Abstract
Aberrations in epigenetic processes, such as histone methylation, can cause cancer. Retinoblastoma binding protein 2 (RBP2; also called JARID1A or KDM5A) can demethylate tri- and dimethylated lysine 4 in histone H3, which are epigenetic marks for transcriptionally active chromatin, whereas the multiple endocrine neoplasia type 1 (MEN1) tumor suppressor promotes H3K4 methylation. Previous studies suggested that inhibition of RBP2 contributed to tumor suppression by the retinoblastoma protein (pRB). Here, we show that genetic ablation of Rbp2 decreases tumor formation and prolongs survival in Rb1+/− mice and Men1-defective mice. These studies link RBP2 histone demethylase activity to tumorigenesis and nominate RBP2 as a potential target for cancer therapy.
Keywords: mouse model, histone methyltransferase, chromatin modifier, neuroendocrine tumor, islet cell tumor
Epigenetic alterations, like genetic alterations, can contribute to tumor initiation and progression (1, 2). Indeed, a number of genes that play roles in chromatin modifications and hence, epigenetic regulation are mutated in human cancers, including mixed-lineage leukemia (MLL1), multiple endocrine neoplasia type 1 (MEN1), and ubiquitously transcribed tetratricopeptide repeat, X chromosome (UTX) (3–6).
The retinoblastoma gene (RB1) tumor suppressor gene is frequently inactivated in a wide variety of cancers (7). The retinoblastoma protein (pRB) inhibits S-phase entry by repressing E2F (7). In addition, pRB promotes senescence and differentiation (8). These latter two activities track closely with the ability of pRB to bind to retinoblastoma binding protein 2 (RBP2; also called JARID1A or KDM5A) rather than to E2F (9). Moreover, RBP2 siRNA is sufficient to promote senescence and differentiation in pRB-defective tumor cells in vitro (9, 10). RBP2 is a histone demethylase capable of demethylating tri- and dimethylated lysine 4 in histone H3 (H3K4me3/2) and repressing gene expression (11–14). It is, therefore, conceivable that deregulation of RBP2 histone demethylase activity contributes to pRB-defective tumor formation.
Epigenetic changes are reversible, suggesting that inhibition of specific enzymes that regulate epigenetic marks would have antitumor effects. In fact, suberoylanilide hydroxamic acid (vorinostat), a histone deacetylase (HDAC) inhibitor, was approved for the treatment of cutaneous T-cell lymphoma (15), and two DNA methyltransferase inhibitors, 5-azacytidine (azacitidine) and 5-aza-2′-deoxycytidine (decitabine), were approved for the treatment of myelodysplastic syndrome (16, 17). RBP2 belongs to a superfamily of 2-oxoglutarate–dependent dioxygenases (18, 19), which can be inhibited with drug-like small molecules (20, 21). We, therefore, used mice carrying null or conditional Rbp2 alleles to further explore potential roles for RBP2 in pRB-defective tumorigenesis. In addition, we tested the hypothesis that loss of RBP2 H3K4 demethylase activity would inhibit tumors driven by loss of the MEN1 tumor suppressor, which is part of an H3K4 methyltransferase complex (6, 22, 23).
Results
Loss of RBP2 Inhibits Proliferation and Induces Senescence.
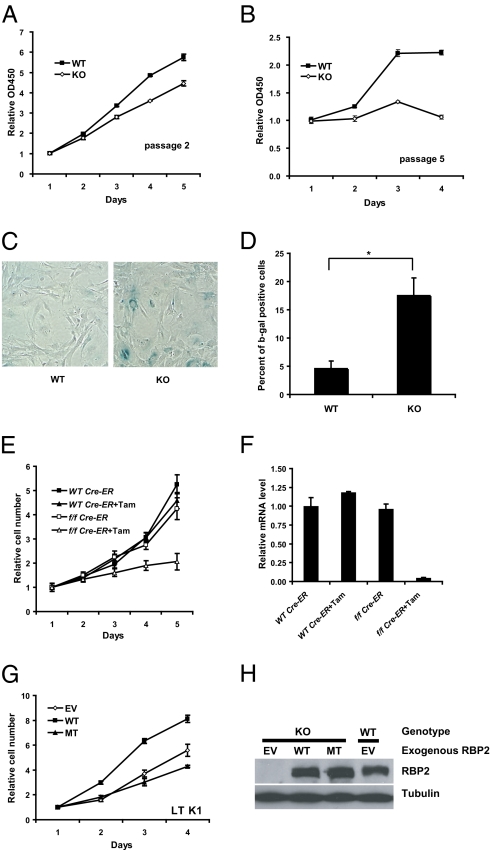
Mouse embryonic fibroblasts (MEFs) derived from Rbp2−/− embryos on a pure genetic background proliferated more slowly than MEFs derived from WT littermate controls, especially when examined at later passages (Fig. 1 A and B). Senescence-associated β-galactosidase (SABG) staining revealed increased staining of late-passage Rbp2−/− MEFs compared with WT control MEFs (Fig. 1 C and D), suggesting that RBP2 loss promotes senescence.
Fig. 1.
Loss of RBP2 inhibits proliferation and induces senescence. (A and B) Proliferation rate of WT and Rbp2−/− (KO) primary MEFs in (A) early and (B) late passages. (C) Senescence-associated β-galactosidase staining of late-passage WT and Rbp2−/− MEFs. (D) Quantitation of β-galactosidase–positive cells of late-passage WT and Rbp2−/− MEFs from three independent experiments; 300 cells of each genotype were counted (*P < 0.02). (E) Proliferation rate of Rbp2f/f;Cre-ER (f/f Cre-ER) and Rbp2+/+;Cre-ER (WT Cre-ER) primary MEFs after a 6-h pulse of tamoxifen (+Tam) compared with untreated MEFs. (F) Real-time RT-PCR analysis of Rbp2 in MEFs in E. Shown are mean values with SEM. (G) Proliferation rate of Rbp2−/− K1 MEFs infected with retroviruses encoding WT RBP2 (WT), RBP2 H483A (MT), or empty vector (EV). (H) Western blot analysis of the MEFs in G.
To study the effect of acute RBP2 inactivation, we created mice that carry a conditional (floxed or f) Rbp2 allele (11) and a transgene encoding a Cre-ER fusion protein, which can be activated by tamoxifen (24). Treatment of Rbp2f/f;Cre-ER MEFs with tamoxifen led to growth arrest, but treatment of Rbp2+/+;Cre-ER control MEFs did not lead to growth arrest (Fig. 1 E and F). Similar results were obtained when RBP2 was acutely deleted in Rbp2f/f MEFs using a retroviral vector encoding Cre recombinase (Fig. S1 A and B). Collectively, these results support the earlier conclusion, obtained with siRNAs, that RBP2 loss impairs proliferation and promotes senescence.
Regulation of Proliferation by RBP2 Is Dependent on Its Histone Demethylase Activity.
Inactivation of p53, using either the SV40 Large T antigen (LT) K1 mutant (25) or a dominant-negative C-terminal fragment of p53 (p53CTF) (26), immortalized Rbp2−/− MEFs, which was evidenced by their ability to be continually passaged in culture and absence of SABG staining; however, it did not correct their proliferation defect relative to similarly immortalized WT MEFs (Fig. S1C and data not shown). The availability of immortalized Rbp2−/− MEFs allowed us to ask whether the proliferation defect in Rbp2−/− cells is caused by loss of RBP2 histone demethylase activity. Reintroduction of WT RBP2, but not the histone demethylase-defective RBP2 H483A mutant (11), into LT K1-immortalized Rbp2−/− MEFs using retroviral vectors rescued the proliferation defect caused by RBP2 loss (Fig. 1 G and H).
Notably, the proliferation defect of Rbp2−/− MEFs was also rescued by inactivation of pRB, achieved with either WT LT (in contrast to LT K1) (Fig. S1D) or Rb1 nullizygosity (Fig. S1 E and F). Rbp2−/−;Rb1−/− primary MEFs did, however, eventually senesce, presumably because of p53 activation. Taken together, these results suggest that the senescence defect caused by RBP2 loss is p53-dependent, whereas the proliferation defect caused by RBP2 loss is pRB-dependent. Moreover, these data, together with earlier studies (9), suggest that RBP2 acts both upstream and downstream of pRB.
Loss of RBP2 Leads to Loss of Stem Cell Markers.
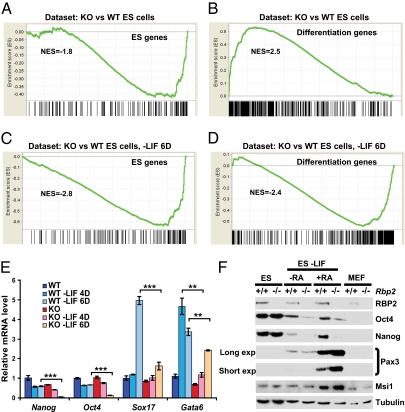
Many developmentally important promoters contain bivalent chromatin, which consists of H3K4me3 and H3K27me3 (27). These marks ensure that the genes are poised for activation or repression on differentiation. Because RBP2 can erase H3K4me3, we asked whether loss of RBP2 affects the maintenance of mouse ES cells. We compared the gene expression profiles of Rbp2f/f and Rbp2−/− ES cells grown either in the presence of leukemia inhibitory factor (LIF) (Fig. 2 A and B), which suppresses differentiation, or 6 d after LIF withdrawal (Fig. 2 C and D), which promotes differentiation, using gene set enrichment analysis (GSEA) (28). GSEA was performed using two previously defined subsets of genes: an ES genes subset that included genes that are highly expressed in undifferentiated ES cells (Fig. 2 A and C) and a differentiation genes subset that included genes that are bound by H3K27me3 and repressed in undifferentiated ES cells but activated 6 d after induction of differentiation (Fig. 2 B and D) (29). These analyses showed that loss of RBP2 down-regulates many genes that are normally highly expressed in ES cells (Fig. 2A) and leads to partial activation of the genes linked to differentiation (Fig. 2B), despite the presence of LIF, suggesting that RBP2 promotes or maintains a stem cell-like phenotype. Consistent with this idea, down-regulation of stem cell markers was more rapid in Rbp2−/− ES cells after LIF withdrawal compared with WT ES cells (Fig. 2C). Nonetheless, transcriptional activation of genes that are normally repressed by LIF was blunted in Rbp2−/− ES cells (Fig. 2D), suggesting that Rbp2−/− ES cells exit the stem cell compartment more rapidly than WT ES cells but are impaired in terms of fully executing a differentiation program.
Fig. 2.
Loss of RBP2 is required for maintenance and proper differentiation of mouse ES cells. (A and B) GSEA analysis of Rbp2f/f (WT) and Rbp2−/− (KO) ES cells using the gene set (A) highly expressed in ES cells (ES genes) or (B) linked to differentiation (differentiation genes). NES, normalized enrichment score. (C and D) GSEA analysis of Rbp2f/f (WT) and Rbp2−/− (KO) ES cells after induction of differentiation by 6 d of LIF withdrawal (−LIF 6D) using the gene set (C) highly expressed in ES cells or (D) linked to differentiation. (E) Real-time RT-PCR analysis of stem cell- and lineage-specific markers of Rbp2f/f (WT) and Rbp2−/− (KO) ES cells before and after differentiation induced by LIF withdrawal (−LIF) as in C and D for 4 (4D) or 6 d (6D; **P < 0.001, ***P < 0.0001). (F) Western blot analysis of stem cell- and neuronal lineage-specific markers of WT and Rbp2−/− (KO) ES cells before and after differentiation in neuronal differentiation assays. RA, retinoic acid; long exp, long exposure; short exp, short exposure.
To further examine this finding, we performed real-time PCR analysis of selected transcripts from the ES cells treated as above. In keeping with the GSEA, Rbp2−/− ES cells prematurely down-regulated the stem cell markers Nanog and Oct4 in response to LIF withdrawal but failed to fully up-regulate the differentiation markers Sox17 and Gata6 (Fig. 2E). Similar findings with respect to Nanog and Oct4 were also observed when WT and Rbp2−/− ES cells were induced to form embryoid bodies (EB) and then treated with retinoic acid (RA) to promote neuronal differentiation (Fig. 2F). In this model, however, Rbp2−/− ES cells displayed enhanced expression of the neuronal markers Pax3 and Msi1 (Fig. 2F). These findings suggest that Rbp2 deficiency down-regulates stem cell markers and promotes differentiation. Similar results were obtained with independently derived ES cell lines.
RBP2 Loss Mitigates Proliferation and Differentiation Abnormalities in pRB-Defective Cells.
Down-regulation of RBP2 using siRNA inhibits the proliferation of pRB-defective tumor cells (9, 10) and restores the ability of Rb1−/− MEFs to differentiate (9). The availability of Rbp2−/− mice allowed us to address the roles of RBP2 without being confounded by siRNA-mediated off-target effects. Through appropriate crosses, we generated WT, Rb1−/−, Rbp2−/−, Rbp2+/−;Rb1−/−, and Rbp2−/−;Rb1−/− embryos. Homozygous loss of Rbp2 impaired the proliferation of Rb1−/− MEFs derived from these littermate embryos (Fig. 3A).
Fig. 3.
Loss of RBP2 inhibits proliferation and promotes differentiation of pRB-defective cells. (A) Proliferation rate of WT, Rbp2−/−, Rbp2+/−;Rb1−/−, and Rbp2−/−;Rb1−/− MEFs. (B) Quantitation of MYHC-positive cells in five representative fields at day 6 of myogenic differentiation. MEFs of the indicated genotypes were infected with an adenovirus expressing MyoD to induce myogenic differentiation. The MYHC-positive cells were also scored for the presence of multiple nuclei. Shown are mean values with SEM from three independent experiments. (C) MYHC expression in MEFs during myogenic differentiation. The cells were fixed and stained with anti-MYHC antibody (red) and counterstained with the nuclear stain DAPI (blue) after growth for the indicated number of days in differentiation media.
Next, WT, Rb1−/−, Rbp2−/−, and Rbp2−/−;Rb1−/− early-passage MEFs were infected with an adenovirus-encoding MyoD and induced to differentiate in differentiation medium. RBP2 status did not influence adenoviral infection efficiency (data not shown). Consistent with previous studies, WT MEFs, but not Rb1−/− MEFs, started to form elongated myocytes 1 d after being placed in differentiation media, and they formed multinucleated myotubes shortly thereafter, which were associated with expression of the late-differentiation marker myosin heavy chain (MYHC). Loss of Rbp2 partially rescued both MYHC expression and formation of multinucleated cells (Fig. 3 B and C). Differentiation of Rbp2−/−;Rb1−/− MEFs was also enhanced after reintroduction of WT pRB or by the pRB variant Δ663, which promotes differentiation despite an inability to bind to E2F or repress E2F-dependent promoters (8) (Fig. S2). This finding suggests that pRB has non-E2F targets in addition to RBP2 that affect differentiation.
Loss of RBP2 Suppresses Tumorigenesis Caused by Deletion of the Rb1 or Men1 Tumor Suppressor Genes.
Although RBP2 regulates proliferation, senescence, and differentiation in vitro, which are processes deregulated in cancer, its potential relevance in transformation in vivo is unknown. We, therefore, asked whether Rbp2 interacts genetically with Rb1 in vivo, exploiting the fact that Rbp2−/− mice in a mixed genetic background are viable and have a normal lifespan (Fig. S3). Rb1−/− embryos die at embryonic day 14.5 (30–32), and Rb1−/− embryos supplied with Rb1+/+ extraembryonic tissues die shortly after birth, possibly because of severe skeletal muscle defects (33, 34). No Rbp2−/−;Rb1−/− pups were born from Rbp2+/−;Rb1+/− intercrosses (Table S1), indicating that Rbp2 loss cannot rescue the embryonic developmental defects caused by Rb1 loss.
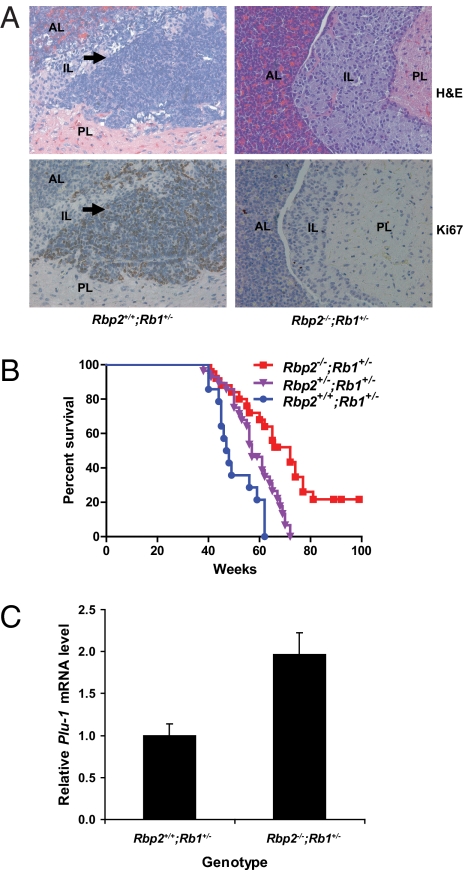
Next, we asked if loss of RBP2 would alter pRB-defective tumorigenesis. Rb1+/− mice develop pituitary and thyroid tumors that are associated with stochastic loss of the second Rb1 allele (30, 35). We, therefore, examined the Rb1+/− progeny of matings between Rbp2+/−;Rb1+/− mice. A limited number of timed necropsies were performed on 28-wk-old mice. As expected, most (3/4) Rb1+/− mice had early pituitary lesions, including small tumors, whereas no abnormalities were detected in the pituitaries of all (4/4) of the Rbp2−/−;Rb1+/− mice (Fig. 4A), suggesting that RBP2 loss suppresses tumor initiation. The remainder of the mice were monitored and killed when they became distressed or moribund because of the development of tumors.
Fig. 4.
Loss of RBP2 suppresses Rb1+/− tumorigenesis in vivo. (A) H&E staining (Upper) and Ki67 staining (Lower) of the pituitary glands of 28-wk-old Rbp2+/+;Rb1+/− (Left) and Rbp2−/−;Rb1+/− (Right) mice. The arrow in Left points to a tiny pituitary tumor in the intermediate lobe. AL, anterior lobe; IL, intermediate lobe; PL, posterior lobe. (B) Kaplan–Meier survival curve comparing Rbp2+/+;Rb1+/− (n = 14), Rbp2+/−;Rb1+/− (n = 28), and Rbp2−/−;Rb1+/− (n = 24) mice (P < 0.0001 for Rbp2−/−;Rb1+/− vs. Rbp2+/+;Rb1+/− and P < 0.01 for Rbp2+/−;Rb1+/− vs. Rbp2+/+;Rb1+/−). (C) Real-time RT-PCR analysis of Plu-1 mRNA in pituitary tumors from 12-mo-old Rbp2−/−;Rb1+/− (n = 4) and Rbp2+/+;Rb1+/− (n = 2) mice. Shown are mean values with SEM.
Importantly, deletion of Rbp2 dramatically extended the life span of Rb1+/− mice (Fig. 4B). The median survival time improved from 47 wk for Rbp2+/+;Rb1+/− mice to 72 wk for Rbp2−/−;Rb1+/− mice. Indeed, some Rbp2−/−;Rb1+/− mice lived up to 2 y, the average life span of WT mice. Importantly, loss of one Rbp2 allele also delayed tumorigenesis and partially extended the life span of Rb1+/− mice (Fig. 4B). Similar results were obtained when the analysis was restricted strictly to littermates (Fig. S4). Notably, all of the Rbp2−/−;Rb1+/− and Rbp2+/−;Rb1+/− mice had microscopic pituitary and/or thyroid tumors at necropsy (Table S2). This finding suggests that RBP2 delays the onset of such tumors or retards their progression rather than preventing tumor initiation.
Mammals have three RBP2 paralogs called PLU-1, SMCX, and SMCY. Plu-1 mRNA levels were significantly increased in pituitary tumors arising in 12-mo-old Rbp2−/−;Rb1+/− mice compared with tumors arising in 12-mo-old Rbp2+/+;Rb1+/− mice (Fig. 4C), suggesting that compensation by Rbp2 paralogs contributes to the eventual formation of pituitary tumors in the Rbp2−/−;Rb1+/− mice.
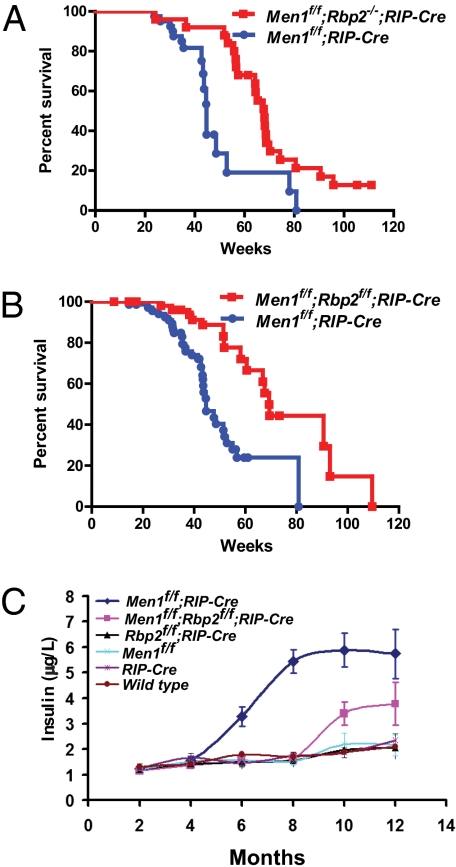
Inactivation of the MEN1 tumor suppressor gene, like inactivation of RB1, leads to formation of neuroendocrine tumors (4, 36, 37). Menin, the MEN1 gene product, is part of a complex that promotes H3K4 methylation, and this activity is diminished by tumor-associated MEN1 mutations (6, 22, 23). We, therefore, reasoned that inactivation of the RBP2 H3K4 demethylase might partially rescue Men1 loss. To this end, we exploited the fact that Men1 inactivation in pancreatic islet cells leads to the development of insulinomas (37), which can be monitored based on changes in circulating insulin levels. Through appropriate matings, we generated Men1f/f;Rbp2+/+, Men1f/f;Rbp2−/−, and Men1f/f;Rbp2f/f mice that also expressed Cre recombinase in their pancreatic islet cells (RIP-Cre) (38). Inactivation of the floxed alleles was confirmed by anti-menin and anti-RBP2 immunohistochemistry (Fig. S5).
Rbp2 inactivation, either systemically (Fig. 5A) or specifically in islet β-cells (Fig. 5B), substantially decreased islet cell tumor burden, which was measured by circulating insulin levels (Fig. 5C), and enhanced survival (Fig. 5 A and B). The median survival for Men1f/f;Rbp2+/+;RIP-Cre mice was 45 wk compared with median survivals of 68 wk for Men1f/f;Rbp2−/−;RIP-Cre mice (Fig. 5A) or 69 wk for Men1f/f;Rbp2f/f;RIP-Cre mice (Fig. 5B), respectively. Inactivation of Rbp2 in islet cells of Men1+/+ mice did not grossly affect islet histology or function, which was determined by gene expression profiling (Fig. S6), circulating insulin (Fig. 5C), and glucose levels (data not shown).
Fig. 5.
Loss of RBP2 suppresses Men1-defective pancreatic islet cell tumorigenesis. (A and B) Kaplan–Meier survival curves comparing (A) Men1f/f;RIP-Cre mice (n = 40) with Men1f/f;Rbp2−/−;RIP-Cre (n = 25; P < 0.005) or (B) Men1f/f;RIP-Cre mice (n = 85) with Men1f/f;Rbp2f/f;RIP-Cre (n = 72; P < 0.0001). (C) Circulating insulin levels in mice with the indicated genotypes.
We also performed timed necropsies on a limited number of Men1f/f;Rbp2+/+;RIP-Cre and Men1f/f;Rbp2f/f;RIP-Cre mice (Fig. 6A). By 2 mo of age, 50% (5/10) of the former exhibited islet cell hyperplasia compared with zero of the latter (0/8) (Table S3). The prevalence of cellular atypia and insulinoma at 4 and 8 mo was dramatically reduced by loss of Rbp2. By 10 mo of age, all (15/15) of the Men1f/f;Rbp2+/+;RIP-Cre mice had insulinomas compared with 2 of 21 Men1f/f;Rbp2f/f;RIP-Cre mice (Table S3). These findings indicate that Rbp2 loss significantly delays the onset of hyperplasia, atypia, and insulinoma in this model.
Fig. 6.
RBP2 loss inhibits proliferation of Men1-defective islets. (A and D) Representative (A) H&E and (D) BrdU staining of pancreata from mice with the indicated genotypes. (Scale bar: 100 μm.) BrdU was administered by i.p. injection to 2-mo-old mice 5 h before sacrifice. (B) Real-time RT-PCR analysis of the indicated mRNAs in pancreatic islets of 12-mo-old RIP-Cre mice (n = 2) and pancreatic islet tumors of 12-mo-old Men1f/f;RIP-Cre mice (n = 2) and Men1f/f;Rbp2f/f;RIP-Cre mice (n = 2). Shown are mean values with SEM. (C) Quantitation of the islet cell BrdU incorporation measured in D.
Notably, insulinomas were observed in some (2/5) 12-mo-old Men1f/f;Rbp2f/f;RIP-Cre mice at necropsy. Comparison of insulinomas from 12-mo-old Men1f/f;Rbp2f/f;RIP-Cre mice with Men1f/f;RIP-Cre mice revealed increased expression of Plu-1 but not Smcx and Smcy after Rbp2 loss (Fig. 6B). This increase, however, was not observed in spleens from 12-mo-old Men1f/f;Rbp2f/f;RIP-Cre mice or pancreatic islets from 2-mo-old Men1f/f;Rbp2f/f;RIP-Cre mice (data not shown). These observations suggest that the eventual formation of insulinomas in Men1f/f;Rbp2f/f;RIP-Cre mice depends on increased levels of PLU-1, perhaps occurring stochastically over time.
To begin to understand the mechanisms underlying these differences, we injected 2-mo-old mice with BrdU and examined their pancreata 5 h later. As expected, BrdU incorporation was increased in the islets of Men1f/f;Rbp2+/+;RIP-Cre mice compared with WT controls (Fig. 6 C and D). This increase was not observed, however, in islets that concurrently lacked Rbp2. We did not observe differences in bulk H3K4 trimethylation by immunohistochemistry (data not shown), possibly reflecting the activity of additional H3K4 methyltransferases and demethylases.
To begin to assess the molecular basis for the effect of Rbp2 loss in attenuating tumorigenesis, we performed mRNA profiling using DNA microarrays on pancreatic islets isolated from 2-mo-old WT, RIP-Cre, Men1f/f;RIP-Cre, Rbp2f/f;RIP-Cre, and Men1f/f;Rbp2f/f;RIP-Cre mice. The gene expression changes caused by deletion of Men1 overlap with those changes reported previously (39) (Dataset S1).
To determine the effects of Rbp2 inactivation on the gene expression changes in Men1-deficient islets, we compared the gene expression changes in Men1f/f;Rbp2f/f;RIP-Cre, Men1f/f;RIP-Cre, and Rbp2f/f;RIP-Cre islets with WT and RIP-Cre control islets. The effects of Men1 deletion on pancreatic islet gene expression were reversed by Rbp2 loss for a number of genes belonging to several classes, including genes involved in signaling, cell cycle, and apoptosis (Fig. 7 A and B). The reversal by Rbp2 deletion of expression changes associated with Men1 deletion in islets was confirmed by real-time RT-PCR (Fig. 7C).
Fig. 7.
Rbp2 loss reverses gene expression changes caused by Men1 loss. (A and B) Microarray data of transcripts that are significantly altered (t test, P < 0.05; fold change > 1.85) in islets from Men1f/f;RIP-Cre mice compared with islets from RIP-Cre control mice and Men1f/f;Rbp2f/f;RIP-Cre mice. (A) Heat map representing relative transcript abundance for the indicated genes (rows) and islet preparations from individual mice (columns). The color scale is based on log2 fold change from the mean signal in RIP-Cre mouse islets. (B) Scatter plot depicting the average fold change of expression of these transcripts in islets from the indicated KO mice compared with RIP-Cre mice. (C) Real-time RT-PCR analysis of the genes marked by red crosses in B and Casp8 and Gata6. Shown are mean values with SEM from at least triplicate experiments.
Discussion
We confirmed that loss of RBP2 impairs proliferation, promotes senescence, and enhances differentiation in vitro. Notably, deletion of Rbp2 was insufficient to rescue the embryonic developmental defects caused by Rb1 loss but significantly suppressed pituitary and thyroid tumorigenesis in Rb1+/− mice and islet cell tumorigenesis after inactivation of Men1 in pancreatic neuroendocrine cells.
The canonical pRB targets are members of the E2F transcription factor family, and suppression of E2F-responsive promoters contributes to cell-cycle control and tumor suppression by pRB (7). pRB also biochemically interacts with a number of chromatin modifiers, including HDACs (40–42), SWI/SNF chromatin remodeling complexes (43, 44), H3K9 methyltransferases Suv39h1 (45) and RIZ1 (46), H4K20 methyltransferase Suv4-20h (47), and DNA methyltransferase 1 (DNMT1) (48). Our findings, together with earlier biochemical and siRNA-derived data, suggest that another pRB-interacting chromatin modifier, RBP2, contributes to tumor suppression by pRB. RBP2 loss inhibits cell proliferation in a pRB-dependent manner, placing RBP2 upstream of pRB. However, RBP2 inhibits senescence and differentiation in pRB-defective tumor cells, and loss of RBP2 inhibits formation of pRB-defective endocrine tumors, suggesting that RBP2 also acts downstream of pRB. In summary, tumor suppression by pRB might involve coordinated regulation of both E2F and RBP2. Consistent with this idea, RBP2 is recruited to E2F target genes during differentiation (49).
It is increasingly clear that alterations in histone methylation play important roles in cancer in general (50, 51). For example, MLL1, a subunit of an H3K4 methyltransferase complex, is frequently translocated in leukemia (52, 53), whereas another H3K4 methyltransferase subunit gene, MEN1, is frequently mutated in endocrine tumors (4, 6, 22, 23). EZH2, the catalytic subunit of an H3K27 methyltransferase polycomb repressive complex 2, is overexpressed in aggressive prostate cancers (54). Finally, copy number changes and intragenic mutations affecting histone methyltransferses and demethylases, such as the UTX H3K27 histone demethylase, are increasingly being identified in cancers (5, 55, 56).
RBP2 is one of four proteins [together with PLU-1 (also known as KDM5B or JARID1B), SMCX (also known as KDM5C or JARID1C), and SMCY (also known as KDM5D or JARID1D)] capable of demethylating trimethylated H3K4 (57). This mark is usually associated with actively transcribed genes and is also found at bivalent domains in association with trimethylated H3K27, which is usually linked to transcriptional repression (27). The paradoxical co-association of both an activating and repressive methylation mark is thought to poise genes to respond to either inhibitory or stimulatory signals linked to differentiation and control of cell fate. Consistent with this idea, bivalent domains seem to be important for both stem cell and cancer biology.
Interestingly, RBP2 has been reported to be translocated in leukemia (58, 59) and overexpressed in gastric cancer (10). A recent study suggested that increased expression of RBP2 promoted a more stem-like phenotype, consistent with our results, and enhanced resistance to anticancer agents (60). PLU-1 is overexpressed in breast (61) and prostate cancers (62), and shRNA-mediated down-regulation of PLU-1 suppresses breast cancer growth in a syngeneic mouse cancer model (63). Interestingly, PLU-1 marks a subpopulation of slow-cycling melanoma cells required for continuous tumor growth (64), and its overexpression in ES cells suppresses differentiation (65). Therefore, PLU-1, like RBP2, might maintain a stem-like phenotype and promote tumorigenesis. Finally, SMCX was recently found to be mutated in a subset of clear cell renal cell carcinomas (5).
Both RB1 and MEN1 have been linked to neuroendocrine tumors. The former is linked to pituitary and thyroid tumors in mice (30, 35) and small cell lung cancer in man (66), whereas the latter is linked to pituitary, parathyroid, and pancreatic islet cell tumors in both species (4, 36, 37). Interestingly, inactivation of Rb1 and Men1 in neuroendocrine tumors arising in Rb1+/−;Men1+/− compound heterozygous mice is mutually exclusive (67, 68), suggesting that Rb1 and Men1 share a critical activity or activities relevant to neuroendocrine tumorigenesis. Our studies suggest that regulation of H3K4 methylation is one such activity.
Enzymes have historically proven to be tractable drug targets. RBP2 and its paralogs PLU-1, SMCX, and SMCY are 2-oxoglutarate–dependent dioxygenases (18, 19). These enzymes can be inhibited with drug-like small organic molecules that act competitively with respect to 2-oxoglutarate, interfere with iron use, or both (20, 21). Our findings suggest that RBP2-inhibitory drugs, should they be developed, would have anticancer activity. Furthermore, elevated expression of PLU-1 in Rbp2 null tumors (Fig. 4C and 6B) suggests that RBP2 inhibitors that also inhibit PLU-1, if they were safe, would be more effective than inhibitors that target RBP2 alone.
Materials and Methods
Mouse Experiments.
Rbp2−/− and Rbp2f/f mice were described previously (11) and backcrossed to C57BL/6 strain for at least five generations. Rbp2+/− mice were intercrossed to generate Rbp2−/− MEFs and WT littermate control MEFs. Rbp2f/f mice were crossed with C57BL/6 chicken β-actin Cre-ER mice (24, 69) to obtain Rbp2+/f;Cre-ER mice. Rbp2+/f;Cre-ER mice were crossed with Rbp2+/f mice to generate Rbp2f/f;Cre-ER MEFs and Rbp2+/+;Cre-ER littermate control MEFs. Rb1+/− mice on a C57BL/6 background (30) were obtained from the National Cancer Institute Mouse Repository. Rb1+/− mice were crossed with Rbp2−/− mice on a mixed 129/SvEv, FVB/N, and C57BL/6 background to obtain Rbp2+/−;Rb1+/− mice. These mice were then intercrossed to generate the experimental cohorts of Rb1+/−, Rbp2+/−;Rb1+/− and Rbp2−/−;Rb1+/− mice.
Men1 conditional KO mice were described previously (6) and maintained on a mixed 129s6, FVB/N, and C57BL/6 background. To specifically delete the Men1 gene in pancreatic islet β-cells, Men1f/f mice were crossed with RIP-Cre transgenic mice (38). The Men1+/f;RIP-Cre mice were crossed with Men1+/f mice to generate Men1f/f;RIP-Cre mice. Men1f/f;Rbp2−/−;RIP-Cre and Men1f/f;Rbp2f/f;RIP-Cre mice were generated by introducing Rbp2 null and floxed alleles into the Men1f/f;RIP-Cre mice through appropriate matings. For in vitro proliferation and differentiation assays, Rb1+/− mice were crossed with Rbp2+/− mice on a pure C57BL/6 background to obtain Rbp2+/−;Rb1+/− mice, which were intercrossed to generate WT, Rbp2−/−, Rb1−/−, Rbp2+/−;Rb1−/−, and Rbp2−/−;Rb1−/− MEFs. Mice and cells carrying Men1 floxed alleles were genotyped using primers described in SI Materials and Methods, and all other mice and cells were genotyped as described (11, 24, 30, 38). All mice were maintained in the research animal facility of the Dana–Farber Cancer Institute and Yale Animal Resources Center in accordance with the National Institutes of Health guidelines. All procedures involving mice were approved by the Institutional Animal Care and Use Committees of the Dana–Farber Cancer Institute and Yale University.
ES Cell Culture and Differentiation.
In Fig. 2 A–E, Rbp2f/f ES cells were isolated from mouse blastocysts after intercrossing Rbp2f/f mice on a pure C57BL/6 background and transiently transfected with pBS500/EF1α-GFPCre plasmid. GFP-positive cells were isolated by FACS and plated at low density. Isolated colonies were then expanded into ES lines. Successful recombination of the Rbp2 locus was confirmed by PCR and Western blot analysis. In Fig. 2F, WT and Rbp2−/− ES cells were isolated from mouse blastocysts after intercrossing Rbp2+/− mice. WT, Rbp2f/f, and Rbp2−/− ES cells were maintained on mitomycin C-treated MEF feeders in standard ES medium: DMEM containing 15% heat-inactivated FBS, 0.1 mM 2-mercaptoethanol, 2 mM l-glutamine, 0.1 mM nonessential amino acid, 1% Embryomax ES cell-qualified nucleosides (100× stock; Chemicon), 1,000 U/mL recombinant LIF (Chemicon), 50 U/mL penicillin/streptomycin.
For differentiation assays, ES cells were passaged at least three times without feeders and maintained on gelatin-coated plates in standard ES culture medium containing LIF. In Fig. 2 A–E, the ES cells were induced to differentiate by removing LIF from culture medium and were harvested at 4 or 6 d after differentiation for analysis. In Fig. 2F, ES cells were induced to differentiate on untreated plates to form EB for 2 d, and were then treated with 1 μM retinoic acid to induce neuronal differentiation for 3 d. After 2 more days on untreated plates, the cells were plated onto gelatin-coated plates and grown for an additional 4 d before Western blot analysis.
Gene Expression Profiling.
Subconfluent Rbp2f/f and Rbp2−/− ES cells were harvested for RNA isolation using the RNeasy mini kit with on-column DNase digestion (Qiagen). Gene expression profiling was performed using Affymetrix GeneChip mouse genome 430 2.0 arrays. Raw gene expression profiling data were analyzed using dChip (70). The two gene sets used for gene set enrichment analysis were described previously (29). The differentiation genes include all genes that are marked by both H3K27me3 and EZH1 in WT ES cells and up-regulated at least threefold 6 d after induction of differentiation by LIF withdrawal. The ES genes are genes highly expressed in pluripotent ES cells compared with differentiated cells.
Pancreatic islets were isolated as described (71). Briefly, 0.25 mg/mL Liberase solution (Roche) in serum-free M199 medium were injected into pancreata through the common bile duct of anesthetized 2-mo-old male mice. The inflated pancreata were incubated at 37 °C for 20 min for digestion before filtered through mesh. Then, islets were purified through histopaque gradient purification and gravity sedimentation. Finally, islets were hand-picked from dark field dishes under a dissecting microscope for RNA isolation using the RNeasy mini kit (Qiagen). Islet RNAs were expression-profiled on Affymetrix GeneChip Mouse Gene 1.0 ST arrays. Raw gene expression profiling data were analyzed using dChip (70). Transcripts were defined to be significantly changed based on a t test P < 0.05. The expression data reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database under accession numbers GSE26446 and GSE26978.
Supplementary Material
Acknowledgments
We thank members of the E.V.B., W.G.K., M.M., S.H.O., and Q.Y. laboratories for their kind help and valuable discussions. We thank Dr. Lili Yamasaki for her valuable comments. We also thank the laboratory members of Dr. David Stern for their kind help. We thank Dr. Ronald DePinho for providing Cre-ER transgenic mice and Dr. Seung Kim for providing RIP-Cre mice. We thank Dr. Edward Fox and the Dana–Farber Cancer Institute microarray core facility and Yale Center for Genome Analysis for gene expression profiling studies. We also thank Harvard rodent histology core, Harvard specialized histopatholology core, Dr. Alexander Vortmeyer, and the Yale research histology facility for histological analysis. This work was supported by National Institutes of Health Grants CA138631 (to E.V.B.), CA076120 (to W.G.K.), and CA16359 (to Yale Comprehensive Cancer Center), the Raymond and Beverly Sackler Fund for the Arts and Sciences (M.M.), the Caring for Carcinoid Foundation (M.M), and the V Scholar Award (to Q.Y.). W.G.K. is a Howard Hughes Medical Institute Investigator and a Doris Duke Distinguished Scientist. Q.Y. is a Breast Cancer Alliance Young Investigator and an Alexander and Margaret Stewart Trust Fellow.
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE26446 and GSE26978).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110104108/-/DCSupplemental.
References
- 1.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 2.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 3.Rowley JD. Rearrangements involving chromosome band 11Q23 in acute leukaemia. Semin Cancer Biol. 1993;4:377–385. [PubMed] [Google Scholar]
- 4.Chandrasekharappa SC, et al. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science. 1997;276:404–407. doi: 10.1126/science.276.5311.404. [DOI] [PubMed] [Google Scholar]
- 5.Dalgliesh GL, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hughes CM, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- 7.Sellers WR, Kaelin WG., Jr Role of the retinoblastoma protein in the pathogenesis of human cancer. J Clin Oncol. 1997;15:3301–3312. doi: 10.1200/JCO.1997.15.11.3301. [DOI] [PubMed] [Google Scholar]
- 8.Sellers WR, et al. Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benevolenskaya EV, Murray HL, Branton P, Young RA, Kaelin WG., Jr Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol Cell. 2005;18:623–635. doi: 10.1016/j.molcel.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 10.Zeng J, et al. The histone demethylase RBP2 Is overexpressed in gastric cancer and its inhibition triggers senescence of cancer cells. Gastroenterology. 2010;138:981–992. doi: 10.1053/j.gastro.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Klose RJ, et al. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007;128:889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 12.Christensen J, et al. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Iwase S, et al. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 14.Hayakawa T, et al. RBP2 is an MRG15 complex component and down-regulates intragenic histone H3 lysine 4 methylation. Genes Cells. 2007;12:811–826. doi: 10.1111/j.1365-2443.2007.01089.x. [DOI] [PubMed] [Google Scholar]
- 15.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: Development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 16.Kaminskas E, Farrell AT, Wang YC, Sridhara R, Pazdur R. FDA drug approval summary: Azacitidine (5-azacytidine, Vidaza) for injectable suspension. Oncologist. 2005;10:176–182. doi: 10.1634/theoncologist.10-3-176. [DOI] [PubMed] [Google Scholar]
- 17.Oki Y, Aoki E, Issa JP. Decitabine—bedside to bench. Crit Rev Oncol Hematol. 2007;61:140–152. doi: 10.1016/j.critrevonc.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 18.Clissold PM, Ponting CP. JmjC: Cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2beta. Trends Biochem Sci. 2001;26:7–9. doi: 10.1016/s0968-0004(00)01700-x. [DOI] [PubMed] [Google Scholar]
- 19.Aravind L, Koonin EV. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001;2:RESEARCH0007. doi: 10.1186/gb-2001-2-3-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivan M, et al. Biochemical purification and pharmacological inhibition of a mammalian prolyl hydroxylase acting on hypoxia-inducible factor. Proc Natl Acad Sci USA. 2002;99:13459–13464. doi: 10.1073/pnas.192342099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Safran M, et al. Mouse model for noninvasive imaging of HIF prolyl hydroxylase activity: Assessment of an oral agent that stimulates erythropoietin production. Proc Natl Acad Sci USA. 2006;103:105–110. doi: 10.1073/pnas.0509459103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yokoyama A, et al. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 23.Yokoyama A, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: A tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 25.Rangarajan A, Hong SJ, Gifford A, Weinberg RA. Species- and cell type-specific requirements for cellular transformation. Cancer Cell. 2004;6:171–183. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 26.Yates KE, Korbel GA, Shtutman M, Roninson IB, DiMaio D. Repression of the SUMO-specific protease Senp1 induces p53-dependent premature senescence in normal human fibroblasts. Aging Cell. 2008;7:609–621. doi: 10.1111/j.1474-9726.2008.00411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 28.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacks T, et al. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 31.Clarke AR, et al. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359:328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- 32.Lee EY, et al. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- 33.de Bruin A, et al. Rb function in extraembryonic lineages suppresses apoptosis in the CNS of Rb-deficient mice. Proc Natl Acad Sci USA. 2003;100:6546–6551. doi: 10.1073/pnas.1031853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu L, et al. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature. 2003;421:942–947. doi: 10.1038/nature01417. [DOI] [PubMed] [Google Scholar]
- 35.Hu N, et al. Heterozygous Rb-1 delta 20/+mice are predisposed to tumors of the pituitary gland with a nearly complete penetrance. Oncogene. 1994;9:1021–1027. [PubMed] [Google Scholar]
- 36.Crabtree JS, et al. A mouse model of multiple endocrine neoplasia, type 1, develops multiple endocrine tumors. Proc Natl Acad Sci USA. 2001;98:1118–1123. doi: 10.1073/pnas.98.3.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crabtree JS, et al. Of mice and MEN1: Insulinomas in a conditional mouse knockout. Mol Cell Biol. 2003;23:6075–6085. doi: 10.1128/MCB.23.17.6075-6085.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herrera PL. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development. 2000;127:2317–2322. doi: 10.1242/dev.127.11.2317. [DOI] [PubMed] [Google Scholar]
- 39.Fontanière S, et al. Gene expression profiling in insulinomas of Men1 beta-cell mutant mice reveals early genetic and epigenetic events involved in pancreatic beta-cell tumorigenesis. Endocr Relat Cancer. 2006;13:1223–1236. doi: 10.1677/erc.1.01294. [DOI] [PubMed] [Google Scholar]
- 40.Brehm A, et al. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 41.Magnaghi-Jaulin L, et al. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 42.Luo RX, Postigo AA, Dean DC. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 43.Dunaief JL, et al. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 44.Strober BE, Dunaief JL, Guha , Goff SP. Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol Cell Biol. 1996;16:1576–1583. doi: 10.1128/mcb.16.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nielsen SJ, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- 46.Kim KC, Geng L, Huang S. Inactivation of a histone methyltransferase by mutations in human cancers. Cancer Res. 2003;63:7619–7623. [PubMed] [Google Scholar]
- 47.Gonzalo S, et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol. 2005;7:420–428. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- 48.Robertson KD, et al. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 49.Lopez-Bigas N, et al. Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol Cell. 2008;31:520–530. doi: 10.1016/j.molcel.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: Histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 52.Milne TA, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 53.Nakamura T, et al. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 54.Varambally S, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 55.van Haaften G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Northcott PA, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41:465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blair LP, Cao J, Zou MR, Sayegh J, Yan Q. Epigenetic regulation by lysine demethylase 5 (KDM5) enzymes in cancer. Cancers (Basel) 2011;3:1383–1404. doi: 10.3390/cancers3011383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Zutven LJ, et al. Identification of NUP98 abnormalities in acute leukemia: JARID1A (12p13) as a new partner gene. Genes Chromosomes Cancer. 2006;45:437–446. doi: 10.1002/gcc.20308. [DOI] [PubMed] [Google Scholar]
- 59.Wang GG, et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459:847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu PJ, et al. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J Biol Chem. 1999;274:15633–15645. doi: 10.1074/jbc.274.22.15633. [DOI] [PubMed] [Google Scholar]
- 62.Xiang Y, et al. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc Natl Acad Sci USA. 2007;104:19226–19231. doi: 10.1073/pnas.0700735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamane K, et al. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 64.Roesch A, et al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dey BK, et al. The histone demethylase KDM5b/JARID1b plays a role in cell fate decisions by blocking terminal differentiation. Mol Cell Biol. 2008;28:5312–5327. doi: 10.1128/MCB.00128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harbour JW, et al. Abnormalities in structure and expression of the human retinoblastoma gene in SCLC. Science. 1988;241:353–357. doi: 10.1126/science.2838909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Loffler KA, et al. Lack of augmentation of tumor spectrum or severity in dual heterozygous Men1 and Rb1 knockout mice. Oncogene. 2007;26:4009–4017. doi: 10.1038/sj.onc.1210163. [DOI] [PubMed] [Google Scholar]
- 68.Matoso A, Zhou Z, Hayama R, Flesken-Nikitin A, Nikitin AY. Cell lineage-specific interactions between Men1 and Rb in neuroendocrine neoplasia. Carcinogenesis. 2008;29:620–628. doi: 10.1093/carcin/bgm207. [DOI] [PubMed] [Google Scholar]
- 69.Minamishima YA, et al. Somatic inactivation of the PHD2 prolyl hydroxylase causes polycythemia and congestive heart failure. Blood. 2008;111:3236–3244. doi: 10.1182/blood-2007-10-117812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: Model validation, design issues and standard error application. Genome Biol. 2001;2:RESEARCH0032. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kulkarni RN, et al. Leptin rapidly suppresses insulin release from insulinoma cells, rat and human islets and, in vivo, in mice. J Clin Invest. 1997;100:2729–2736. doi: 10.1172/JCI119818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.