Abstract
Platensimycin (PTM) and platencin (PTN) are potent and selective inhibitors of bacterial and mammalian fatty acid synthases and have emerged as promising drug leads for both antibacterial and antidiabetic therapies. Comparative analysis of the PTM and PTN biosynthetic machineries in Streptomyces platensis MA7327 and MA7339 revealed that the divergence of PTM and PTN biosynthesis is controlled by dedicated ent-kaurene and ent-atiserene synthases, the latter of which represents a new pathway for diterpenoid biosynthesis. The PTM and PTN biosynthetic machineries provide a rare glimpse at how secondary metabolic pathway evolution increases natural product structural diversity and support the wisdom of applying combinatorial biosynthesis methods for the generation of novel PTM and/or PTN analogues, thereby facilitating drug development efforts based on these privileged natural product scaffolds.
Keywords: antibiotic, metabolic pathway engineering, biosynthetic gene cluster, ent-copalyl diphosphate, terpene synthase
Infectious disease is the second leading cause of death worldwide, and the growing number of antibiotic-resistant microbes threatens to worsen this problem; only two previously undescribed classes of antibiotics have been introduced into the clinic since the 1960s (1, 2). Diabetes affects nearly 24 million people in the United States, and current therapies suffer from serious limitations (3). Platensimycin (PTM) and platencin (PTN) are recently discovered natural products (4) that are potent and selective inhibitors of bacterial (5, 6) and mammalian (7) fatty acid synthases. Remarkably, they have emerged as promising drug leads for both antibacterial (5, 6, 8, 9) and antidiabetic (7) therapies. The efficacy of PTM and PTN in treating bacterial infections (5, 6), including those that are resistant to commercially available drugs, and the efficacy of PTM in treating diabetes and related metabolic disorders (7) have been demonstrated in mouse models.
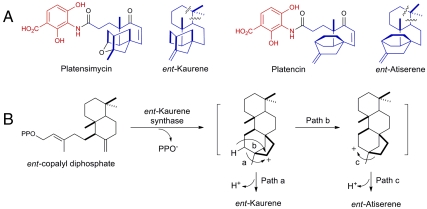
Structurally, PTM and PTN are composed of two distinct moieties—a substituted benzoic acid and an aliphatic cage moiety joined together by a flexible propionamide chain (Fig. 1A) (10, 11). Stable isotope feeding studies (12, 13) suggest that (i) the benzoic acid moiety of PTM and PTN is derived from a C4 intermediate from the Krebs cycle and a C3 intermediate from glycolysis, similar to the recently characterized grixazone biosynthetic pathway (14) and other natural products (see SI Appendix, Fig. S1B), and (ii) the aliphatic cage moieties of PTM and PTN both originate from the methylerythritol phosphate (MEP) pathway (15, 16) for terpene biosynthesis. Their unique carbon scaffolds point toward an ent-kaurene intermediate in PTM biosynthesis (12) and an ent-atiserene intermediate in PTN biosynthesis (13) (Fig. 1A) and diterpenoid natural products of both ent-kaurene and ent-atiserene origin are well known (SI Appendix, Fig. S1 C and D). Although numerous terpene synthase genes have been cloned from eukaryotes, only a few have been cloned from prokaryotes (17–19). The only ent-kaurene synthase of bacterial origin was reported in 2009 (20), and no gene or enzyme of eukaryotic or prokaryotic origin for ent-atiserene biosynthesis has ever been reported. Interestingly, ent-kaurene synthase-catalyzed biosynthesis of ent-kaurene from ent-copalyl diphosphate (ent-CPP) can produce ent-atiserene as a minor metabolite (21). Minor mutations to terpene synthases in general (22) and CPP-utilizing terpene synthases in particular (21, 23, 24) are also known to alter product specificity. These observations, together with the fact that no ent-atiserene synthase is known, has become the basis of the current proposal that ent-kaurene synthase might control the biosynthesis of both ent-kaurene and ent-atiserene-derived diterpenoid natural products (Fig. 1B).
Fig. 1.
PTM and PTN and their proposed biosynthetic relationship to ent-kaurene and ent-atiserene. (A) Structures of PTM and PTN with the 3-amino-2,4-dihydroxybenzoic acid moiety common to PTM and PTN shown in red and with the ent-kaurene and ent-atiserene-derived diterpenoid moieties of PTM and PTN shown in blue. Wavy lines indicate bonds broken during biosynthesis en route to PTM and PTN. (B) Commonly accepted proposal for ent-kaurene synthase-catalyzed formation of ent-kaurene (path a) and ent-atiserene (paths b and c) from ent-copalyl diphosphate.
PTM was originally isolated from Streptomyces platensis MA7327, a strain found in a soil sample collected in South Africa (5, 10), whereas PTN was first isolated from Streptomyces platensis MA7339, a strain found in a soil sample collected in Spain (6, 11); we subsequently established MA7327 as a PTM and PTN dual producer (25). Because the structural differences between PTM and PTN lie in their diterpene moieties, most likely resulting from catalytic specificity of key terpene synthases, the distinct chemical profiles of these closely related strains present a unique opportunity to investigate the evolutionary and molecular mechanisms of how terpene synthases control regiochemistry and dictate the fate of the ultimate cyclization products in terpenoid biosynthesis (15–24). Here we now report a comparative study of PTM and PTN biosynthesis in S. platensis MA7327 and MA7339. Our findings revealed that the divergence of PTM and PTN biosynthesis is controlled by dedicated ent-kaurene and ent-atiserene synthases, the latter of which represents a previously undescribed pathway for diterpenoid biosynthesis.
Results and Discussion
Cloning, Sequencing, and Annotation of the PTM and PTN Biosynthetic Gene Cluster from S. platensis MA7327 and MA7339.
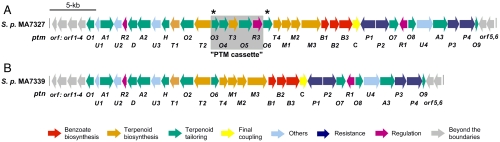
We first cloned the PTM and PTN dual biosynthetic gene cluster from the MA7327 strain (named ptm cluster hereafter), and a 47-kb contiguous DNA was sequenced, revealing 43 ORFs (Fig. 2A) (SI Appendix). A combination of bioinformatics and in vivo gene inactivation was used to define the ptm cluster boundaries (SI Appendix), and the 36 ORFs within the ptm cluster (i.e., from ptmO1 to ptmO9) could be assigned putative roles in the biosynthesis, resistance, or regulation of PTM and PTN based on their homology to enzymes of known function (Table 1).
Fig. 2.
Genetic organization of the PTM and PTN biosynthetic gene clusters. (A) The ptm cluster encoding PTM and PTN dual production from S. platensis MA7327. (B) The ptn cluster encoding PTN production from S. platensis MA7339. Genes are color-coded to highlight their predicted functions in PTM and PTN biosynthesis, resistance, and regulation. The PTM cassette that endows the ptm cluster the capacity for dual PTM and PTN production is shaded in gray and is absent from the ptn cluster. The asterisks denote the location of the 1-kb repeats in the ptm gene cluster. Functional annotations of the genes within the ptm and ptn clusters are summarized in Table 1.
Table 1.
Deduced function of ORFs from the ptm gene cluster in S. platensis MA7327 and the ptn gene cluster from MA7339
| Gene | #AAs* | Identity | Gene | #AAs | Protein homolog (accession #) | Homology (%identity/%similarity) | Proposed function |
| orf1-4 | orf 1-4 | ORFs beyond the upstream boundary | |||||
| ptnO1 | 254 | 0.97 | ptmO1 | 254 | DitI (AAD21071) | 35/48 | dehydrogenase |
| ptnU1 | 184 | 0.96 | ptmU1 | 184 | SSEG_08609(YP_002208525) | 68/73 | unknown |
| ptnA1 | 434 | 0.96 | ptmA1 | 434 | PaaK (CAD76926) | 24/40 | acyl-CoA synthetase |
| ptnU2 | 236 | 0.96 | ptmU2 | 236 | SSEG_08597(YP_002208498) | 47/64 | unknown |
| ptnR2 | 133 | 0.96 | ptmR2 | 133 | SSEG_08605(YP_002208515) | 65/76 | regulatory (DNA binding protein) |
| ptnD | 385 | 0.97 | ptmD | 385 | DitF (AAD21068) | 27/40 | thiolase |
| ptnA2 | 522 | 0.98 | ptmA2 | 522 | Fcs (CAC18323) | 30/42 | acyl-CoA synthetase |
| ptnU3 | 360 | 0.99 | ptmU3 | 360 | BarH (AAN32982) | 28/44 | hydrolase |
| ptnH | 284 | 0.94 | ptmH | 284 | Hsd4B (ABW74860) | 46/61 | 2-enoyl acyl-CoA hydratase |
| ptnT1 | 308 | 0.96 | ptmT1 | 308 | Neut_1128 (YP_747348) | 26/42 | ent-atiserene synthase |
| ptnO2 | 441 | 0.98 | ptmO2 | 440 | HctG (AAY42399) | 27/46 | P-450 monooxygenase |
| ptnT2 | 533 | 0.98 | ptmT2 | 533 | ent-Cdps (BAD86797) | 44/57 | ent-copalyl diphosphate synthase |
| ptmO3 | 285 | TfdA (ACF35485) | 27/38 | α-ketogluterate dependent dioxygenase | |||
| ptmO4 | 388 | CaiA (CAA5211) | 26/43 | long-chain acyl CoA dehydrogenase | |||
| ptmT3 | 309 | KSB (Q39548) | 19/36† | ent-kaurene synthase | |||
| ptmO5 | 430 | CphP (BAG16627) | 46/57 | P-450 monooxygenase | |||
| ptmR3 | 355 | MAV_2666 (YP_881857) | 30/42 | regulatory (hypothetical kinase) | |||
| ptnO6 | 280 | 0.96 | ptmO6 | 280 | TfdA (ACF35485) | 26/37 | α-ketogluterate dependent dioxygenase |
| ptnT4 | 348 | 0.99 | ptmT4 | 348 | Ggdps (BAB07816) | 66/77 | geranylgeranyl diphosphate synthase |
| ptnM1 | 448 | 0.94 | ptmM1 | 448 | IspH (AAL38655) | 51/69‡ | HMBDP reductase (MEP pathway) |
| ptnM2 | 366 | 0.99 | ptmM2 | 366 | PlaT5 (ABB69755) | 77/84 | HMBDP synthase (MEP pathway) |
| ptnM3 | 587 | 0.97 | ptmM3 | 587 | PlaT6 (ABB69756) | 62/71 | DXP synthase (MEP pathway) |
| ptnB1 | 277 | 0.98 | ptmB1 | 277 | GriI (BAF36651) | 63/76 | ADHOHP synthase |
| ptnB2 | 368 | 0.98 | ptmB2 | 368 | GriH (BAF36650) | 71/81 | 3,4-AHBA synthase |
| ptnB3 | 396 | 0.95 | ptmB3 | 396 | MhbM (AAW63416) | 29/45 | flavin-dependent benzoate hydroxylase |
| ptnC | 291 | 0.96 | ptmC | 291 | NfNAT (3D9W_A) | 27/37 | amide synthase (N-acetyl transferase) |
| ptnP1 | 474 | 0.96 | ptmP1 | 474 | PhlA (AAB48109) | 23/34§ | putative resistance |
| PhlB (AAB48107) | 21/36¶ | unknown | |||||
| ptnP2 | 414 | 0.99 | ptmP2 | 414 | PhlC (AAB48108) | 22/35 | putative resistance |
| ptnO7 | 301 | 0.95 | ptmO7 | 301 | ORF27 (BAD66689) | 43/57 | oxidoreductase |
| ptnR1 | 238 | 0.99 | ptmR1 | 238 | KorSA (CAA79637) | 16/32 | transcriptional regulator |
| ptnO8 | 278 | 0.99 | ptmO8 | 278 | PsfG (ACA09736) | 37/55 | dehydrogenase |
| ptnU4 | 596 | 0.95 | ptmU4 | 596 | QbsK (AAL65280) | 31/42 | acyl-CoA transferase |
| ptnA3 | 536 | 0.95 | ptmA3 | 537 | Fcs (CAC18323) | 28/40 | acyl-CoA synthetase |
| ptnP3 | 405 | 0.96 | ptmP3 | 405 | TiKAS II (1J3N_A) | 46/63 | putative resistance |
| ptnP4 | 520 | 0.97 | ptmP4 | 520 | Pep (AAG31689) | 30/48 | efflux pump |
| ptnO9 | 111 | 0.99 | ptmO9 | 111 | FdxA (P24496) | 74/83 | ferredoxin |
| orf5-7 | orf5-7 | ORFs beyond downstream boundary |
*AAs, amino acids.
†Values from alignment of full-length PtmT3 with residues 393–773 of KSB.
‡Values from alignment of full-length IspH with residues 124–448 of PtmM1.
§Values from alignment of full-length PhlA with residues 1–334 of PtmP1.
¶Values from alignment of full-length PhlB with residues 335–474 of PtmP1.
We next cloned the PTN biosynthetic gene cluster from the MA7339 strain (named ptn cluster thereafter), and a 41-kb contiguous DNA was sequenced, revealing 38 ORFs (Fig. 2B) (SI Appendix), of which 31 ORFs (i.e., from ptnO1 to ptnO9) were similarly annotated to encode PTN production (Table 1). The ptm and ptn clusters are strikingly similar in both sequence conservation and overall organization (Fig. 2). The most significant difference between the two gene clusters is the complete absence of a 5.4-kb DNA fragment, termed the “PTM cassette,” from the ptn gene cluster, which consists of five genes: ptmO3, O4, T3, O5, and R3 (Fig. 2).
Confirmation of PTM and PTN Biosynthesis Sharing a Common Pathway for 3-Amino-2,4-Dihydroxybenzoic Acid (ADHBA) Biosynthesis.
The ptm and ptn gene clusters contain three genes predicted to encode the biosynthesis of the ADHBA moiety of PTM and PTN (Fig. 2 and Table 1). PtmB1/PtnB1 and PtmB2/PtnB2 are homologous to GriI and GriH, respectively, which have been shown recently to catalyze the synthesis of 3,4-aminohydroxybenzoic acid (AHBA) from aspartate 4-semialdehyde (ASA) and dihydroxyacetone phosphate (DHAP) in grixazone biosynthesis (14). Following ptmB1/ptnB1 and ptmB2/ptnB2 in the same operon is ptmB3/ptnB3, encoding an enzyme homologous to flavin-dependent benzoate hydroxylases. PtmB3/PtnB3 serves as a candidate hydroxylating AHBA to afford the ADHBA moiety that is incorporated into PTM and PTN (Fig. 3 A and D). To experimentally support this proposal, we inactivated ptmB2 in MA7327 to afford the ΔptmB2 mutant strain SB12006 (SI Appendix). When cultured under conditions that yield PTM and PTN with the wild-type MA7327 strain as a positive control, SB12006 failed to produce PTM or PTN, but accumulated two new metabolites (Fig. 3D, I and II), identified as platensic and platencinic acids by mass and NMR spectroscopic analyses (SI Appendix). SB12006 can be chemically complemented with the addition of AHBA to the fermentation medium (SI Appendix), restoring PTM and PTN production (Fig. 3D, III) as would be expected for its intermediacy in PTM and PTN biosynthesis (Fig. 3 A and C).
Fig. 3.
A unified pathway featuring dedicated ent-kaurene and ent-atiserene synthases that channel the common precursor ent-copalyl diphosphate to PTM and PTN biosynthesis in S. platensis MA7327 and MA7339. (A) The common ADHBA moiety from ASA and DHAP. (B) The most advanced common intermediate ent-copalyl diphosphate (ent-CPP) for the terpenoid moieties via the MEP pathway. (C) The ent-kaurene synthase PtmT3 and ent-atiserene synthases PtmT1 and PtnT1 catalyzed divergence of ent-CPP en route to PTM and PTN with coupling between the terpenoid and benzoate moieties as the last step. (D) Inactivation of selected genes in S. platensis MA7327 supporting the proposed pathway. HPLC chromatograms of I, MA7327 wild type; II, SB12006 (i.e., ΔptmB2 mutant); III, SB12006 fermented with supplementation of AHBA; IV, SB12007 (i.e., ΔptmT1 mutant); V, SB12008 (i.e., ΔptmT3 mutant). (E) Production of PTN by expressing the ptn cluster from MA7339 in S. lividans K4-114 and PTM by expressing the PTM cassette from MA7327 in S. platensis MA7339. Extracted ion (m/z at 442 for the [PTM + H]+ ion in blue and m/z at 426 for the [PTN + H]+ ion in red) chromatograms from liquid chromatography–mass spectrometry (LC-MS) analyses of I, PTN standard; II PTM standard; III, MA7339 wild type [the two peaks shown in blue are platencin A1 and A3, which have been characterized previously from MA7339 and have the same molecular weight as PTM (26)]; IV, SB12606 (i.e., S. lividans K4-114/pBS12619); V, SB12604 (i.e., MA7339/pBS12603). HPLC chromatograms of the same analyses with UV detection at 240 nm are provided in SI Appendix. PTM (♦); PTN (●); platensic acid (◊); platencinic acid (○).
Discovery of PtmT3 and PtmT1/PtnT1 as Dedicated ent-Kaurene and ent-Atiserene Synthases That Channel the Common Diterpene Biosynthetic Intermediate ent-CPP into the PTM and PTN Scaffolds.
In agreement with previous isotopic labeling studies (12, 13), a number of genes, ptmM1/ptnM1, M2, and M3, from the MEP pathway (15, 16) were found in the ptm and ptn gene clusters; all localized to a putative operon with the geranylgeranyl diphosphate (GGPP) synthases PtmT4/PtnT4 (Fig. 2 and Table 1), ensuring that sufficient amounts of GGPP are available for PTM and PTN biosynthesis. Both ptm and ptn clusters also encode the ent-CPP synthases PtmT2/PtnT2 (Fig. 2 and Table 1), supporting ent-CPP as the most advanced common diterpene intermediate for PTM and PTN biosynthesis (Fig. 3B). Strikingly, although the ptm cluster features a canonical ent-kaurene synthase, PtmT3, consistent with the proposal that ent-kaurene synthase could catalyze the cyclization of ent-CPP to afford both ent-kaurene and ent-atiserene, accounting for the PTM and PTN dual production in the MA7327 strain, ptmT3 is localized within the PTM cassette, which is absent from the ptn cluster (Fig. 2). The fact that the ptn cluster lacks a PtmT3 homolog challenges the current proposal for ent-kaurene and ent-atiserene biosynthesis (Fig. 1B), demonstrating that PTN biosynthesis must have an alternative pathway, independent of an ent-kaurene synthase, for ent-atiserene production.
To explore this mechanistic dilemma of how the common precursor ent-CPP is channeled into PTM and PTN biosynthesis, we first inactivated ptmT3 in MA7327 to afford the ΔptmT3 mutant strain, SB12008 (SI Appendix). Under the conditions where the wild-type produces both PTM and PTN, SB12008 produces PTN exclusively, and the PTN titer in SB12008 is approximately 9-fold greater than that observed in the wild type (Fig. 3D, V). This result excluded PtmT3 from playing a role in PTN biosynthesis and provided experimental evidence supporting the functional assignment of PtmT3 as an ent-kaurene synthase dedicated to PTM biosynthesis. The fact that the PTN titer is improved in SB12008 is consistent with the proposal of an independent ent-atiserene synthase that, in the absence of the competing PtmT3, efficiently channels all ent-CPP precursors to PTN biosynthesis (Fig. 3C).
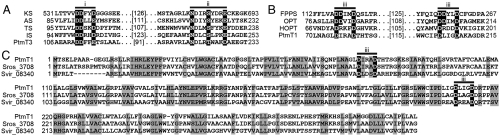
Because no ent-atiserene synthase has ever been reported, initial bioinformatics examination of the genes within either the ptm or ptn cluster to search for an ent-atiserene synthase using known terpene synthases as points of reference (17–19) failed to yield any apparent candidate. We subsequently focused on PtmT1/PtnT1 as the most likely candidates based more on the necessity of an ent-atiserene synthase for PTN biosynthesis rather than on bioinformatics data (Fig. 2 and Table 1). PtmT1/PtnT1 fall into the UbiA family of aromatic prenyltransferases (26) on the basis of BLAST analyses but lack the canonical “DDXXD/E” and “NSE/DTE” motifs characteristic of type I terpene synthases (17–19, 27) (Table 1 and Fig. 4 A and B). To investigate the possible roles that PtmT1/PtnT1 may play in PTN biosynthesis, we inactivated ptmT1 in MA7327 to yield the ΔptmT1 mutant strain SB12007 (SI Appendix). Under the PTM and PTN dual production conditions with the MA7327 wild type as a positive control, SB12007 completely failed to produce PTN, instead producing only PTM (Fig. 3D, IV). This result unambiguously established the function of PtmT1/PtnT1 in PTN biosynthesis. In contrast to the current proposal that ent-kaurene synthases catalyze the formation of both ent-kaurene and ent-atiserene from ent-CPP en route to these two subfamilies of diterpenoid natural products (Fig. 1B), PTM and PTN biosyntheses are controlled by dedicated ent-kaurene (PtmT3) and ent-atiserene (PtmT1/PtnT1) synthases that channel the common ent-CPP precursor to the characteristic PTM and PTN scaffolds, respectively (Fig. 3C). PtmT1/PtnT1 therefore represent the first ent-atiserene synthases discovered, unveiling a previously undescribed pathway for the biosynthesis of diterpenoid natural products from the common precursor ent-CPP.
Fig. 4.
Primary sequence alignments of PtmT1 and PtmT3 with characterized terpene synthases and homologues of unknown functions. (A) PtmT3 contains the two conserved metal-binding active-site motifs, DDxxD/E (motif i) and NSE/DTE (motif ii), present in characterized type-I terpene synthases. KS, ent-kaurene synthase (Q39548); AS, aristolochene synthase (AAF13264); TS, trichodiene synthase (AAN05035); IS, epi-isozizaene synthase (Q9K499); PtmT3, ent-kaurene synthase (ACO31279). Given in parentheses are GenBank accession numbers. The numbers between the active site motifs indicate the number of amino acids. (B) PtmT1 contains two atypical putative active site motifs, deviated from the two canonical DDxxD (motif iii) motifs present in characterized prenyltransferases. FPPS, farnesyl pyrophosphate synthase (P08836); OPT, octoprenyltransverase (1V4E_B); HOPT, HBA oligoprenyltransferase (AEE59373); PtmT1, ent-atiserene synthase (ACO31274). (C) PtmT1 shows high sequence homology, including the two deviated putative active site motifs DxxxD (motif iii), to proteins of unknown function uncovered in genome sequencing projects. Sros_3708 (ACZ86631), annotated as a hypothetic protein in Streptosporangium roseum DSM43021. Svir_08340 (ACU95894), annotated a 4-hydroxybenzoate prenyltransferase-like prenyltransferase in Saccharomonospora viridis DSM 43017.
Finally, we demonstrated the production of PTN by overexpressing the cloned ptn cluster in selected heterologous Streptomyces hosts. We have previously shown that ptmR1 and ptnR1 are transcriptional regulators, and upon inactivation of ptmR1 in MA7327 (25) or ptnR1 in MA7339 (28), respectively, we have yielded mutant strains that significantly overproduce PTN, PTM, or both. We isolated a plasmid pBS12615 that harbors the entire ptn cluster from MA7339, inactivated ptnR1 within the ptn cluster to yield the expression plasmid pBS12619, and introduced pBS12619 into a PTN-nonproducing heterologous host Streptomyces lividans K4-114 (29) to afford the recombinant strain SB12606 (SI Appendix). Fermentation of SB12606 under the same PTN production conditions used for the wild-type MA7339 resulted in the production of PTN (Fig. 3E, I, III, and IV), whose identity was confirmed by HPLC and mass spectrometric analyses with comparison to an authentic standard (SI Appendix). This result eliminated any concerns that components of the MA7339 genome beyond the ptn cluster might contribute to PTN biosynthesis and unambiguously established that the genes within the cloned ptn cluster are sufficient to support PTN production, further supporting the functional assignment of PtnT1 as a dedicated ent-atiserene synthase (Fig. 3C).
Evolutionary Relationship Between the PTM and PTN Biosynthetic Machineries.
Comparative analysis of PTM and PTN dual production in MA7327 and PTN production in MA7339 reveals a rare event of metabolic pathway evolution caught in action. The high level of sequence conservation is striking, indicating a recent divergence of these two gene clusters on an evolutionary timeline. As both the 16S rDNA sequences of the strains are similar (SI Appendix) and the genes surrounding the clusters are conserved, it appears that the two clusters diverged orthologously along with the two strains. An approximately 1-kb duplicated region flanking the PTM cassette in the ptm gene cluster provides two plausible mechanisms to explain the evolutionary relationship between the nearly identical ptm and ptn gene clusters (Fig. 2A) (SI Appendix). In the most likely scenario, a genetic duplication in a predecessor of the ptm gene cluster produced the 1-kb repeats and subsequent homologous recombination resulted in a loss of the PTM cassette to yield the ptn gene cluster. Deletion of ptmT3 from MA7327 thereby generating the ΔptmT3 mutant strain SB12008 represents an analogous conversion of a dual producer to a PTN specific producer (Fig. 3D, V). Alternatively, the ptn gene cluster could predate the ptm gene cluster; introduction of a mobile genetic element containing the five-gene PTM cassette into the ptn cluster across ptnO6 via homologous recombination would have led to the evolution of a dual-producing strain. We demonstrated the latter scenario by introducing the PTM cassette into MA7339, thereby converting it into a PTM producer. We constructed an expression plasmid pBS12603, in which the expression of the PTM cassette is under the control of the constitutive promoter ErmE* and introduced this construct into MA7339 to afford the recombinant strain SB12604. SB12604 was then cultured under standard PTM production conditions with both the MA7339 wild-type and empty vector-containing SB12605, as controls (SI Appendix). PTM was readily produced by SB12604 but could not be detected in crude extracts of MA7339 or SB12605 cultures (Fig. 3E, II, III, and V); the identity of PTM produced by SB12604 was verified by mass and NMR analyses (SI Appendix). These findings unambiguously establish that the PTM cassette carries all the activities necessary to endow the PTN machinery with the ability to produce PTM and that, under the control of the ErmE* promoter, it can efficiently channel the common precursors into PTM biosynthesis, mimicking the evolutionary relationship between the PTN and PTM biosynthetic machineries.
ent-Kaurene and ent-atiserene represent two major scaffolds of a large family of diterpenoid natural products, whose biosynthetic relationship is largely unknown (15–24). Comparative analysis of the PTM and PTN dual biosynthetic machinery from MA7327 and the PTN biosynthetic machinery from MA7339 revealed a dedicated ent-kaurene synthase, PtmT3, for PTM biosynthesis and dedicated ent-atiserene synthases, PtmT1 and PtnT1, for PTN biosynthesis (Fig. 3); ent-atiserene synthase represents a previously undescribed pathway for biosynthesis of diterpenoid natural products. PtmT1 and PtnT1 homologs can be identified in other sequenced genomes (Fig. 4C), suggesting that they may represent a previously undescribed family of previously unappreciated diterpene synthases. The PTM and PTN biosynthetic machineries provide a rare opportunity to explore how secondary metabolic pathway evolution increases natural product structural diversity and support the wisdom of applying combinatorial biosynthesis methods for the generation of unique PTM and/or PTN analogues, thereby facilitating drug development efforts based on these privileged natural product scaffolds.
Methods
The PTM and PTN biosynthetic gene clusters were cloned from S. platensis MA7327 (5, 10) and MA7339 (6, 11), respectively. Targeted inactivation of orf4, orf5, ptmB2, ptmO1, ptmT1, and ptmT3 in S. platensis MA7327 was carried out by following the λRED-mediated PCR-targeting mutagenesis method (30). Genetic manipulation of PTM biosynthesis in S. platensis MA7327 (25) and PTN biosynthesis in S. platensis MA7339 (28) in vivo followed previously described procedures. Heterologous expression of the ptn gene cluster from S. platensis MA7339 was carried out in S. lividans K4-114 (29). Conversion of the PTN-producing S. platensis MA7339 strain into the PTM and PTN dual-producing S. platensis SB12604 was accomplished by introduction of the PTM cassette from S. platensis MA7327 into S. platensis MA7339. Fermentation of S. platensis wild-type and recombinant strains for PTM and PTN production as well as the production and isolation of PTM and PTN biosynthetic intermediates was performed as described previously, and their structures were confirmed by HPLC analysis with authentic standards as well as mass spectrometry and 1H and 13C NMR analyses (25, 28).
Materials, methods, and detailed experimental procedures are provided in SI Appendix. Included in SI Appendix are Tables S1 and S2 and Figs. S1–S13. Table S1 contained strains and plasmids and Table S2 summarized all the primers used in this study. Fig. S1 highlighted the structures of PTM and PTN with biosynthetically related natural products. Figs. S2 and Figs. S3 supported the determination of the ptm cluster boundaries. Fig. S4 confirmed the ΔptmB2 genotype in SB12006. Figs. S5 and S6 and Figs. S7 and S8 show the 1H and 13C NMR spectra of platensic acid and platencinic acid, respectively, isolated from SB12006. Fig. S9 confirmed the genotypes of ΔptmT1 in SB12007 and ΔptmT3 in SB12008. Fig. S10 depicted possible routes of ptm and ptn cluster evolution. Fig. S11 shows the 1H NMR spectrum of PTM isolated from SB12604. Fig. S12 summarizes partial alignment of 16S rDNA sequences from S. platensis MA7327 and MA7339. Fig. S13 examined the phenotypes of recombinant SB12604, SB12605, and SB12606 strains by HPLC analysis with UV detection at 240 nm in comparison with authentic PTM and PTN as well as the wild-type S. platensis MA7339 strain.
Supplementary Material
Acknowledgments.
We thank Dr. Sheo B. Singh, Merck Research Laboratories, Rahway, NJ, for providing the S. platensis MA7327 and MA7339 wild-type strains, the Analytical Instrumentation Center of the School of Pharmacy, University of Wisconsin–Madison for support in obtaining MS and NMR data, and the John Innes Center, Norwich, United Kingdom, for providing the REDIRECT Technology kit. This work was supported in part by National Institutes of Health (NIH) Grant AI079070. M.J.S was supported in part by NIH Predoctoral Training Grant GM08505.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database, http://www.ncbi.nlm.nih.gov/genbank/ [accession nos. FJ655920 (ptm cluster) and CG398492 (ptn cluster)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106919108/-/DCSupplemental.
References
- 1.Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430:242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boucher HW, et al. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 3.Nathan DM, et al. American Diabetes Association. European Association for Study of Diabetes. Medical management of hyperglycemia in type 2 diabetes: A consensus algorithm for the initiation and adjustment of therapy: A consensus statement of the American Diabetes Association and the European Association for the Study of Diabetes. Diabetes Care. 2009;32:193–203. doi: 10.2337/dc08-9025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Genilloud O, et al. Current approaches to exploit actinomycetes as a source of novel natural products. J Ind Microbiol Biotechnol. 2011;38:375–389. doi: 10.1007/s10295-010-0882-7. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, et al. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature. 2006;441:358–361. doi: 10.1038/nature04784. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, et al. Discovery of platencin, a dual FabF and FabH inhibitor with in vivo antibiotic properties. Proc Natl Acad Sci USA. 2007;104:7612–7616. doi: 10.1073/pnas.0700746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu M, et al. Antidiabetic and antisteatotic effects of the selective fatty acid synthase (FAS) inhibitor platensimycin in mouse models of diabetes. Proc Natl Acad Sci USA. 2011;108:5378–5384. doi: 10.1073/pnas.1002588108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brinster S, et al. Type II fatty acid synthesis is not a suitable antibiotic target for Gram-positive pathogens. Nature. 2009;458:83–86. doi: 10.1038/nature07772. [DOI] [PubMed] [Google Scholar]
- 9.Balemans W, et al. Essentiality of FASII pathway for Staphylococcus aureus. Nature. 2010;463:E3. doi: 10.1038/nature08667. discussion E4. [DOI] [PubMed] [Google Scholar]
- 10.Singh SB, et al. Isolation, structure, and absolute stereochemistry of platensimycin, a broad spectrum antibiotic discovered using an antisense differential sensitivity strategy. J Am Chem Soc. 2006;128:11916–11920. doi: 10.1021/ja062232p. [DOI] [PubMed] [Google Scholar]
- 11.Jayasuriya H, et al. Isolation and structure of platencin: A FabH and FabF dual inhibitor with potent broad-spectrum antibiotic activity. Angew Chem Int Ed Engl. 2007;46:4684–4688. doi: 10.1002/anie.200701058. [DOI] [PubMed] [Google Scholar]
- 12.Herath K, Attygalle AB, Singh SB. Biosynthetic studies of platencin. Tetrahedron Lett. 2008;49:5755–5758. [Google Scholar]
- 13.Herath KB, Attygalle AB, Singh SB. Biosynthetic studies of platensimycin. J Am Chem Soc. 2007;129:15422–15423. doi: 10.1021/ja0758943. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki H, Ohnishi Y, Furusho Y, Sakuda S, Horinouchi S. Novel benzene ring biosynthesis from C3 and C4 primary metabolites by two enzymes. J Biol Chem. 2006;281:36944–36951. doi: 10.1074/jbc.M608103200. [DOI] [PubMed] [Google Scholar]
- 15.Kuzuyama T, Seto H. Diversity of the biosynthesis of the isoprene units. Nat Prod Rep. 2003;20:171–183. doi: 10.1039/b109860h. [DOI] [PubMed] [Google Scholar]
- 16.Dairi T. Studies on biosynthetic genes and enzymes of isoprenoids produced by actinomycetes. J Antibiot. 2005;58:227–243. doi: 10.1038/ja.2005.27. [DOI] [PubMed] [Google Scholar]
- 17.Christianson DW. Structural biology and chemistry of the terpenoid cyclases. Chem Rev. 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- 18.Cao R, et al. Diterpene cyclases and the nature of the isoprene fold. Proteins. 2010;78:2417–2432. doi: 10.1002/prot.22751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bohlmann J, Meyer-Gauen G, Croteau R. Plant terpenoid synthases: Molecular biology and phylogenetic analysis. Proc Natl Acad Sci USA. 1998;95:4126–4133. doi: 10.1073/pnas.95.8.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrone D, et al. Gibberellin biosynthesis in bacteria: Separate ent-copalyl diphosphate and ent-kaurene synthases in Bradyrhizobium japonicum. FEBS Lett. 2009;583:475–480. doi: 10.1016/j.febslet.2008.12.052. [DOI] [PubMed] [Google Scholar]
- 21.Xu M, Wilderman PR, Peters RJ. Following evolution’s lead to a single residue switch for diterpene synthase product outcome. Proc Natl Acad Sci USA. 2007;104:7397–7401. doi: 10.1073/pnas.0611454104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshikuni Y, Ferrin TE, Keasling JD. Designed divergent evolution of enzyme function. Nature. 2006;440:1078–1082. doi: 10.1038/nature04607. [DOI] [PubMed] [Google Scholar]
- 23.Wilderman PR, Peters RJ. A single residue switch converts abietadiene synthase into a pimaradiene specific cyclase. J Am Chem Soc. 2007;129:15736–15737. doi: 10.1021/ja074977g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrone D, Xu M, Fulton DB, Determan MK, Peters RJ. Increasing complexity of a diterpene synthase reaction with a single residue switch. J Am Chem Soc. 2008;130:5400–5401. doi: 10.1021/ja710524w. [DOI] [PubMed] [Google Scholar]
- 25.Smanski MJ, Peterson RM, Rajski SR, Shen B. Engineered Streptomyces platensis strains that overproduce antibiotics platensimycin and platencin. Antimicrob Agents Chemother. 2009;53:1299–1304. doi: 10.1128/AAC.01358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bräuer L, Brandt W, Schulze D, Zakharova S, Wessjohann LA. Structural model of the membrane-bound aromatic prenyltransferase UbiA from E. coli. Chembiochem. 2008;9:982–992. doi: 10.1002/cbic.200700575. [DOI] [PubMed] [Google Scholar]
- 27.Aaron JA, Lin X, Cane DE, Christianson DW. Structure of epi-isozizaene synthase from Streptomyces coelicolor A3 (2), a platform for new terpenoid cyclization templates. Biochemistry. 2010;49:1787–1797. doi: 10.1021/bi902088z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu Z, et al. Engineering of Streptomyces platensis MA7339 for overproduction of platencin and congeners. Org Lett. 2010;12:1744–1747. doi: 10.1021/ol100342m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ziermann R, Betlach MC. Recombinant polyketide synthesis in Streptomyces: Engineering of improved host strains. BioTechniques. 1999;26:106–110. doi: 10.2144/99261st05. [DOI] [PubMed] [Google Scholar]
- 30.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.