MD. Mitochondria are morphologically and functionally heterogeneous within cells. EMBO J. 2002;21:1616–1627. doi: 10.1093/emboj/21.7.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wikstrom JD, et al. beta-Cell mitochondria exhibit membrane potential heterogeneity that can be altered by stimulatory or toxic fuel levels. Diabetes. 2007;56:2569–2578. doi: 10.2337/db06-0757. [DOI] [PubMed] [Google Scholar]
- 23.Distelmaier F, et al. Life cell quantification of mitochondrial membrane potential at the single organelle level. Cytometry A. 2008;73:129–138. doi: 10.1002/cyto.a.20503. [DOI] [PubMed] [Google Scholar]
- 24.Jakobs S, Stoldt S, Neumann D. Light microscopic analysis of mitochondrial heterogeneity in cell populations and within single cells. Adv Biochem Eng Biotechnol. 2011;124:1–19. doi: 10.1007/10_2010_81. [DOI] [PubMed] [Google Scholar]
- 25.Marc P, et al. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 2002;3:159–164. doi: 10.1093/embo-reports/kvf025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quenault T, Lithgow T, Traven A. PUF proteins: Repression, activation and mRNA localization. Trends Cell Biol. 2011;21:104–112. doi: 10.1016/j.tcb.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 27.Chacinska A, et al. Mitochondrial translocation contact sites: Separation of dynamic and stabilizing elements in formation of a TOM-TIM-preprotein supercomplex. EMBO J. 2003;22:5370–5381. doi: 10.1093/emboj/cdg532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schleyer M, Neupert W. Transport of proteins into mitochondria: Translocational intermediates spanning contact sites between outer and inner membranes. Cell. 1985;43:339–350. doi: 10.1016/0092-8674(85)90039-x. [DOI] [PubMed] [Google Scholar]
- 29.Rassow J, et al. Translocation arrest by reversible folding of a precursor protein imported into mitochondria. A means to quantitate translocation contact sites. J Cell Biol. 1989;109:1421–1428. doi: 10.1083/jcb.109.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jascur T, Goldenberg DP, Vestweber D, Schatz G. Sequential translocation of an artificial precursor protein across the two mitochondrial membranes. J Biol Chem. 1992;267:13636–13641. [PubMed] [Google Scholar]
- 31.Kanamori T, Nishikawa S, Shin I, Schultz PG, Endo T. Probing the environment along the protein import pathways in yeast mitochondria by site-specific photocrosslinking. Proc Natl Acad Sci USA. 1997;94:485–490. doi: 10.1073/pnas.94.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hackenbrock CR. Chemical and physical fixation of isolated mitochondria in low-energy and high-energy states. Proc Natl Acad Sci USA. 1968;61:598–605. doi: 10.1073/pnas.61.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perkins G, et al. Electron tomography of neuronal mitochondria: Three-dimensional structure and organization of cristae and membrane contacts. J Struct Biol. 1997;119:260–272. doi: 10.1006/jsbi.1997.3885. [DOI] [PubMed] [Google Scholar]
- 34.Dekker PJ, et al. The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J. 1997;16:5408–5419. doi: 10.1093/emboj/16.17.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herrmann JM, Köhl R. Catch me if you can! Oxidative protein trapping in the intermembrane space of mitochondria. J Cell Biol. 2007;176:559–563. doi: 10.1083/jcb.200611060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stengel A, Benz JP, Soll J, Bölter B. Redox-regulation of protein import into chloroplasts and mitochondria: Similarities and differences. Plant Signal Behav. 2010;5:105–109. doi: 10.4161/psb.5.2.10525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Regev-Rudzki N, Battat E, Goldberg I, Pines O. Dual localization of fumarase is dependent on the integrity of the glyoxylate shunt. Mol Microbiol. 2009;72:297–306. doi: 10.1111/j.1365-2958.2009.06659.x. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt O, et al. Regulation of mitochondrial protein import by cytosolic kinases. Cell. 2011;144:227–239. doi: 10.1016/j.cell.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 39.Kolmakov K, et al. Red-emitting rhodamine dyes for fluorescence microscopy and nanoscopy. Chem Eur J. 2010;16:158–166. doi: 10.1002/chem.200902309. [DOI] [PubMed] [Google Scholar]
- 40.Wurm CA, Neumann D, Schmidt R, Egner A, Jakobs S. Sample preparation for STED microscopy. Methods Mol Biol. 2010;591:185–199. doi: 10.1007/978-1-60761-404-3_11. [DOI] [PubMed] [Google Scholar]
- 41.Harke B, et al. Resolution scaling in STED microscopy. Opt Express. 2008;16:4154–4162. doi: 10.1364/oe.16.004154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.